Weak Intermolecular Interactions in a Series of Bioactive Oxazoles
Abstract
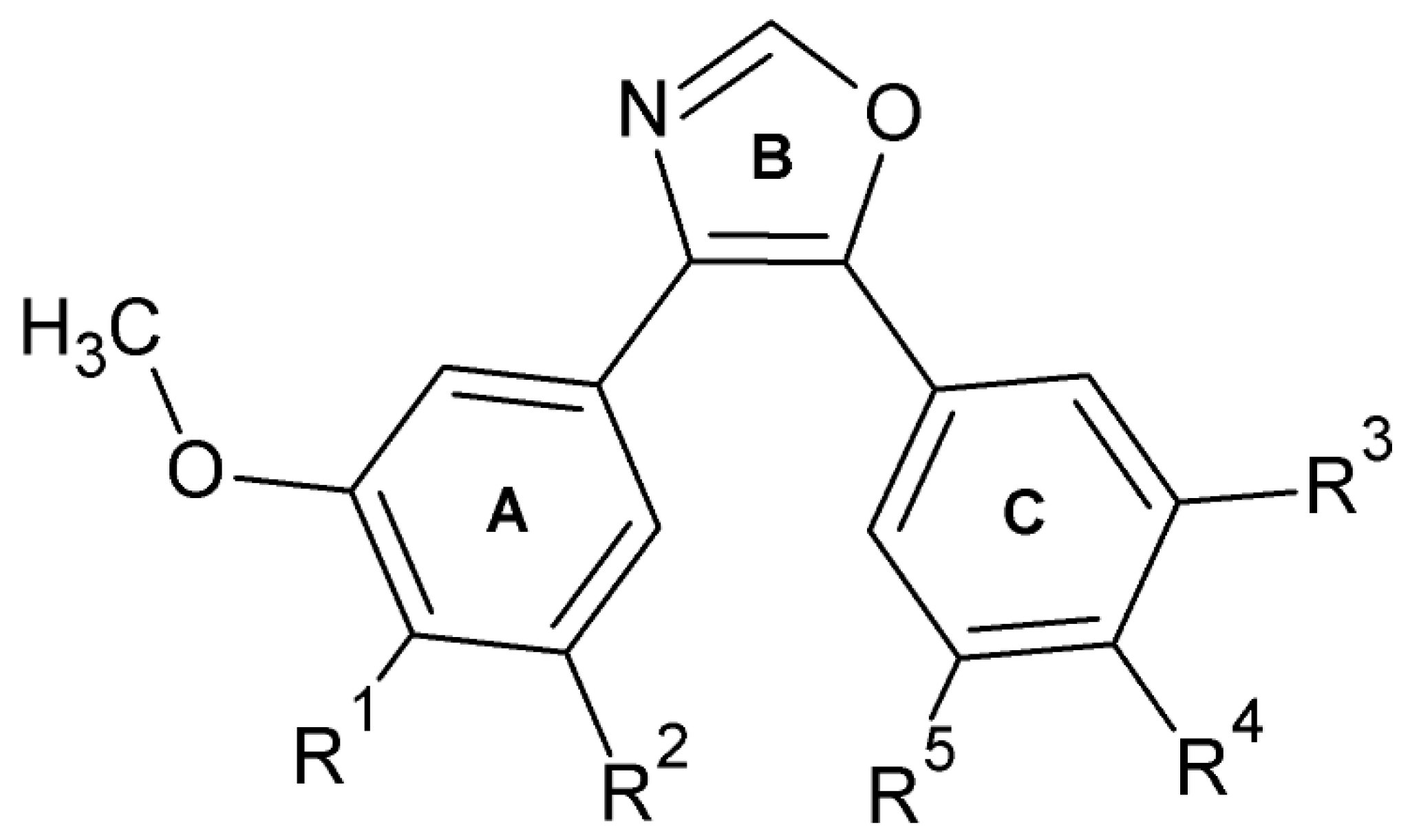
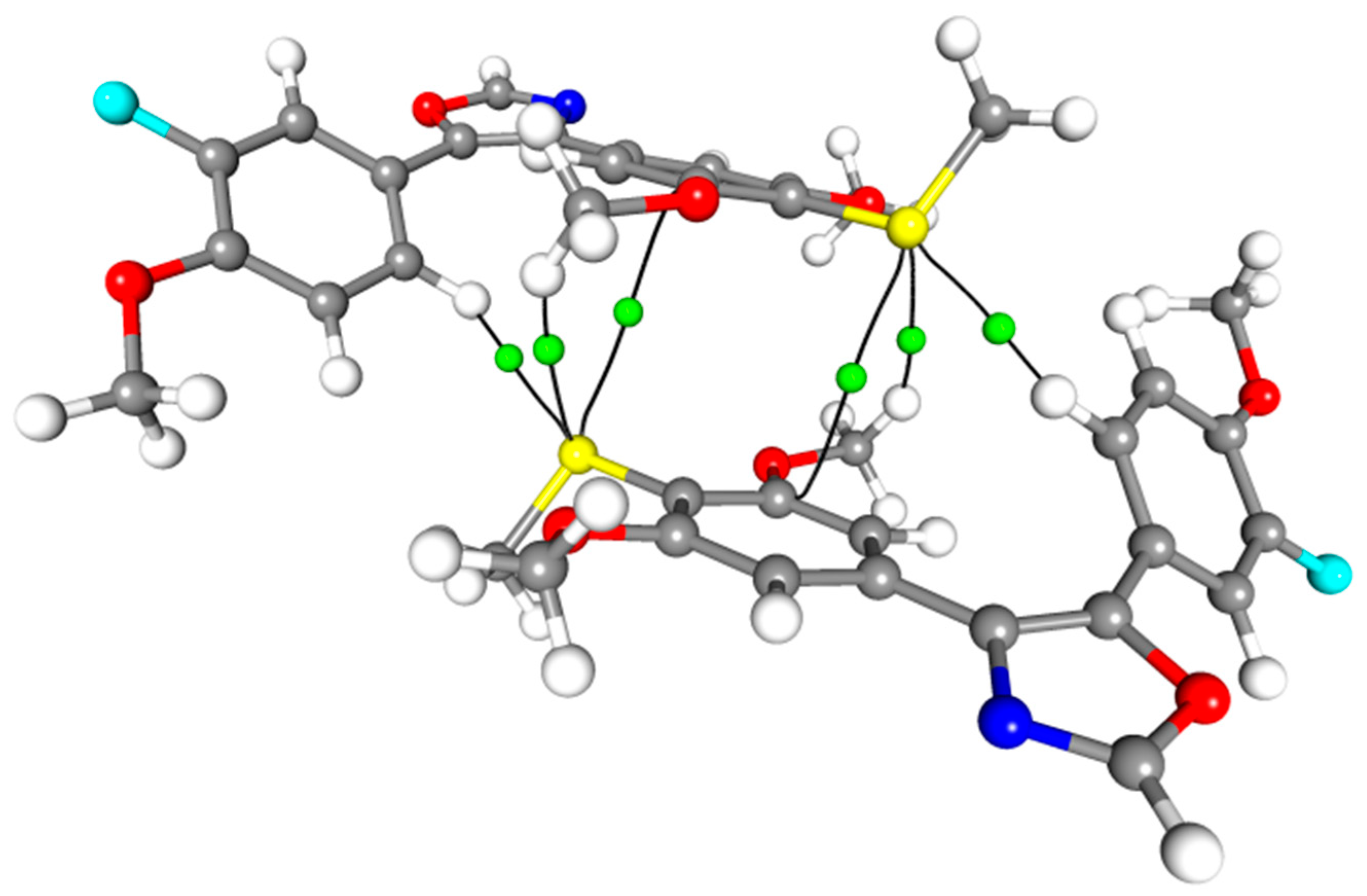
1. Introduction
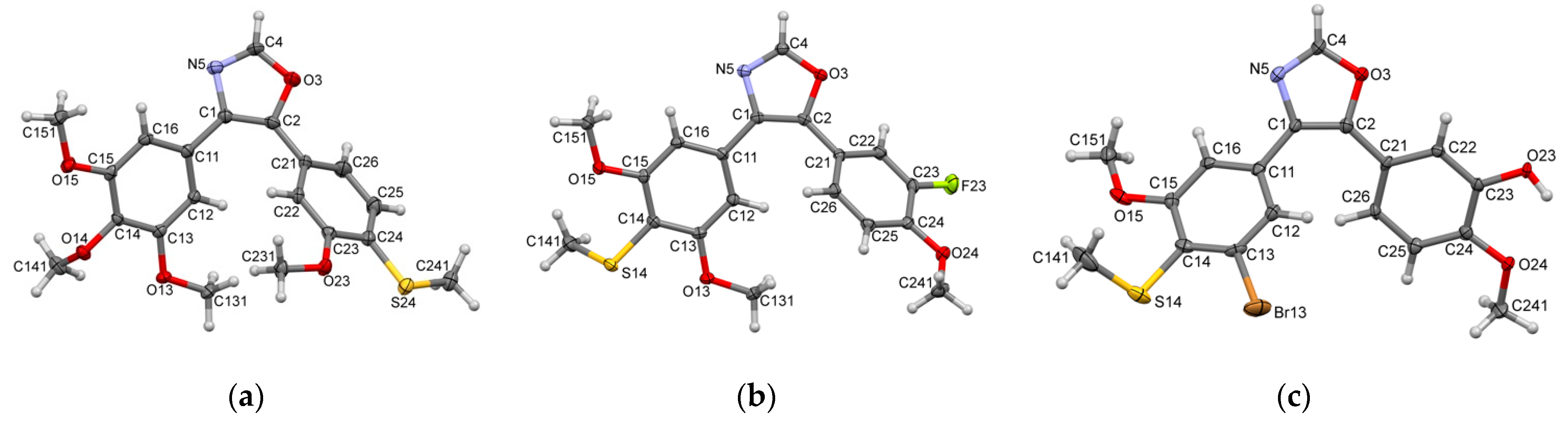
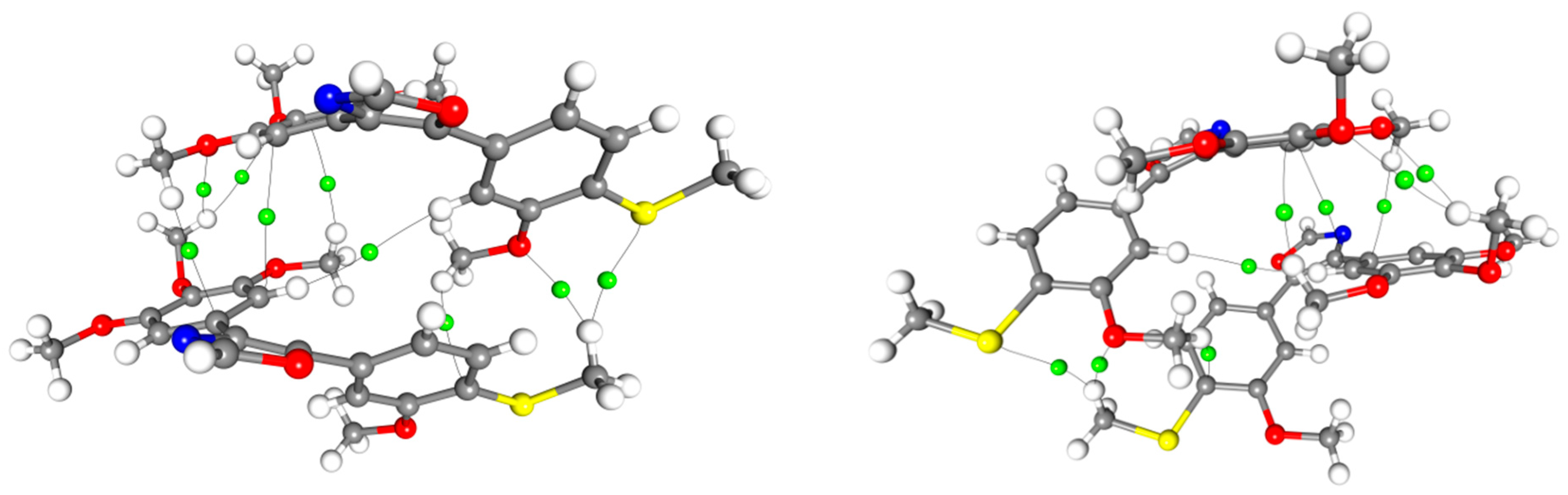
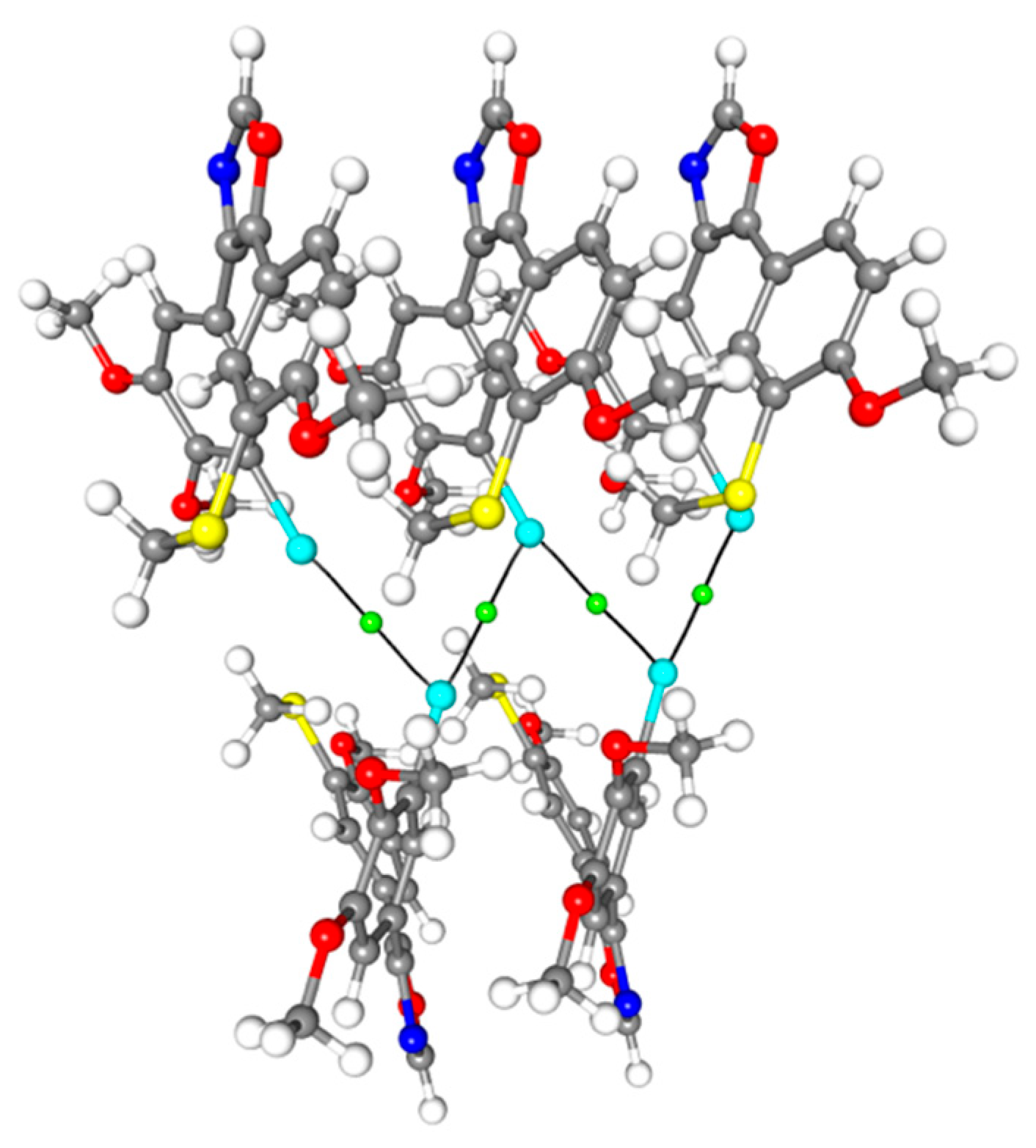
2. Results
3. Materials and Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Dunitz, J.D. Phase transitions in molecular crystals from a chemical viewpoint. Pure Appl. Chem. 1991, 63, 177–185. [Google Scholar] [CrossRef]
- Desiraju, G.R. The Crystal as a Supramolecular Entity. In Perspectives in Supramolecular Chemistry 2; Desiraju, G.R., Ed.; Wiley and Sons: Chichester, UK, 1996. [Google Scholar]
- Desiraju, G.R. Hydrogen bridges in crystal engineering: Interactions without borders. Acc. Chem. Res. 2002, 35, 565–573. [Google Scholar] [CrossRef] [PubMed]
- Metrangolo, P.; Neukirch, H.; Pilati, T.; Resnati, G. Halogen bonding based recognition processes: A world parallel to hydrogen bonding. Acc. Chem. Res. 2005, 38, 386–395. [Google Scholar] [CrossRef] [PubMed]
