Abstract
Lake Mariout is one of the polluted coastal marine ecosystems in Egypt which is considered to be a reservoir of serious effluents from different anthropogenic activities. Such selective pressure enforces indigenous microbial populations to acquire new advantageous themes. Thus, in this study, two Streptomyces strains were screened, from Lake Mariout’s sediment for bioreduction of 5 mM AgNO3. Both strains were identified molecularly; their biochemical and physiological characterization revealed their ability to secrete bioactive metabolites with antagonistic activity. The cultural and incubation conditions influencing AgNPs productivity were evaluated. Subsequently, the physicochemical properties of the biofabricated AgNPs were pursued. UV-Vis spectroscopy detected surface plasmon resonance at range 458–422 nm. XRD indicated crystalline, pure, face-centered cubic AgNPs; EDX demonstrated strong silver signal at 3.5 keV. Besides, FT-IR and TGA analysis unveiled self-stabilization and functionalization of AgNPs by bioorganic molecules. However, electron microscopy micrographs depicted numerous uniform spherical AgNPs (1.17–13.3 nm). Potent bactericidal and fungicide activity were recorded by zone of inhibition assay at 50 μg/mL. Further, the antibiofilm activity was exerted in a dose-dependent manner. Moreover, the conjugation of AgNPs with the crude bioactive metabolites of both bionanofactories ameliorated the antimicrobial potency, reflecting a synergistic efficiency versus examined pathogens (free-living and biofilm).
1. Introduction
Lake Mariout is one of the major lagoons in Egypt, its shore occupied by aquaculture and fishers. Topographically, it is located in Alexandria city, with coordinates that lie between latitude 31°9′11″ (N), longitude 29°53′55″ (E); it covers 50 km2 area with water depth range 0.6–1.5 m [1]. Based on artificial embankments, this shallow brackish lake is composed of four basins separated by rods, including main basin, south basin, east basin and the aquaculture basin. Through three main inflows (El-Qalaa, El-Umum and El-Nubariya) and other small outfalls from water treatment plants and the effluent drains from the petrochemical area, the lake receives substantial industrial, agricultural and municipal discharges that degrade the quality of the lake water [2,3]. Presently, several investigations have assessed its water quality by determining several parameters such as eutrophication status, heavy metals content, microbial diversity, salinity and inorganic matter contents as well [1,4,5]. However, the heavy metals concentration in Lake Mariout sediments seemed to be higher than those recorded in other Egyptian lakes and so it was referred to as the most anthropogenically polluted and eutrophic wetland in the Nile Delta [5].
Nonetheless, such intensive doses of heavy metals in conjugation with other anthropogenic organic and inorganic pollutants compel the indigenous living communities (microbial, planktonic, aquatic fauna) to acquire new additive functionalities for maintaining their stability either by resistance or resilience. Microbially, this adaptation phenomenon mediates via physiological modifications (induction/derepression of enzymes) and genetic mutation mechanisms that guarantee their coping with such a wide range of stressors [6,7]. Interestingly, several physiological adaptation strategies could be followed by microbes, including extracellular precipitation, intracellular bioaccumulation, cell surface biosorption, enzymatic oxidation/reduction and metal efflux [8]. It is worth mentioning that such bacterial means for metal detoxification participate mainly in the bottom-up approach of nanoparticles (NPs) preparation. The biological synthesis of metal NPs is deemed to be an essential building pillar of green nanotechnology applications. Bacteria-mediated synthesis of NPs, which is classified among such green synthesis approaches, poses the potential to substitute physicochemical means. It is characterized by an environmentally friendly nature, biocompatibility behavior, energy saving and lowering the risk of global warming [9,10,11].
By the virtue of small dimensions (≤100 nm) and high surface to volume ratio, magnetic and catalytic properties of metallic NPs and their oxides, including Ag, Au, Zn, Fe, etc., find their way into distinct bactericidal formulations for different applications such as food packaging, plant disease management, water disinfection and skin care products [12,13]. Recently, the combination of different antibiotics with biogenic metals NPs enhanced their antagonistic performance which is considered an upsurge opportunity to improve adjuvant or combination therapy against multidrug-resistant (MDR) pathogens [14,15,16].
Remarkably, actinomycetes are a prominent, phenotypically diverse clade among bacterial phyla which attract special biotechnological interest. Arguably, this filamentous bacterial group is an enormously important producer for powerful functional bioactive metabolites with a vast range of biological activities such as antimicrobial, antiviral, insecticides, herbicides, anticancer, immunomodulators and probiotic activity. Besides, it secretes a diverse array of enzymes which degrade and transform xenobiotic compounds and insoluble organic polymers into their simple substituents [17]. Moreover, it is resilient in extreme, hostile and contaminated ecosystems, representing an effective agent in the bioremediation process. Whereas, it produces spores, different types of metal chelators, metal uptake systems and extracellular polymeric substances (EPS) to adapt to such excessive pressure [17]. Recently, it was also listed as bionanofactories by endowing simultaneous reduction and functionalization for the as-synthesized NPs in one single pot and without additional successive steps in a cost-effective way. Whereas, it reduces metal salt and converts it to its nanostructure in oxidation–reduction reaction via their biomolecules, meanwhile encompassing the as-synthesized NPs and serving as capping and functionalizing agents [9,10,18,19].
Apparently, the polluted marine habitat could be considered to be a rich source for screening and isolation of unique actinomycetes strains with multicharacteristic features such as exhibiting new efficient bioactive metabolites, withstanding exceedingly high concentrations of pollutants and biosynthesis of NPs. Accordingly, selection pressure in polluted Lake Mariout met this multitarget aim. In the light of the above, our investigation focused on screening, isolation and characterization of AgNPs-producing actinomycetes isolates. Thereafter, the culture conditions influencing AgNPs synthesis were optimized. Further, the study was extended to examine the antibacterial, antifungal and antibiofilm efficiency of AgNPs alone and in combination with crude secondary bioactive metabolites.
2. Results
2.1. Screening, Isolation and Identification of AgNPs-Synthesizing Actinomycetes
The current investigation focused on the biosynthesis of AgNPs by actinomycetes isolates screened from one of the most polluted marine ecosystems in Alexandria. Only two isolates designated as EM1 and EM2 were obtained based on their capability to reduce 5 mM of AgNO3 to AgNPs. The isolates were subjected to taxonomic identification by the PCR amplification of 16S rDNA gene (approximately 720 bp), Blastn analysis, pairwise and multiple sequence alignment, which revealed ≥99% identity with the sequences of Streptomyces fulvissimus EM1 and Streptomyces mediolani EM2 and were deposited in NCBI GenBank under accession numbers KY964506 and KY964507, respectively. Phylogenetic relationship between selected strains and the closely related species was represented using the neighbor-joining (NJ) method as illustrated in Figure 1.
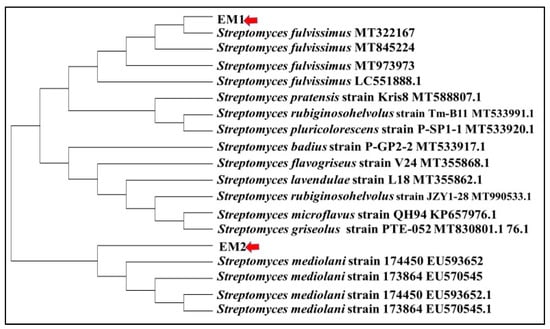
Figure 1.
Phylogenetic tree of S. fulvissimus EM1 and S. mediolani EM2 based on 16S rRNA sequences analysis representing the relationship between two strains and other members of the Streptomyces spp.
2.2. Characterization of AgNPs-Synthesizing Actinomycetes Strains
2.2.1. Cultural and Morphological Characteristics
The development of pigments, morphology, forms of aerial hyphae and growth of vegetative hyphae of both selected strains were examined in different synthetic and complex media, which were tabulated in Table 1. Generally, both strains exhibited versatility in aerial, and substrate mycelia growth ranged from poor, moderate to good growth.