- Biot, N.; Bonifazi, D. Chalcogen-bond driven molecular recognition at work. Coord. Chem. Rev. 2020, 413, 213243. [Google Scholar] [CrossRef]
- Politzer, P.; Murray, J.S.; Clark, T. σ-Hole bonding: A physical interpretation. Top. Curr. Chem. 2015, 358, 19–42. [Google Scholar] [PubMed]
- Hunter, C.A.; Lawson, K.R.; Perkins, J.; Urch, C.J. Aromatic interactions. J. Chem. Soc. Perkin Trans. 2001, 2, 651–669. [Google Scholar] [CrossRef]
- Matta, C.F.; Hernandez-Trujillo, J.; Tang, T.-H.; Bader, R.F.W. Hydrogen-hydrogen bonding: A stabilizing interaction in molecules and crystals. Chem. Eur. J. 2003, 9, 1940–1951. [Google Scholar] [CrossRef]
- Dunitz, J.D.; Gavezzotti, A. Molecular recognition in organic crystals: Directed intermolecular bonds or nonlocalized bonding? Angew. Chem. Int. Ed. Engl. 2005, 44, 1766–1787. [Google Scholar] [CrossRef]
- Dunitz, J.D. Intermolecular atom-atom bonds in crystals? IUCrJ 2015, 2, 157–158. [Google Scholar] [CrossRef]
- Thakur, T.S.; Dubey, R.; Desiraju, G.R. Intemolecular atom-atom bonds in crystals—A chemical perspective. IUCrJ 2015, 2, 159–160. [Google Scholar] [CrossRef]
- Lecomte, C.; Espinosa, E.; Matta, C.F. On atom-atom ‘short contact’ bonding interactions in crystals. IUCrJ 2015, 2, 161–163. [Google Scholar] [CrossRef]