Table 1.
Cultural characterization of S. fulvissimus EM1 and S. mediolani EM2 on different media.
The morphological properties of strains under study were visualized by SEM (Figure 2). As observed, well-developed aerial, vegetative hyphae were fragmented to smooth surface, long chain, small rods, non-flagellated spores. The micro morphological observations recorded the absence of both sclerotic granules and sporangia.
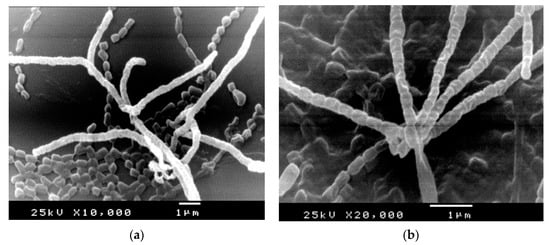
Figure 2.
SEM micrographs showing morphology of aerial hyphae, fragmentation and rod-shape spores of S. fulvissimus EM1 (a) and S. mediolani EM2 (b).
2.2.2. Physiological Characteristics
There were common physiological features between S. fulvissimus EM1 and S. mediolani EM2, including growth in the pH range of 6.5–8.5 with optimum at pH 7. Good growth was exhibited in temperature range between 20–37 °C with the optimum temperature at 30 °C; both strains tolerated NaCl up to 5% with optimum growth at 2% salt, encompassing them in intermediate salt tolerance category. Besides, the growth was inhibited in the presence of phenol (0.1%) and was not influenced by sodium azide (0.01%). In addition, both strains assimilated different carbon sources such as D-glucose, starch, galactose, fructose, sucrose; both failed to assimilate citrate, mannose, xylose and cellulose. Regarding the biochemical characteristics, both strains displayed positive response to oxidase, catalase, urease and nitrate reductase. However, both recorded negative results for hydrogen sulfide production, gelatin liquefaction, DNase, cellulase and hydrolysis of protein and lipid. Generally, the loss and gain of biochemical capabilities depend on surrounding environment that influence the loss of some genes on account of the other to adapt the environmental stress [20,21].
2.2.3. Characterization of Cell Wall Amino Acids
Chemotaxonomic analysis of S. fulvissimus EM1 showed the existence of LL- 2, 6 Diaminopimelic acid (LL-DAP) along with aspartic acid, leucine, valine, glycine, alanine, cystine, tyrosine, histidine, threonine and glycine, which ranged from trace to fair in the cell wall hydrolysate. Whereas, the cell-wall composition of S. mediolani EM2 contained LL- 2, 6 Diaminopimelic acid (LL-DAP) along with histidine and tyrosine. The findings of the work are consistent with [22,23].
2.2.4. Screening of Bioactive Compounds
The antagonistic activity of strains S. fulvissimus EM1 and S. mediolani EM2 was tested against various pathogens via streak plate approach. The antimicrobial activities of both strains were varied by suppressing the pathogenic microbes in different degrees. As displayed in Figure 3AI–II, there was no observed growth of the test pathogens, except P. aeruginosa, after 24 h near to the streaking of S. fulvissimus EM1 growth line, reflecting positive and high-score antimicrobial activity. That suggested a wide-spectrum nature for its bioactive compound. However, there was lower inhibitory effect of the bioactive metabolites produced by S. mediolani EM2, indicated by presence of growth of the test pathogens in the entire streak line, except E. coli (Figure 3BI–II).
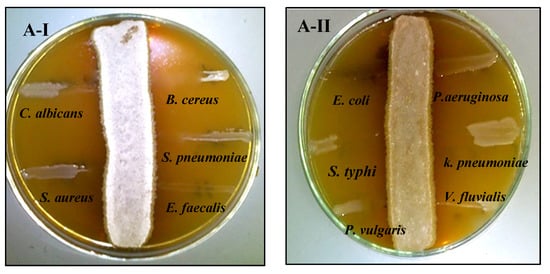
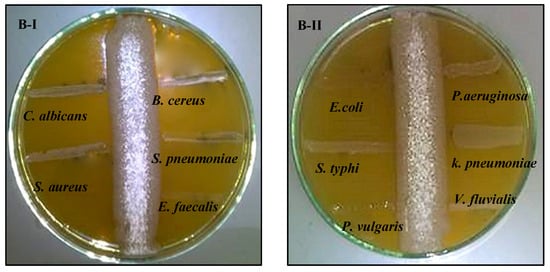
Figure 3.
Antagonistic activity of S. fulvissimus EM1 (AI–II) and S. mediolani EM2 (BI–II) against pathogenic Gram-positive, Gram-negative bacteria and yeast.
2.3. Synthesis of AgNPs and Evaluation of Factors Enhancing Their Productivity
The preliminary assessment of silver precursor reduction and the consequent AgNPs production by both actinomycetes strains were identified by the change of color of the culture media and the change of actinomycetes pellets from yellow to black. Besides, no change in color was observed with negative control medium (without biomass) and biomass control medium (without AgNO3). The productivity of AgNPs was affected by several factors such as carbon sources, nitrogen sources, temperature, pH and RPM as illustrated in Figure 4a–e. These factors did not only support bacterial multiplication, but also enhanced the production and governed the rate of enzyme activity affecting the synthesis of silver nanoparticles [2,24].The data depicted graphically in Figure 4a revealed that the best carbon sources capable of promoting AgNPs biosynthesis by S. fulvissimus EM1 were complex organic groups including malt extract, beef extract and yeast extract; whereas, malt extract, glucose and sucrose were considered the optimum carbon sources for enhancing AgNPs productivity by S. mediolani EM2. Regarding the nitrogen sources (Figure 4b), ammonium nitrate, potassium nitrate, tyrosine and peptone improved biosynthesis of AgNPs by both strains. Other nitrogen sources such as amino acid, in particular, glutamic acid and glycine, showed negative impact on AgNPs production. Meanwhile, the optimum pH and temperature that uplifted AgNPs productivity were recorded at 7 and 30 °C for both strains (Figure 4c,d). Above and below these ranges, the productivity decreased. Moreover, the effect of the agitation speed in the production of AgNPs could be arranged in the following descending order; for both strains, 200 > 150 > 100 > 50 (Figure 4e), which was coincident with [25].
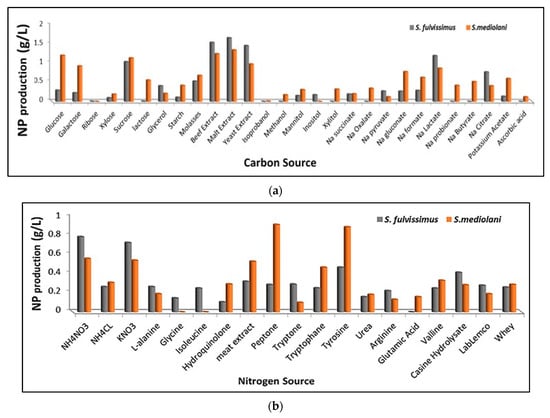
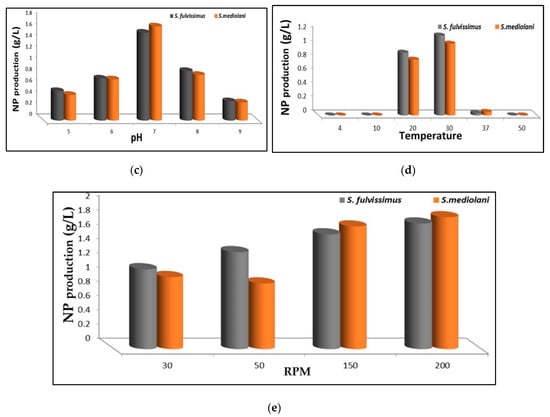
Figure 4.