- Dominiak, P.M.; Makal, A.; Mallinson, P.R.; Trzcińska, K.; Eilmes, J.; Grech, E.; Chruszcz, M.; Minor, W.; Woźniak, K. Continua of interactions between pairs of atoms in molecular crystals. Chem. Eur. J. 2006, 12, 1941–1949. [Google Scholar] [CrossRef]
- Mallinson, P.R.; Smith, G.T.; Wilson, C.C.; Grech, E.; Woźniak, K. From weak interactions to covalent bonds: A continuum in the complexes of 1,8-bis(dimethylamino)naphthalene. J. Am. Chem. Soc. 2003, 125, 4259–4270. [Google Scholar] [CrossRef]
- Espinosa, E.; Lecomte, C.; Molins, E. Experimental electron density overlapping in hydrogen bonds: Topology vs. energetics. Chem. Phys. Lett. 1999, 300, 745–748. [Google Scholar] [CrossRef]
- Spackman, M.A. Hydrogen bond energetics from topological analysis of experimental electron densities: Recognising the importance of the promolecule. Chem. Phys. Lett. 1999, 301, 425–429. [Google Scholar] [CrossRef]
- Gatti, C.; May, E.; Destro, R.; Cargnoni, F. Fundamental properties and nature of CH··O interactions in crystals on the basis of experimental and theoretical charge densities. The case of 3,4-bis(dimethylamino)-3-cyclobutene-1,2-dione (DMACB) crystal. J. Phys. Chem. A 2002, 106, 2707–2720. [Google Scholar] [CrossRef]
- Bader, R.F.W. Atoms in Molecules: A Quantum Theory; Clarendon Press: Oxford, UK, 1990. [Google Scholar]
- Gavezzotti, A. Are crystal structures predictable? Acc. Chem. Res. 1994, 27, 309–314. [Google Scholar] [CrossRef]
- Gavezzotti, A.; Fillippini, G. Geometry of the intermolecular X-H···Y (X, Y = N, O) hydrogen bond and the calibration of empirical hydrogen-bond potentials. J. Phys. Chem. 1994, 98, 4831–4837. [Google Scholar] [CrossRef]
- Macrae, C.F.; Sovago, I.; Cottrell, S.J.; Galek, P.T.A.; McCabe, P.; Pidcock, E.; Platings, M.; Shields, G.P.; Stevens, J.S.; Towler, M.; et al. Mercury 4.0: From visualization to analysis, design and prediction. J. Appl. Crystallogr. 2020, 53, 226–235. [Google Scholar] [CrossRef]
- Turner, M.J.; McKinnon, J.J.; Wolff, S.K.; Grimwood, D.J.; Spackman, P.R.; Jayatilaka, D.; Spackman, M.A.; CrystalExplorer17. University of Western Australia. 2017. Available online: http://crystalexplorer.scb.uwa.edu.au/ (accessed on 29 January 2021).