Effect of different carbon sources (a), nitrogen sources (b), pH (c), temperature (d) and RPM (e) on enhancing AgNPs productivity.
2.4. Activity Determination and Molecular Detection of Nitrate Reductase Enzyme
In this study, the presence of NR enzyme was confirmed at biochemical and molecular levels. The activity of the enzyme was detected during all stages of the growth and synthesis process, where it was recorded as 53 and 71.2 µmole/min/mL after 12 h incubation for S. fulvissimus EM1 and S. mediolani EM2, respectively. It reached the maximum activity at 42 and 54 h by 212 and 371.2 µmole/min/mL for S. fulvissimus EM1 and S. mediolani EM2, respectively. At molecular level, NR enzyme was successfully detected on 1.5% agarose gel at 650 bp for both strains (Figure 5).
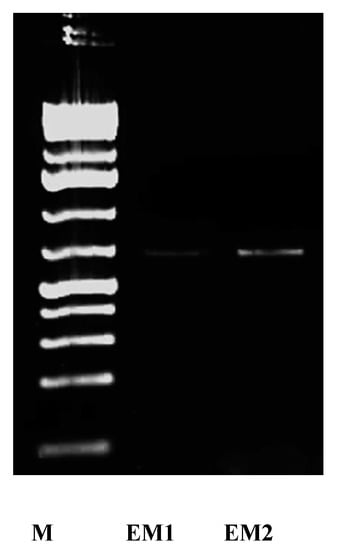
Figure 5.
Agarose gel of the PCR products of NR gene of strain S. fulvissimus EM1 and S. mediolani EM2; Lane M: 1 Kb ladder mix (GeneRuler™ Fermentase).
2.5. Production and Physicochemical Characterization of Biosynthesized AgNPs
Under selected optimized growth conditions, including carbon source (malt extract and beef extract), nitrogen source (ammonium nitrate and peptone) and neutral pH, the highest reduction of 5 mM of AgNO3 by S. fulvissimus EM1 and S. mediolani EM2, respectively, was achieved. Both cultures were incubated under 200 rpm and 30 °C. In this bottom-up approach, the silver salt was reduced into their respective NPs. The purified AgNPs were subjected to the following techniques to identify their physicochemical properties.
2.5.1. UV-Vis Spectroscopy
As a preliminary step to screen the optical and electronic features of examined nanoparticles, UV-Vis spectroscopy was employed. As demonstrated in Figure 6, a single surface plasmon resonance (SPR) band was localized at 458 and 422 nm for S. fulvissimus EM1 and S. mediolani EM2, respectively. Remarkably, the blackening of the cultural solution and pellets arose from excitation of longitudinal surface plasmon resonance (SPR), which is a unique feature for any material with a metallic nature. Nonetheless, it is affected by solution chemistry and synthesis method as reported by [26]. Likewise, [27,28] mentioned that the optimal peaks for green-synthesized AgNPs were located between 400–460 nm, which agreed with our results.
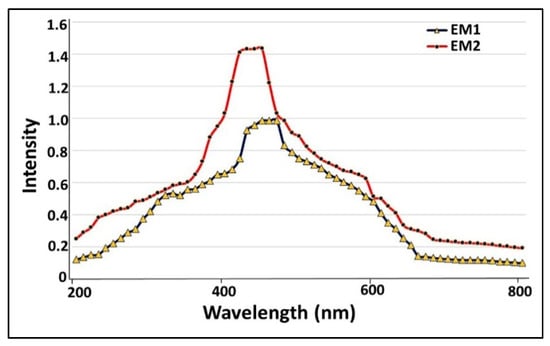
Figure 6.
UV-Vis spectroscopy of microbially synthesized AgNPs by strains S. fulvissimus EM1 and S. mediolani EM2.
2.5.2. Energy Dispersive X-ray Analysis (EDX)
The X-ray microanalysis gives qualitative as well as quantitative insinuation of elements that were involved in the fabrication of NPs. The elemental profile of the as-prepared AgNPs confirmed that silver is the main constituent element (Figure 7a,b). As shown, a strong unique elemental peak was noticed at 3.5 keV with weight percentages around 65.8%, which was ascribed to the SPR of the Ag nanocrystals. Besides, other elements such as P and S were detected in a considerable weight percentage which could be attributed to other microbial biomolecules tightly conjugated with AgNPs. The presence of such elements was commonly observed, particularly in green synthesized NPs, which suggested that they provide NPs with stabilization and functionalization [29,30].
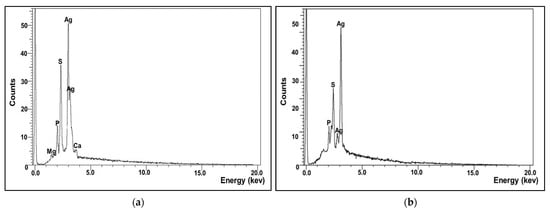
Figure 7.
Elemental composition analysis of microbially synthesized AgNPs by strains S. fulvissimus EM1 (a) and S. mediolani EM2 (b).
2.5.3. X-ray Diffraction (XRD)
The phase identity and crystalline structure of as-prepared AgNPs were verified by X-ray diffractogram. Figure 8a,b displayed four intense peaks at 2θ = 38.2°, 44.4°, 64.6° and 77.5° which correspond to hkl of (111, 200, 220 and 311) planes of face-centered cubic silver. These peaks matched with the standard pattern of JCPDS 0.4–0.783 [16]. Nonetheless, a little shift in the peak positions from XRD and small background at 2θ range 20–30° were shown, reflecting the presence of microbial proteinaceous residues associated with crystalline AgNPs. Evidently, our results are consistent with those obtained by [31,32].
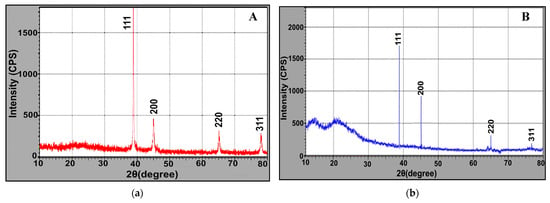
Figure 8.
XRD (a,b) analysis of microbially synthesized AgNPs by strains S. fulvissimus EM1 and S. mediolani EM2, respectively.
2.5.4. Fourier-Transform Infrared (FT-IR)
This is a vibrational study applied to characterize the surface chemistry and identify the functional groups associated with AgNPs biosynthesized by both marine isolates under study. FT-IR spectrum of the freeze-dried powder of purified biogenic AgNPs is displayed in Figure 9a,b. As evident from the figure, there is the existence of common bands in the region of 3700 cm−1 which could be ascribed to stretching vibrations of O–H groups of water molecule as referenced by [33]. The vibration bands at 3444 and 3387 cm−1 could be attributed to primary amines (NH2) [34]; the carboxylic acid (OH) stretch was located at 2936 cm−1 [35]. However, peaks at 2356 cm−1 could be assigned to C-H asymmetric stretching vibration for aliphatic groups [36]. Meanwhile, the spectral peaks in around 1644, 1636 and 1744 cm−1 implied the presence of –C=C bond [36]. For the fingerprint region (600–1500 cm−1), a number of sharp bands were clearly observed. The absorbance bands centered at 1436, 1414 and 1412 cm−1 could be attributed to C-O stretch; the absorbance peaks at wavenumber 1119 cm−1 indicated the C–N stretching [37] vibration of primary aliphatic amines [38]. However, the incidence of band at 572 cm−1 revealed the (S–S) stretch band of protein and/or P–O–C groups in phospholipids [31,38]. Other studies reported that peaks at lower field in range 400–700 cm−1 reflected the metallic nature of any examined sample [39]. Generally, FT-IR study reflected the binding of protein, carbohydrates and phospholipids with AgNPs which contributed considerably to maintaining stability of AgNPs, by acting as capping and functionalizing agents and subsequently preventing them from agglomeration [40]. Apparently, the results of this study coincided with others [31,38,41].