- Stefański, T.; Mikstacka, R.; Kurczab, R.; Dutkiewicz, Z.; Kucińska, M.; Murias, M.; Zielińska-Przyjemska, M.; Cichocki, M.; Teubert, A.; Kaczmarek, M.; et al. Design, synthesis, and biological evaluation of novel combretastatin A-4 thio derivatives as microtubule targeting agents. Eur. J. Med. Chem. 2018, 144, 797–816. [Google Scholar] [CrossRef]
- Groom, C.R.; Bruno, I.J.; Lightfoot, M.P.; Ward, S.C. The Cambridge Structural Database. Acta Crystallogr. Part B 2016, 72, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Kalman, A.; Parkanyi, L.; Argay, G. Classification of the isostructurality of organic molecules in the crystalline state. Acta Crystallogr. Part B 1993, 49, 1039–1049. [Google Scholar] [CrossRef]
- Rutherford, J.S. On comparing lattice parameters among isostructural molecular crystals. Acta Chim. Hung. 1997, 134, 395–405. [Google Scholar]
- Rigaku. CrysAlisPro; Rigaku Oxford Diffraction Ltd.: Oxford, UK, 2013. [Google Scholar]
- Sheldrick, G.M. SHELXT—Integrated space-group and crystal-structure determination. Acta Crystallogr. Part A 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Part C 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Guillot, B.; Viry, L.; Guillot, R.; Lecomte, C.; Jelsch, C. Refinement of proteins at subatomic resolution with MOPRO. J. Appl. Crystallogr. 2001, 34, 214–223. [Google Scholar] [CrossRef]
- Domagala, S.; Fournier, B.; Liebschner, D.; Guillot, B.; Jelsch, C. An improved experimental databank of transferable multipolar atom models—ELMAM2. Construction details and applications. Acta Crystallogr. Part A 2012, 68, 337–351. [Google Scholar] [CrossRef]
- Przybył, A.K.; Grześkiewicz, A.M.; Kubicki, M. Weak interactions in the structures of newly synthesized (-)-cytisine amino acid derivatives. Crystals 2021, 11, 146. [Google Scholar] [CrossRef]

| 1 | 2 | 3 | 4 | 5 | 6A | 6B | 7 | 8 | 9 | CSDB | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| C1-C2 | 1.355(4) | 1.350(6) | 1.367(2) | 1.363(5) | 1.3595(19) | 1.362(4) | 1.363(4) | 1.354(11) | 1.361(3) | 1.354(5) | 1.357(14) 1.348(15) |
| C1-N5 | 1.403(3) | 1.387(6) | 1.408(2) | 1.409(4) | 1.4099(17) | 1.401(4) | 1.401(4) | 1.423(10) | 1.397(3) | 1.404(5) | 1.398(10) 1.393(12) |
| C2-O3 | 1.385(3) | 1.381(5) | 1.3906(18) | 1.395(4) | 1.3899(16) | 1.391(3) | 1.391(3) | 1.376(9) | 1.378(3) | 1.387(5) | 1.296(15) 1.294(15) |
| O3-C4 | 1.352(3) | 1.346(5) | 1.3504(19) | 1.346(4) | 1.3550(16) | 1.347(4) | 1.353(4) | 1.350(10) | 1.356(4) | 1.345(4) | 1.350(16) 1.359(18) |
| C4-N5 | 1.287(4) | 1.256(6) | 1.292(2) | 1.287(5) | 1.2874(18) | 1.276(4) | 1.275(4) | 1.286(11) | 1.276(4) | 1.288(5) | 1.387(11) 1.379(13) |
| C2-C1-N5 | 108.8(3) | 109.0(4) | 108.62(13) | 108.5(3) | 108.51(12) | 108.9(3) | 109.0(2) | 109.5(8) | 109.1(2) | 108.8(4) | 108.6(6) 109.5(11) |
| C1-C2-O3 | 107.8(2) | 107.1(5) | 107.25(13) | 107.0(3) | 107.57(12) | 106.8(3) | 106.8(3) | 107.2(8) | 107.2(2) | 107.1(3) | 107.4(6) 107.4(11) |
| C2-O3-C4 | 103.9(2) | 103.3(4) | 104.54(12) | 104.9(3) | 104.36(10) | 104.4(2) | 104.2(2) | 104.5(7) | 104.0(2) | 104.9(3) | 104.9(8) 104.7(9) |
| O3-C4-N5 | 115.3(3) | 116.2(5) | 115.06(14) | 114.8(3) | 114.85(12) | 115.3(3) | 115.4(3) | 116.1(9) | 115.5(3) | 114.5(4) | 114.0(11) 113.9(10) |
| C1-N5-C4 | 104.3(2) | 104.3(4) | 104.50(13) | 104.9(3) | 104.68(11) | 104.6(3) | 104.5(3) | 102.6(8) | 104.1(2) | 104.7(3) | 105.1(8) 104.6(9) |
| C12-C13-C14 | 120.5(3) | 119.3(5) | 120.42(14) | 121.3(3) | 120.62(13) | 121.5(3) | 121.3(3) | 121.8(8) | 121.7(4) | 122.9(4) | |
| C13-C14-C15 | 119.7(3) | 119.2(5) | 119.60(14) | 118.4(3) | 118.46(12) | 117.9(2) | 118.4(2) | 119.9(9) | 118.8(2) | 116.8(4) | |
| C14-C15-C16 | 120.1(3) | 120.6(5) | 120.28(14) | 120.8(3) | 121.24(12) | 120.8(3) | 120.3(3) | 116.7(10) | 119.4(3) | 121.0(4) | |
| C22-C23-C24 | 121.1(3) | 122.6(5) | 119.76(14) | 120.2(3) | 123.31(13) | 119.7(3) | 120.0(3) | 117.3(9) | 120.7(2) | 119.6(4) | |
| C23-C24-C25 | 118.2(3) | 116.2(4) | 119.60(14) | 119.3(3) | 117.34(13) | 119.7(3) | 119.9(3) | 121.5(9) | 118.4(2) | 120.6(4) | |
| C2-C1-C11-C12 | −26.2(5) | −16.8(9) | −13.0(3) | −40.5(6) | −18.1(3) | −7.1(6) | −20.8(6) | −30.5(15) | −28.6(10) 144.9(13) | −39.6(7) | |
| C2-C1-C11-C16 | 154.4(3) | 162.5(5) | 168.67(16) | 142.1(4) | 163.19(14) | 173.6(4) | 161.2(4) | 152.9(10) | 146.6(6) −54(2) | 144.2(5) | |
| N5-C1-C11-C12 | 149.3(3) | 163.3(5) | 163.65(14) | 140.2(4) | 158.95(13) | 171.5(3) | 157.7(3) | 148.1(8) | 143.7(7) −22(2) | 141.0(4) | |
| N5-C1-C11-C16 | −30.1(4) | −17.4(7) | −14.6(2) | −37.3(5) | −19.78(19) | −7.8(5) | −20.4(5) | −28.5(13) | −41.1(9) 139.7(13) | −35.2(6) | |
| C1-C2-C21-C22 | −36.1(6) | −28.4(9) | 147.51(19) | 154.5(4) | 143.26(17) | 139.7(5) | 145.3(4) | −22.2(18) | −30.5(5) | 153.8(5) | |
| C1-C2-C21-C26 | 142.4(4) | 155.0(6) | −38.3(3) | −26.5(7) | −38.2(3) | −40.5(7) | −34.9(6) | 162.0(12) | 151.6(4) | −28.3(8) | |
| O3-C2-C21-C22 | 139.8(3) | 149.4(5) | −35.70(19) | −28.2(5) | −39.97(18) | −37.5(5) | −31.5(4) | 157.1(8) | 145.1(3) | −28.9(6) | |
| O3-C2-C21-C26 | −41.7(4) | −27.2(7) | 138.51(14) | 150.8(3) | 138.57(13) | 142.3(3) | 148.3(3) | −18.7(13) | −32.9(4) | 149.0(4) | |
| A/B | 28.36(14) | 17.04(18) | 14.51(6) | 39.20(10) | 19.42(7) | 7.3(3) | 20.56(19) | 30.8(3) | 35.4(2) | 37.53(11) | |
| B/C | 39.72(14) | 27.75(11) | 38.28(5) | 28.08(16) | 39.63(4) | 38.46(14) | 32.64(18) | 20.2(4) | 32.79(17) | 29.31(18) | |
| A/C | 51.51(11) | 32.47(12) | 47.42(4) | 51.84(11) | 47.83(4) | 38.69(1) | 39.00(14) | 42.5(3) | 49.6(2) | 51.54(13) | |
| A/OMe3 | 6.5(2) | 4.7(3) | 3.15(14) | 8.3(2) | 13.09(15) | 2.0(7) | 5.6(5) | ||||
| A/O(S)Me4 | 73.27(13) | 81.2(3) | 80.09(13) | 50.39(13) | 60.10(5) | 86.7(2) | 69.44(15) | 79.4(5) | 79.6(3) | 50.25(16) | |
| A/OMe5 | 0.9(2) | 6.6(3) | 4.09(15) | 9.0(3) | 4.84(17) | 8.7(5) | 3.5(5) | 2.5(7) | 5.6(7) | 7.3(3) | |
| B/OMe3 | 12.3(3) | 3.24(18) | 1.3(5) | 6.19(18) | |||||||
| B/SMe4 | 1.2(2) | 85.6(2) | 1.2(2) | ||||||||
| B/OMe4 | 0.80(16) | 1.3(5) | 4.42(17) | 3.10(17) | 1.4(5) | 2.3(11) | 2.3(5) |
| Atom1 | Atom2 | Gcp | Vcp | DEN | LAP | X···Y | H···Y | X-H···Y | Pix | HF B3LYP |
|---|---|---|---|---|---|---|---|---|---|---|
| C16 | C12 i | 4.35 | −2.77 | 0.01834 | 0.218 | 3.742 | −78.9 | −69.1 −67.4 | ||
| C14 | H13C i | 8.55 | −6.18 | 0.03675 | 0.401 | 3.717 | 2.90 | 131 | ||
| O15 | H14B i | 10.27 | −6.41 | 0.02888 | 0.519 | 3.447 | 2.78 | 119 | ||
| O14 | H14B i | 3.86 | −2.43 | 0.01651 | 0.194 | 4.173 | 3.14 | 158 | ||
| H15C | C11 i | 8.86 | −7.02 | 0.0442 | 0.393 | 3.980 | 2.90 | 170 | ||
| H22 | H12 i | 0.98 | −0.58 | 0.00581 | 0.051 | 3.39 | ||||
| O23 | H24A i | 14.83 | −10.65 | 0.05047 | 0.698 | 3.452 | 2.53 | 142 | ||
| H23C | C24 i | 12.94 | −10.26 | 0.05547 | 0.574 | 3.550 | 2.69 | 136 | ||
| S24 | H24A i | 9.07 | −6.53 | 0.03773 | 0.427 | 3.894 | 2.96 | 144 | ||
| H13A | O13 ii | 12.71 | −8.13 | 0.03529 | 0.634 | 3.259 | 2.74 | 109 | −40.9 | −53.7 −46.1 |
| O13 | H23B ii | 2.3 | −1.28 | 0.00743 | 0.122 | 4.084 | 3.37 | 124 | ||
| H13A | O14 ii | 7.86 | −4.98 | 0.02567 | 0.394 | 3.613 | 2.84 | 128 | ||
| H14A | O23 ii | 12.93 | −8.69 | 0.04038 | 0.63 | 3.429 | 2.63 | 130 | ||
| H14A | S24 ii | 6.97 | −5.34 | 0.03615 | 0.316 | 4.137 | 3.07 | 167 | ||
| H23A | O14 iii | 22.51 | −17.79 | 0.077 | 0.999 | 3.308 | 2.34 | 147 | −11.2 | −30.6 −24.2 |
| H14B | O14 iii | 3.86 | −2.43 | 0.01651 | 0.194 | |||||
| H4 | N5 iv | 19.54 | −13.48 | 0.05478 | 0.94 | 3.413 | 2.48 | 143 | −5.9 | −27.1 −22.7 |
| R | E_ele | E_pol | E_dis | E_rep | E_tot |
|---|---|---|---|---|---|