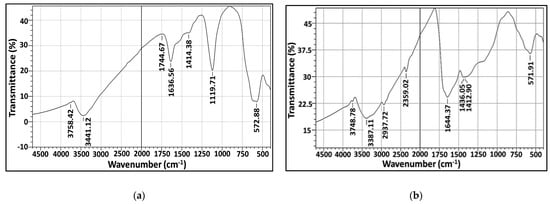
Figure 9.
FT-IR (a,b) analysis of microbially synthesized AgNPs by strains S. fulvissimus EM1 and S. mediolani EM2, respectively.
2.5.5. Thermogravimetric Analysis
TGA was conducted to study the behavior of biofabricated AgNPs at higher temperature through following up the change in their mass with temperature, which ultimately unveils their moisture content, thermal stability and the amount of organic layer surrounding them. The thermogravimetric curve represented in Figure 10a,b illustrates visible weight loss in four phases process during heating to 1000 °C in a controlled N2 atmosphere. First, the initial weight loss of AgNPs ranged from 3.5 to 4.5%, as was observed between 30 °C and 280 °C for S. fulvissimus EM1 and S. mediolani EM2, respectively. Such loss could be attributed to the evaporation of water molecules attached to AgNPs. In the second phase, 18.7 and 15.5% weight was lost between 280 °C and 620 °C for S. fulvissimus EM1 and S. mediolani EM2, respectively. The maximum weight loss by 31% was observed for both strains detected at the third phase between 620 and 770 °C, which was ascribed to the degradation of the organic residues such as proteins, polysaccharides and phospholipids conjugated to AgNPs. The final phase extended from 770 to 985 °C with weight loss assessed at 2.5%.
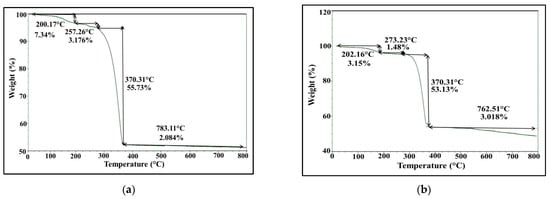
Figure 10.
TGA (a,b) analysis of microbially synthesized AgNPs by strains S. fulvissimus EM1 and S. mediolani EM2, respectively.
2.5.6. Scanning and Transmission Electron Microscopy (SEM and TEM)
The morphology and size of AgNPs, besides their dispersion uniformity, were visualized by SEM and TEM micrographs. As elucidated in Figure 11, AgNPs biosynthesized by both strains appeared as numerous opaque electrons of uniform spherical NPs with particle size ranging from 1.17 nm to 13.3 nm in slight aggregates. As observed, AgNPs synthesized intracellularly and extracellularly. After extraction, they appeared well dispersed in agreement with [31,42].
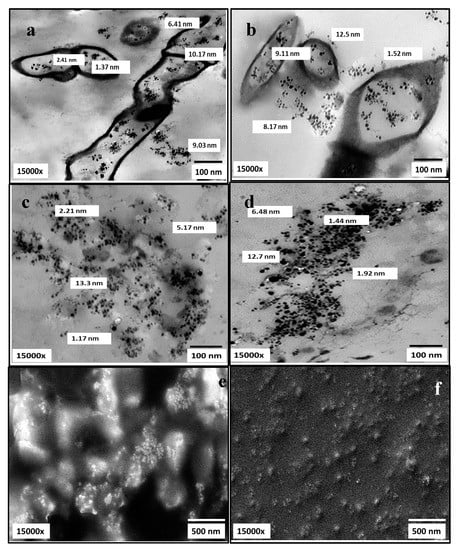
Figure 11.
Electron microscopy micrographs (TEM; a–d) and (SEM; e,f) illustrating AgNPs producing nanofactories S. fulvissimus EM1 and S. mediolani EM2 (a,b) and AgNPs size and shape after extraction.
2.6. Application of Biosynthesized AgNPs
2.6.1. Antimicrobial Activity of Biosynthesized AgNPs against Free-Living Pathogens
In this study, the inhibitory potential of the as-prepared AgNPs was examined against various microbial groups by well diffusion assay, as listed in Table 2. Notably, the highest sensitivity to AgNPs was recorded by B. cereus by 0.8 ± 0.1 cm; however, the lowest sensitivity and the highest resistance were represented by P. vulgaris by 0.2 ± 0.0 cm. Hence, the image data revealed the sensitivity order of examined free-living pathogens against biosynthesized AgNPs as summarized as B. cereus ˃ E. faecalis ˃ S. aureus ˃ E. coli ˃ S. typhi = P. aeruginosa = K. pneumoniae ˃ P. vulgaris. Generally, the as-prepared AgNPs exerted higher suppression potential in the Gram-positive bacterial group relative to the Gram-negative group. More so, the fungicide potency of the biogenic AgNPs was displayed versus C. albicans, A. bracelleuse and Alternaria sp. Obvious zones of mycostasis were noted by 1.2 ± 0.05, 0.8 ± 0.05 and 0.8 ± 0.0 cm, respectively, Table 2.

Table 2.
Antimicrobial activity of biologically synthesized AgNPs and synergistic effect in combination with crude bioactive metabolites of S. fulvissimus EM1 and S. mediolani EM2 against wide spectrum of free-living microbes.
2.6.2. In Vitro Antibiofilm Efficiency of Biosynthesized AgNPs
Biofilm is a complicated structure of microbiome in which the microbial cells aggregate in mucilaginous-like matrix composed of polysaccharides, eDNA and proteins. It could be colonized in different biotic and abiotic surfaces, including natural, medical, industrial and food-processing devices which subsequently represent serious issues. By such a complex form of growth pattern, it could be recalcitrant to nutrient starvation, osmolarity, pH changes, mechanical forces and antibiotics [43]. As referenced by [44], it could tolerate up to 1000 times more than free-living pathogens by exerting multiple resistance mechanism. Hence, the employment of NPs as an antibiofilm is being unambiguously considered as an alternative solution. In this investigation, AgNPs were utilized in suppression of the biofilm synthesis by biofilm-producing prokaryotes (P. vulgaris and B. cereus) representing Gram-negative and Gram-positive classes, respectively. Moreover, C. albicans was examined as biofilm-forming eukaryotes. Obviously, remarkable progressive inhibition of biofilm formation was noticed with increasing of AgNPs concentrations (Table 3). Similar behavior of antibiofilm activity was observed in free-living, where Gram-positive biofilm exhibited higher susceptibility than Gram-negative biofilm against different concentrations of tested AgNPs. In fact, about 44.7 ± 2.8% of B. cereus biofilm was suppressed at 50 μg/mL and reached 91.7 ± 3.5% at 150 μg/mL; whereas, 21.5 ± 1.8 and 68.4 ± 4.1% were inhibited by the biofilm of P. vulgaris at exact concentrations, respectively. However, about 85.4% survival percentage was recorded by the biofilm of C. albicans at the lowest examined concentration.

Table 3.
Antibiofilm behavior of biologically synthesized AgNPs and synergistic effect in combination with crude bioactive metabolites of S. fulvissimus EM1 and S. mediolani EM2 against biofilm-forming microbes.
2.6.3. Synergistic Antimicrobial-Antibiofilm Activities of AgNPs Combined with Crude Metabolite of Selected Strains
Herein, the extracellular bioactive metabolites of both actinomycetes strains S. fulvissimus EM1 and S. mediolani EM2 were screened previously in Section 2.2.4. The combination between such crude bioactive metabolites and the biosynthesized AgNPs (50 and 100 μg/mL) was examined toward planktonic (Table 1) and biofilm (Table 2) lifestyles, respectively. As illustrated, with the selected AgNPs concentration, such combination strongly boosted antibacterial, anticandidal, antifungal and antibiofilm activity. It enhanced antagonistic activity among free-living pathogens by the range of 1.125-fold to 2.5-fold. Moreover, it significantly improved antibiofilm activity from 39.3 ± 3.3% to 71.8 ± 5.1 and 66.9 ± 3.4% toward P. vulgaris for combination with S. fulvissimus EM1 and S. mediolani EM2 crude metabolites, respectively. Whereas, the inhibition in biofilm formation by B. cereus increased significantly from 65.5 ± 2.1% to 93.4 ± 3.7 and 88.1 ± 3.9% for combination with S. fulvissimus EM1 and S. mediolani EM2 crude metabolites, correspondingly. Regarding C. albicans, the inhibition percentage increased by more than two-fold via such combination. Notably, the combination with crude metabolite excreted by strain S. fulvissimus EM1 displayed higher biocide potency than that shown by S. mediolani EM2.
3. Discussion
The emergence of multidrug-resistant pathogens (MDR) is an even more serious threat that is directly related to upsurges in mortality rate among severe nosocomial infections. Such a phenomenon appeared today as a result of globalization, an increase in growing population and immoderate, uncontrolled and multiple use of antibiotics and chemotherapeutics which ultimately restricted antibiotic therapy. Subsequently, success in finding new proceedings and new agents is the decisive solution for this issue. Therefore, exploring advantageous microbial groups with improved physiological capabilities from unique environmental habitats is urgently required. Additionally, the hybridization between nanotechnology and antimicrobial therapy is a promising approach to defeat this threat. Remarkably, the actinomycetes are classified among the most essential producers of efficient bioactive metabolites. As reported by [39], they produced more than 45% of known pharmaceutical products; additionally, they were categorized among nanoparticle producers, thus occupying a prominent site in both medical and biotechnological sectors.
Hence, the results of the present study were deemed characteristic where two marine strains were isolated from high salinity and contaminated lake and have the ability to produce active metabolites along with AgNPs. Thereafter, different nutritional parameters were evaluated to enhance the productivity of AgNPs. As carbon sources, malt extract and beef extract were selected; whereas, ammonium nitrate and peptone were utilized as nitrogen sources, in neutral pH and at 30 °C, to accelerate the reduction of 5 mM of AgNO3 by S. fulvissimus EM1 and S. mediolani EM2, respectively. Remarkably, several studies recorded the urgent requirement of actinomycetes for highly nutritive substrates that support metabolic reduction of the metals to their nanoform counterparts [45]. Reference [2] found that peptone-containing medium was the most significant nutritive factor for AgNPs production from S. viridodiastaticus. Besides, in agreement with our results, [2,24], declared that the internal environment of living cells is believed to be nearly neutral, so the activity of bacteria decreased as the pH deviates from neutral conditions. Reference [46] reported that the maximum AgNPs production was achieved at 25 °C, while at high temperature (40 °C), the enzyme activity decreased and hence the synthesis of AgNPs slowed down. However, References [47,48], found that optimum temperature for enzyme activity that enhanced AgNPs production was in the range of 20–30 °C.
However, the nitrate reductase enzyme, key enzyme that regulated the AgNPs formation, was determined successfully at molecular and biochemical levels. Interestingly, NADH-dependent nitrate reductase (NR) was proposed by various investigations to be involved substantially in NPs synthesis (Au, Fe, Cu, etc.) as a catalyzing biomolecule [49]. In general, the enzyme transforms nitrate to nitrite and the electron shuttle is stimulated to reduce the dissociated metal ions to their nanoform counterparts, in a continuous cycle of oxidation reduction reactions [50]. It is noteworthy to mention that the degenerated primers designed by [51] succeeded in detecting narG gene in several bacterial genera, including Thermus spp., Streptomyces, Corynebacterium, Mycobacterium, Bacillus spp. Staphylococcus spp., Brucella spp. and Ralstonia spp., which inferred the versatility of nitrate reductase function. It could be expressed variously for certain different functions in different organisms at different growth circumstances. Under microaerophilic conditions, it mediated the oxidation of NADH or any carbon source to generate proton gradient that enabled the production of ATP in Campylobacter jejuni [52]. Moreover, it catalyzed nitrate respiration in denitrification process anaerobically in Paracoccus denitrificans PD1222 [53]. Whereas, it dissipated excess electrons generated from growth on electron-rich substrates, under aerobic conditions, to maintain redox homeostasis in Rhodobacter sphaeroides [54]. Further, it played a crucial role in scavenging low concentrations of nitrate in Shewanella spp. [55].
By different characterization approaches, including UV-Vis spectroscopy, XRD, EDX, FT-IR, TGA, SEM and TEM, the optical, structural, elemental and morphological properties of the biosynthesized AgNPs were described. The biosynthesized AgNPs exhibited uniform spherical shape with slight aggregation associated with bioorganic molecules which eventually led to delaying their volatilization, stabilization and functionalization as implied by EDX, FT-IR and TGA. Such results matched those reported by [14,56,57,58]. Upon recruitment as antimicrobial agent, AgNPs displayed considerable and significant clear zones of inhibition in all examined pathogens, which were deemed good and effective as long as the zone exceeded 1 mm as highlighted by [59]. Notably, their effectiveness was more evident among Gram-positive group than Gram-negative one. That could be assigned to the structural variations of the outer cell wall. Whereas, Gram-positive bacteria were recognized by a thick peptidoglycan layer (20–80 nm) in the cell wall which exerts higher permeability than that of Gram-negative, which represents 7–8 nm. That makes Gram-positive bacteria more susceptible to biocidal agents. On the other hand, Gram-negative bacteria are characterized by the presence of an additional external lipopolysaccharide layer which might accumulate AgNPs in aggregates, preventing their entrance to the interior of the cell. In addition, Gram-negative bacteria pose a powerful multiple efflux pump system that externally expels any detrimental agents [59,60]. Remarkably, the efficiency of NPs versus pathogens was influenced by the microbial physiology, its metabolism, NPs dose, degree of contact and their diffusion rate. Apparently, our results match those reported elsewhere [16,60,61]. Moreover, the antifungal potency was pronounced against examined C. albicans, A. bracelleuse and Alternaria sp. Such fungicide efficacy could be explained by high damage of glycoprotein–glucan–chitin cross linkage which is the main constituent of the fungi cell wall. Consequently, the penetrated AgNPs caused metabolic collapse of the cellular biochemistry [11,31].
Interestingly, the higher surface area (surface/volume ratio) accompanied by small particle size NPs (10–80 nm) is considered the decisive factor in the biocidal activity. Such features enable a faster dissolution rate and more tight binding with the microbial cell which eventually lead to efficient cytotoxicity [62]. The antimicrobial activity of NPs could be summarized in the following steps. Whereas, it tightly attaches to the microbial cells, causing pits and holes in the cell wall, weakening membrane integrity and creating osmotic imbalance. Once NPs get inside the cell, it dissolves quickly in cytoplasmic solution, generating Ag ions that uplift reactive oxygen species level (ROS) and increase massive oxidative stress. Additionally, AgNPs interact selectively with active sites of biomolecules, causing protein malfunctioning and DNA disorder [40,63].
On the other hand, AgNPs showed antibiofilm capability in a dose-dependent manner, as the inhibition percentage decreases with continuous elevation in the treatment dosage. Our results concurred with the findings of [64,65]. The antibiofilm potency of biofabricated AgNPs could be exerted in multisuccessive stages begun by inhibition of the planktonic forms (initial stage of biofilm), followed by inhibition of exopolysaccharide formation which adheres to and aggregates the sessile cells (second stage), then passing through to blocking the quorum sensing activity [63].