| 5.13 | −18.7 −19.2 | −10.0 −5.9 | −90.3 −90.3 | 46.7 58.2 | −69.1 −67.4 |
| 7.84 | −25.4 −21.0 | −9.4 −5.7 | −46.4 −46.4 | 24.7 33.5 | −53.7 −46.1 |
| 8.78 | −20.3 −16.6 | −7.3 −4.2 | −18.0 −18.0 | 13.6 19.5 | −30.6 −24.2 |
| 11.39 | −21.9 −18.8 | −4.0 −3.1 | −12.2 −12.2 | 10.9 16.3 | −27.1 −22.7 |
| Atom1 | Atom2 | Gcp | Vcp | DEN | LAP | X···Y | H···Y | X-H···Y | pixel | HF |
|---|---|---|---|---|---|---|---|---|---|---|
| C2 | C15 i | 4.51 | −2.93 | 0.01966 | 0.224 | 4.382 | −89.8 | −66.4 | ||
| C11 | H26 i | 9.44 | −6.67 | 0.03728 | 0.448 | 3.733 | 2.85 | 139 | ||
| O3 | S14 i | 7.59 | −5 | 0.02788 | 0.374 | 3.538 | ||||
| H12 | C16 i | 6.21 | −3.89 | 0.02169 | 0.313 | 3.766 | 3.05 | 124 | ||
| H12 | H15C i | 2.9 | −1.98 | 0.01708 | 0.14 | 2.73 | ||||
| H13C | H16 i | 11.03 | −7.61 | 0.03883 | 0.531 | 2.28 | ||||
| H13C | N5 i | 12.59 | −10.2 | 0.05647 | 0.55 | 3.651 | 2.66 | 151 | ||
| H14B | C23 i | 7.24 | −5.36 | 0.03481 | 0.335 | 3.953 | 2.98 | 150 | ||
| H15B | H24C i | 7.2 | −4.85 | 0.02851 | 0.351 | 2.46 | ||||
| C22 | O15 i | 14.28 | −10.14 | 0.04821 | 0.677 | 3.096 | ||||
| H22 | S14 i | 6.18 | −4.27 | 0.02755 | 0.297 | 4.095 | 3.17 | 143 | ||
| H23 | N5 ii | 56.52 | −82.77 | 0.27482 | 1.112 | 2.763 | 1.83 | 156 | −39.6 | −60.0 |
| O23 | H15B ii | 12.04 | −8.8 | 0.0461 | 0.561 | 3.620 | 2.60 | 155 | ||
| O23 | H16 ii | 21.77 | −15.63 | 0.06353 | 1.025 | 3.216 | 2.38 | 132 | ||
| O24 | H4 ii | 15.58 | −10.72 | 0.04758 | 0.75 | 3.211 | 2.63 | 113 |
| Atom1 | Atom2 | Gcp | Vcp | DEN | LAP | X···Y | H···Y | X-H···Y | pixel | HF |
|---|---|---|---|---|---|---|---|---|---|---|
| C13 | C15 i | 7.73 | −5.48 | 0.03329 | 0.366 | 3.490 | −105.0 | −67.8 | ||
| C4 | N5 i | 2.08 | −1.22 | 0.0089 | 0.108 | 4.083 | ||||
| C16 | H15B i | 11.62 | −9.11 | 0.05108 | 0.519 | 3.673 | 2.69 | 150 | ||
| H12 | H23B i | 7.18 | −5.23 | 0.0336 | 0.335 | 2.29 | ||||
| Br13 | C231 i | 8.05 | −5.3 | 0.02884 | 0.397 | 3.636 | ||||
| Br13 | O14 i | 9.33 | −6.44 | 0.03524 | 0.449 | 3.505 | ||||
| H14C | O14 i | 14.14 | −11.04 | 0.05703 | 0.633 | 3.593 | 2.52 | 171 | ||
| H14C | O15 i | 9.03 | −5.54 | 0.02533 | 0.459 | 3.463 | 2.87 | 115 | ||
| C22 | H23B i | 8.91 | −6.4 | 0.03717 | 0.419 | 3.900 | 2.89 | 155 | ||
| C24 | S23 i | 7.59 | −5.42 | 0.03341 | 0.358 | 3.674 | ||||
| C25 | C23 i | 6.72 | −4.71 | 0.02986 | 0.321 | 3.600 | ||||
| H24B | O24 i | 7.52 | −5.33 | 0.03276 | 0.356 | 3.815 | 2.80 | 155 | ||
| Br13 | Br13 ii | 12.28 | −9.6 | 0.0526 | 0.549 | 3.564 | −14.2 | −14.4 | ||
| Br13 | H23A ii | 8.14 | −5.2 | 0.02691 | 0.407 | 3.838 | 3.19 | 118 | ||
| O14 | H23A ii | 4.73 | −3.05 | 0.01997 | 0.235 | 4.057 | 3.00 | 165 | ||
| H14A | S23 ii | 3.24 | −1.99 | 0.01374 | 0.165 | 4.232 | 3.51 | 125 |
| Atom1 | Atom2 | Gcp | Vcp | DEN | LAP | X···Y | H···Y | X-H···Y | pixel | HF |
|---|---|---|---|---|---|---|---|---|---|---|
| H14C | O3 i | 2.97 | −1.78 | 0.01199 | 0.153 | 4.188 | 3.22 | 148 | −76.0 | −72.9 |
| C15 | H22 i | 6.48 | −4.37 | 0.02686 | 0.315 | 3.803 | 2.99 | 131 | ||
| N5 | C12 i | 6.97 | −4.66 | 0.02749 | 0.341 | 3.568 | ||||
| H14C | H22 i | 10.3 | −7.12 | 0.03753 | 0.495 | 2.25 | ||||
| C4 | H13A i | 13.33 | −9.18 | 0.04339 | 0.642 | 3.467 | 2.76 | 122 | ||
| C13 | S14 ii | 4.69 | −3.13 | 0.02168 | 0.229 | 3.886 | −54.5 | −54.0 | ||
| H13C | S14 ii | 11.01 | −8.54 | 0.04862 | 0.495 | 3.886 | 2.89 | 142 | ||
| H26 | S14 ii | 11.53 | −8.35 | 0.04412 | 0.54 | 3.693 | 2.86 | 133 |
| Compound | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| Formula | C20H21NO5S | C20H20BrNO5S | C19H19NO5S | C19H19NO5S | C19H18FNO4S |
| Formula weight | 387.44 | 466.34 | 373.41 | 373.41 | 375.40 |
| Crystal system | triclinic | monoclinic | triclinic | orthorhombic | triclinic |