Notably, several investigations have reported the enhancement of NPs efficiency in the combination of standard antimicrobial agents, hence, overcoming troubleshooting of multidrug resistance [15,16]. Therefore, the combination between the biogenic AgNPs and the crude bioactive metabolites of S. fulvissimus EM1 and S. mediolani EM2 was assessed and showed enhanced biocidal efficacy. Broadly, the highest antagonistic activity of such a combination could be attributed to the synergetic or additive effect of both constituents that disturb the microbial cell, frustrating its capability to mutate its genome for tolerating such condensed antimicrobial dose. Moreover, such a combination between bioactive metabolites and AgNPs might be directed concurrently to multiple active sites in the microbial cell, generating multiple damages such as cell wall deterioration, DNA/RNA inhibition and protein denaturation. That conclusion paves the way for promising avenues for its utilization in adjuvant therapy to overcome multiple antibiotic-resistant phenomena. Likewise, References [14,15,16] discussed similar findings. All of them found that the enhanced antagonistic activity of AgNPs was supported with certain antibiotics. Nonetheless, the exact constituent that prohibits the microbial growth in the crude bioactive metabolites of S. fulvissimus EM1 and S. mediolani EM2 will be optimized, extracted, purified and identified in an ongoing study. Generally, the current study implied the promising capability of AgNPs conjugated with crude metabolites in the management of infectious diseases, drug development and prevention of bacterial colonization.
4. Materials and Methods
4.1. Collection of Samples
Samples of water and sediments were collected from different places at Lake Mariout (main basin). The water samples were homogenously mixed, transported in sterile bottles, the different sediment samples were thoroughly mixed and placed in sterile plastic bags, then the samples were placed in ice until they were transferred to the laboratory. The samples were then stored at 4 °C before the actinomycetes were isolated.
4.2. Screening and Isolation of AgNPs-Producing Actinomycetes
Actinomycetes that have the capability to synthesize AgNPs were screened from the collected samples by serial dilution method on starch–nitrate medium, with the following components (g/L): 20 starch, 0.5 K2HPO4, 1 KNO3, 0.5 MgSO4.7H2O, 0.01 FeSO4 and 15 agar (for solid medium), pH 7. The medium was supplemented by different concentrations of AgNO3 (1.5, 3 and 5 mM). The inoculated plates were incubated at 30 °C for 7 days [39]. The actinomycetes isolates that had the ability to synthesize AgNPs caused blackening in media as a result of AgNO3 reduction. The selected colonies with different morphological characteristics were isolated and purified by streaking on the modified International Streptomyces Project 2 agar medium (ISP2). These isolated strains were regularly subcultured and stored on agar slants at 4 °C and hyphal fragments were preserved in 20% glycerol (v/v) at −80 °C until further use.
4.3. Molecular Identification of Actinomycetes Isolates Synthesizing AgNPs
The actinomycetes pellets of two isolates were obtained by their cultivation in shake flask of starch nitrate broth at 150 rpm at 30 °C for 4 days. Cells were collected by centrifugation and washed successive times with sterile phosphate buffer (pH 7). The bacterial genomic DNA of the selected isolates was extracted according to protocol followed by [15]. PCR amplicon was conducted using genomic DNA (0.1 μg) as a template and 20 pmol/μL of commercially synthesized 16S universal primer pairs 27f (5′AGA GTT TGA TCC TGG AG3′) and 1492r (5′TAC GGC TAC CTT GTT ACG ACT3′) in a 25 μL reaction volume containing 1 unit of Taq (Thermo Scientific), 15 μL of buffer and 2 μL of 10 mM dntp mix (Thermo Scientific). The PCR conditions began at 94 °C for 5 min as initial denaturation step, followed by 35 cycles of 94 °C for 20 s, 55 °C for 1 min and 72 °C for 1 min; the final extension step was at 72 °C for 10 min. Volumes of 5 µl of the PCR products were examined by loading on 1% agarose gel. The amplified PCR products were purified with GenEluateTM PCR Clean-Up Kit (Sigma) and sequenced. The phylogenetic position was inquired by comparing the procured sequences with the database sequences of NCBI. The sequences were deposited in the GenBank to obtain corresponding accession numbers. Evolutionary tree was inferred using the neighbor-joining technique by MEGA 6 software package.
4.4. Characterization of the Actinomycetes Isolates Synthesizing AgNPs
4.4.1. Cultural Characteristics on Different Media
The macro-morphological and cultural characteristics of the selected isolates were studied by inoculation in the sterile International Streptomyces Project (ISP media): ISP-1 (casein yeast extract agar), ISP-2 (yeast extract, malt extract agar), ISP-4 (inorganic salt starch agar), ISP-5 (glycerol asparagine agar), ISP-6 (peptone yeast extract iron agar), ISP-7 (tyrosine agar). Moreover, their cultural behavior on LB, NB, glycerol-asparagine, casein-nitrate, starch-nitrate, starch-casein agar, Kuster’s agar and Bennet’s agar were tested [66,67]. Media were sterilized and poured into sterile plats. After solidifying the media, the culture of the selected isolates was streaked onto the medium surface and incubated for 7 days at 30 °C. Morphological characteristics such as colony characteristics, aerial hyphae type, vegetative hyphae growth, spore formation and pigments excretion were observed.
4.4.2. Morphological Characteristics
The micro-morphological features of selected actinomycetes isolates, including hyphae fragmentation, spore chain, spore ornamentation and presence of sporangia were investigated and visualized by scanning electron microscope. At culture age 14 day, the specimens were prepared, fixed in glutaraldehyde (3%, v/v), washed and post-fixed in 1.5% osmium tetroxide for 2 h. The samples were washed, dehydrated by ethanol (40–100%), coated with gold and examined at 15–20 kV by SEM-JEOL JEM-1230-Japan.
4.4.3. Physiological Characteristics
The growth of selected isolates under different pH (3, 5, 7, 9, 10 and 11), temperatures (10, 20, 30, 40 and 50 °C), salinity (1, 2, 3, 5, 7, 10 and 20%) was examined. The growth pattern in the presence of 0.1% phenol and sodium azide was recorded. However, the biochemical activities exhibited by both strains were determined by assimilation of different carbon sources and some enzymes. The carbon utilization test was performed using ISP-9 supplemented with 1% of the filter-sterilized D-glucose, sucrose, fructose, D-maltose, D-galactose, citrate, mannose, xylose, cellulose and starch. The mineral salt agar supplemented with starch, Tween 20, gelatin, casein, DNA, nitrate and urea was employed to examine the presence of amylase, lipase, gelatinase, protease, DNase, nitrate reductase and urease, respectively; besides, Kligler’s Iron Agar (KIA) was used to determine hydrogen sulfide production. Furthermore, oxidase and catalase were investigated according to [67].
4.4.4. Characterization of Cell Wall Amino Acids
The chemical composition of the isolates cell wall was determined. Diaminobimlic acid isomers have been determined as described by [68].
4.4.5. Screening of Bioactive Compounds
The perpendicular streaking method was employed to detect the presence of bioactive metabolites against some pathogens (Pseudomonas aeruginosa (ATCC 15442), Escherichia coli (ATCC 25922), Salmonella typhimurium (ATCC 14028), Klebsiella pneumonia (ATTC 700603), Proteus vulgaris (ATCC-8427), Vibrio fluvialis (ATCC 33809), Bacillus cereus (ATCC 33019), Staphylococcus aureus (ATCC 29213), Enterococcus faecalis (ATCC 29212), Streptococcus pneumoniae (ATCC 6303) and Candida albicans (ATCC 10231)). Briefly, the pure actinomycetes isolates were inoculated in a single line streak on nutrient agar and incubated at 30 °C for 4 days for the development of diffusible antimicrobial secondary metabolite. Thereafter, a single streak was inserted perpendicular to the actinomycetes line by each test pathogen in a parallel position to each other. The plates were incubated for 24 h at 37 °C [69].
4.5. Synthesis of AgNPs and Evaluation of Factors Enhancing Their Productivity
The biosynthesis process was initiated by inoculating 7 discs (9 mm diameter) taken from 72 h cultures of selected isolates in starch nitrate broth supplemented with 5 mM of AgNO3. The inoculated cultures were incubated for 120 h at 30 °C in an orbital shaker at 150 rpm. For enhancing the productivity of AgNPs, different parameters were scrutinized, including temperatures (4, 10, 20, 30, 40 and 50 °C), pH (5, 6, 7, 8 and 9), agitation (30, 50, 150 and 200 rpm), carbon sources (monosaccharides: ribose, xylose, glucose, galactose; disaccharides: sucrose, lactose; polysaccharides: starch; complex organic: beef extract, malt extract, yeast extract; sugar alcohols: isopropanol, methanol, glycerol, xylitol, mannitol, inositol; organic salts: citrate, oxalate, lactate, formate, succinate, butyrate, pyruvate, propionate; agricultural wastes: molasses) and nitrogen sources (inorganic salts: NH4NO3, NH4Cl, KNO3, urea; amino acids: L-alanine, L-glutamic acid, glycine, iso-leucine, valline, arginine, l-tyrosine, tryptophan; complex organic compounds: peptone, Lab-Lemco, casein hydrolysate, meat extract; dairy product wastes: whey). The incubation was followed as described formerly. The AgNPs were harvested by centrifugation, washed and extracted from the cells through conjugation of chemical lysis and physical disruption by sonication for 10 min at 40–60% amplitude and frequency of 20 kHz with 0.6 s pulse rate. The lysed suspension was vortexed vigorously for 5 min to obtain a homogeneous mixture. The extracted NPs were weighted and subjected to physicochemical analysis after centrifugation, washing and drying.
4.6. Activity Determination and Molecular Detection of Nitrate Reductase Enzyme
Under optimized conditions, the nitrate reductase activity for both isolates was assessed, as it was considered to be the key enzyme governing the bioreduction process [70,71]. In brief, the pellets of bionanofactories were harvested, washed and disrupted using mild osmotic shock. The obtained slurry was centrifuged at 12,000 rpm, 10 °C for 20 min. The supernatant was utilized as crude enzyme in a reaction mixture containing 0.2 M phosphate buffer (pH 7.0), 20 mM KNO3, 5 mM Benzyl Viologen and 10 mM sodium dithionite. After incubation at 30 °C for 30 min, the reagent A (2% sulfanilic acid) and B (0.2% N,N-dimethyl-1-naphthylamine) were added to the reaction mixture for detecting nitrite liberated as a result of nitrate reduction. The pink color was measured spectrophotometrically at 540 nm. Nitrite standard curve was performed to determine the concentration of nitrite. Remarkably, one unit of nitrate reductase represented the amount of enzyme that catalyzes the production of 1 μmol of nitrite/min or 1 μmol of nitrate reduced/min under standard assay conditions [70].
Moreover, the nitrate reductase enzyme was detected at molecular level using specific primers narG-1960f (5′ TAYGTSGGSCARGARAA 3′) and narG-2650r (5′ TTYTCRTACCABGTBGC 3′) which correspond to the large subunit of nitrate reductase enzyme (narG) [27]. The reaction mixture of 50 μL contained 2 μL of 25 ng DNA, 5 μL of 10× PCR buffer, 2 μL of 10 mM deoxynucleotide triphosphate (dATP, dCTP, dGTPand dTTP) mix, 10 μL (20 pmol) of primers and 1 U of Taq-polymerase; the thermocycling conditions included 5 min at 95 °C, 35 cycles of 1 min at 94 °C, 30 s at 60 °C, 45 s at 72 °C, 10 min at 72 °C. The amplified products were visualized on 1.5% (w/v) agarose gels stained with ethidium bromide (0.125 μg/mL) along with 1 Kb ladder mix (GeneRuler™ Fermentase) as a DNA marker and photographed in the MultiImage light cabinet.
4.7. Production and Physicochemical Characterization of Biosynthesized AgNPs
The absorption properties of the as-synthesized AgNPs were assessed at room temperature by Labomed model UV-Vis spectrophotometer. The maximum surface plasmon resonance (SPR) was detected in a wavelength range of 200–800 nm, as a first step for monitoring AgNO3 reduction. The fingerprint identification of AgNPs was determined using X-ray diffractometer (Shimadzu 7000, USA) that operates with Cu Kα radiation tube (λ = 1.5406 Å). The voltage and electric current were 30 kV and 30 mA, respectively; the scanning rate 2°/min for 2θ angular range of 2° to 80°. The obtained XRD patterns were compared with JCPDS database to analyze the data. The elemental contents of examined AgNPs samples were conducted by EDX detector connected with SEM- JEOL JEM-1230, Japan. However, TEM and SEM-JEOL JEM-1230-Japan, operating at 200 kV, were used to describe the morphology and size of as-prepared AgNPs. In addition, the surface chemistry and functional molecules associated with the biosynthesized AgNPs were detected, after washing, drying, grinding with KBr and formulation into pellets, by Shimadzu FT*IR-8400 S, Japan. The FT-IR spectrum was scanned in the region of 4000 to 400 cm−1 wave number at a resolution of 4 cm−1. Moreover, the thermal behavior of biosynthesized AgNPs toward increasing in temperature was analyzed by TGA-50H, Shimadzu (Japan). The experiment was performed in a nitrogen atmosphere at temperature range of 35–1000 °C and with heating rate of 20 °C/min [64,65].
4.8. Application of Biosynthesized AgNPs
4.8.1. Antimicrobial Activity of Biosynthesized AgNPs
The antimicrobial activity of the biosynthesized AgNPs was examined versus various microbial pathogenic groups by well-diffusion method (zone of inhibition, ZOI). The examined procaryotic groups included Gram-negative bacteria (P. aeruginosa (ATCC 15442), E. coli (ATCC 25922), P. vulgaris (ATCC-8427), K. pneumoniae (ATTC 700603) and S. typhimurium (ATCC 14028)), Gram-positive bacteria (B. cereus (ATCC 33019), S. aureus (ATCC 29213) and E. faecalis (ATCC 29212)); however, the eukaryotic pathogens encompassed yeast C. albicans (ATCC 10231) and molds (Aspergillus brasiliensis (ATTC -16404) and Alternaria sp.). About 1 × 108 CFU/mL of freshly prepared pathogens was spread uniformly on the Müller-Hinton agar using sterile cotton swab. The sterile cork-borer (10 mm) was used to make even punctures which were loaded by 50 μg/mL of biologically synthesized AgNPs in distilled water. The bacterial plates were incubated at 37 °C for 24 h; whereas, fungal plates were incubated at 25 ± 2 °C for 4–5 days. Negative control wells were run in parallel, containing distilled water. Upon the end of incubation, the plates were investigated for the presence of inhibition zone surrounding each well, which were measured and expressed in centimeters (cm) [65].
4.8.2. In Vitro Antibiofilm Efficiency of Biosynthesized AgNPs
The inhibitory activity of the biosynthesized AgNPs was evaluated using the microtiter plate assay (MTP). In 96-well flat bottom polystyrene titer plates, about 1 × 106 CFU/mL fresh cultures of P. vulgaris (ATCC-8427), B. cereus (ATCC 33019) and C. albicans (ATCC 10231) were inoculated separately in each well containing 100 μL of Tryptone Soy Broth (TSB). About 50 μL of AgNPs solution (50, 100 and 150 μg/mL) was added to each well. Positive and negative controls were examined simultaneously, containing bacterial suspension and sterile media only, respectively; the plates were incubated for 24 h at 37 °C. After incubation, the plates were processed according to the procedure described by [65]. The absorbance of the attached cells was assessed at 595 nm by a microtiter ELISA reader (Tecan Infinite M200, Switzerland), and the biofilm inhibition percentage was calculated by the following formula:
where A represents the absorbance of the positive control and A0 the absorbance of the AgNPs-treated well.