| Space group | P-1 | P21/c | P-1 | P212121 | P-1 |
| a(Å) | 5.1299(4) | 12.3841(4) | 7.7068(3) | 8.3865(4) | 7.8698(8) |
| b(Å) | 11.4060(10) | 7.7755(4) | 10.2384(3) | 11.1560(6) | 10.4011(8) |
| c(Å) | 16.0097(13) | 21.2517(7) | 11.9461(4) | 19.1503(9) | 12.0236(7) |
| α(°) | 97.245(7) | 90 | 108.544(3) | 90 | 72.165(6) |
| β(°) | 94.098(7) | 101.623(3) | 94.602(3) | 90 | 88.285(6) |
| γ(°) | 99.063(7) | 90 | 100.912(3) | 90 | 68.924(8) |
| V(Å3) | 913.68(13) | 2004.42(14) | 867.47(5) | 1791.70(15) | 870.64(13) |
| Z | 2 | 4 | 2 | 4 | 2 |
| Dx(g cm−3) | 1.408 | 1.545 | 1.430 | 1.384 | 1.432 |
| F(000) | 408 | 952 | 392 | 784 | 392 |
| μ(mm−1) | 0.210 | 2.186 | 0.218 | 0.211 | 0.221 |
| Reflections: | |||||
| collected | 17518 | 19235 | 16633 | 6970 | 11038 |
| unique (Rint) | 4187 (0.0778) | 3463 (0.0708) | 3837 (0.0227) | 3324 (0.0439) | 3493 (0.0246) |
| with I > 2σ(I) | 2504 | 1493 | 3234 | 2876 | 3168 |
| R(F) [I > 2σ(I)] | 0.0620 | 0.0535 | 0.0369 | 0.0436 | 0.0325 |
| wR(F2) [I > 2σ(I)] | 0.1073 | 0.1045 | 0.0917 | 0.0986 | 0.0806 |
| R(F) [all data] | 0.1234 | 0.1402 | 0.0467 | 0.0594 | 0.0362 |
| wR(F2) [all data] | 0.1303 | 0.1117 | 0.0956 | 0.1052 | 0.0831 |
| Goodness of fit | 1.022 | 0.991 | 1.037 | 1.050 | 1.032 |
| Flack parameter | −0.07(6) | ||||
| max/min Δ (e·Å−3) | 0.45/−0.38 | 0.41/−0.53 | 0.48/−0.31 | 0.23/−0.26 | 0.27/−0.27 |
| CCDC number | 2040699 | 2040698 | 2040700 | 2040701 | 2040702 |
| Compound | 6 | 7 | 8 | 9 | |
| Formula | C20H21NO4S | C19H18BrNO4S | C19H18BrNO4S | C18H16BrNO4S | |
| Formula weight | 371.44 | 436.31 | 436.31 | 422.29 | |
| Crystal system | triclinic | monoclinic | monoclinic | orthorhombic | |
| Space group | P-1 | P21/c | P21/c | P212121 | |
| a(Å) | 8.97622(14) | 12.4359(9) | 19.8673(3) | 8.2803(3) | |
| b(Å) | 9.83530(14) | 4.1836(3) | 4.79320(10) | 11.3223(2) | |
| c(Å) | 21.8875(3) | 17.9998(12) | 20.3846(4) | 19.3117(4) | |
| α(°) | 101.0102(13) | 90 | 90 | 90 | |
| β(°) | 94.2346(13) | 103.988(7) | 105.164(2) | 90 | |
| γ(°) | 90.0267(12) | 90 | 90 | 90 | |
| V(Å3) | 1891.36(5) | 908.70(11) | 1873.59(6) | 1810.51(8) | |
| Z | 4 | 2 | 4 | 4 | |
| Dx(g cm−3) | 1.304 | 1.595 | 1.547 | 1.549 | |
| F(000) | 784 | 444 | 888 | 856 | |
| μ(mm−1) | 1.728 | 2.402 | 4.244 | 2.408 | |
| Reflections: | |||||
| collected | 36875 | 9380 | 16185 | 10218 | |
| unique (Rint) | 36875 (0.0160) | 3151 (0.0907) | 3535 (0.0291) | 3269 (0.0307) | |
| with I > 2σ(I) | 32098 | 1829 | 3067 | 2882 | |
| R(F) [I > 2σ(I)] | 0.0656 | 0.0593 | 0.0392 | 0.0336 | |
| wR(F2) [I > 2σ(I)] | 0.1879 | 0.0605 | 0.1147 | 0.0755 | |
| R(F) [all data] | 0.0722 | 0.1341 | 0.0479 | 0.0415 | |
| wR(F2) [all data] | 0.1879 | 0.0703 | 0.1206 | 0.0782 | |
| Goodness of fit | 1.024 | 0.965 | 1.045 | 1.045 | |
| Flack parameter | −0.002(4) | ||||
| max/min Δ (e·Å−3) | 0.57/−0.32 | 0.40/−0.32 | 0.89/−0.82 | 0.66/−0.37 | |
| CCDC number | 2040703 | 2040704 | 2040705 | 2040706 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grześkiewicz, A.M.; Stefański, T.; Kubicki, M. Weak Intermolecular Interactions in a Series of Bioactive Oxazoles. Molecules 2021, 26, 3024. https://doi.org/10.3390/molecules26103024
Grześkiewicz AM, Stefański T, Kubicki M. Weak Intermolecular Interactions in a Series of Bioactive Oxazoles. Molecules. 2021; 26(10):3024. https://doi.org/10.3390/molecules26103024
Chicago/Turabian StyleGrześkiewicz, Anita M., Tomasz Stefański, and Maciej Kubicki. 2021. "Weak Intermolecular Interactions in a Series of Bioactive Oxazoles" Molecules 26, no. 10: 3024. https://doi.org/10.3390/molecules26103024
APA StyleGrześkiewicz, A. M., Stefański, T., & Kubicki, M. (2021). Weak Intermolecular Interactions in a Series of Bioactive Oxazoles. Molecules, 26(10), 3024. https://doi.org/10.3390/molecules26103024