Inhibition percentage of biofilm = [(A − A0/A) × 100]
4.8.3. Synergistic Antimicrobial-Antibiofilm Activities of AgNPs Combined with Crude Metabolite of Selected Isolates
The combination of AgNPs and the crude bioactive metabolites excreted by selected isolates was evaluated against the previously mentioned pathogens, both prokaryotes and eukaryotes, either free-living or biofilm. The crude bioactive metabolites were prepared by inoculating actinomycetes isolates separately in 75 mL of ISP2 media containing the following ingredients (g/L); malt extract, 10.0; yeast extract, 4.0; dextrose, 4.0. The final pH was adjusted to be 7.0 ± 0.2. The flasks were incubated at 30 °C for 4 days in an orbital shaker at 150 rpm. At the end of the incubation period, the cultures were collected by centrifugation at 12,000 rpm for 20 min. The obtained cell-free supernatant, which represents the crude bioactive metabolite, was filter sterilized with a 0.22 μm syringe filter. The combination was employed by thorough suspending of 50 and 100 μg/mL of AgNPs in the sterile crude bioactive metabolites rather than distilled water for free-living and biofilm, correspondingly. All the procedures for well diffusion method and microtiter plate assay were accomplished as previously mentioned in details [15,16].
4.9. Data Analysis
All results displayed in this investigation were represented by the means of three independent replicates ± standard error of the mean (SEM). The antimicrobial and antibiofilm activity data were subjected to analysis of variance (ANOVA) by GraphPad Prism software. Tukey post hoc was employed to analyze the mean difference comparison between the treatments. In all analyzed data, a probability level of p ≤ 0.05 was considered for the significance of differences between values.
5. Conclusions
This study sheds light on the biosynthesis of AgNPs via two Streptomycetes sp. strains isolated from Lake Mariout in a simple, less expensive, efficient and eco-friendly approach. It is anticipated that isolation of actinomycetes from a polluted marine ecosystem will be considered to be useful in the discovery of characteristic strains that exhibit a dual and simultaneous role as secondary metabolites producer and bionanofactory. The impact of different nutritional parameters on productivity of AgNPs was examined. The synthesized AgNPs under optimum conditions were characterized using UV-Vis spectroscopy, XRD, EDX, FT-IR, TGA, SEM and TEM. By applying as antimicrobial agent, the biosynthesized AgNPs exhibited promising antagonistic activity versus wide spectrum of Gram-positive and Gram-negative bacteria. AgNPs’ recorded ZOI ranged from 0.2 ± 0.0 cm by P. vulgaris to 0.8 ± 0.1 cm by B. cereus. The sensitivity order of examined free-living pathogens against biosynthesized AgNPs is summarized as B. cereus ˃ E. faecalis ˃ S. aureus ˃ E. coli ˃ S. typhi = P. aeruginosa = K. pneumoniae ˃ P. vulgaris. Additionally, powerful antifungal and antibiofilm efficacy were observed. By combination of AgNPs with secondary bioactive metabolites of both strains, the antagonistic capability increased by 1.125-fold to 2.5-fold for free-living pathogens and more than two-fold for biofilm and fungal pathogens, reflecting potential application in overcoming microbial threats.
Author Contributions
Conceptualization, M.E. and A.I.; methodology, H.E.-s. and E.I.; software H.E.-s. and E.I.; validation, M.E., A.I., H.E.-s. and E.I.; formal analysis, M.E., A.I., H.E.-s. and E.I.; investigation, H.E.-s. and E.I.; resources, M.E. and A.I.; data curation, M.E. and A.I.; writing—original draft preparation, M.E. and A.I.; writing—review and editing, M.E. and A.I.; visualization, M.E., A.I., H.E.-s. and E.I.; supervision, M.E., A.I. and E.F.; project administration, M.E., A.I. and F.A.; funding acquisition, F.A. All authors have read and agreed to the published version of the manuscript.
Funding
Taif University Researchers Supporting Project number (TURSP - 2020/222), Taif University, Taif, Saudi Arabia.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding authors.
Acknowledgments
The authors would like to thank the Deanship of Scientific Research at Taif University for funding this work through Taif University Researchers Supporting Project number (TURSP - 2020/222), Taif University, Taif, Saudi Arabia.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of the compounds are not available from the authors.
References
- El Zokm, G.M.; Tadros, H.R.Z.; Okbah, M.A.; Ibrahim, G.H. Eutrophication assessment using TRIX and Carlson’s indices in lake mariout water, Egypt. Egypt. J. Aquat. Biol. Fish. 2018, 22, 321–349. [Google Scholar] [CrossRef]
- Mohamedin, A.; El-Naggar, N.E.-A.; Hamza, S.S.; Sherief, A. Green Synthesis, Characterization and Antimicrobial Activities of Silver Nanoparticles by Streptomyces viridodiastaticus SSHH-1 as a Living Nanofactory: Statistical Optimization of Process Variables. Curr. Nanosci. 2015, 11, 640–654. [Google Scholar] [CrossRef]
- Ahmed, S.M. Impact of different pollutants on some water quality aspects of Lake Mariout, Egypt. Egypt. J. Aquac. 2021, 10, 17–37. [Google Scholar] [CrossRef]
- Donia, N.; Bahgat, M. Water quality management for Lake Mariout. Ain Shams Eng. J. 2016, 7, 527–541. [Google Scholar] [CrossRef]
- Abu El-Magd, S.A.; Taha, T.; Pienaar, H.; Breil, P.; Amer, R.; Namour, P. Assessing heavy metal pollution hazard in sediments of Lake Mariout, Egypt. J. Afr. Earth Sci. 2021, 176, 104116. [Google Scholar] [CrossRef]
- Azarbad, H.; Van Gestel, C.A.; Niklińska, M.; Laskowski, R.; Röling, W.F.M.; Van Straalen, N.M. Resilience of soil microbial communities to metals and additional stressors: DNA-based approaches for assessing “stress-on-stress” responses. Int. J. Mol. Sci. 2016, 17, 933. [Google Scholar] [CrossRef] [PubMed]
- Ford, T. Pollutant effects on the microbial ecosystem. Environ. Health Perspect. 1994, 102, 45–48. [Google Scholar] [CrossRef] [PubMed]
- Benmalek, Y.; Fardeau, M.-L. Isolation and characterization of metal-resistant bacterial strain from wastewater and evaluation of its capacity in metal-ions removal using living and dry bacterial cells. Int. J. Environ. Sci. Technol. 2016, 13, 2153–2162. [Google Scholar] [CrossRef]
- Alsharif, S.M.; Salem, S.S.; Abdel-Rahman, M.A.; Fouda, A.; Eid, A.M.; Hassan, S.E.-D.; Awad, M.A.; Mohamed, A.A. Multifunctional properties of spherical silver nanoparticles fabricated by different microbial taxa. Heliyon 2020, 6, e03943. [Google Scholar] [CrossRef] [PubMed]
- Salem, S.S.; El-Belely, E.F.; Niedbała, G.; Alnoman, M.M.; Hassan, S.E.-D.; Eid, A.M.; Shaheen, T.I.; Elkelish, A.; Fouda, A. Bactericidal and In-Vitro Cytotoxic Efficacy of Silver Nanoparticles (Ag-NPs) Fabricated by Endophytic Actinomycetes and Their Use as Coating for the Textile Fabrics. Nanomaterials 2020, 10, 2082. [Google Scholar]
- Soliman, A.M.; Abdel-Latif, W.; Shehata, I.H.; Fouda, A.; Abdo, A.M.; Ahmed, Y.M. Green Approach to Overcome the Resistance Pattern of Candida spp. Using Biosynthesized Silver Nanoparticles Fabricated by Penicillium chrysogenum F9. Biol. Trace Elem. Res. 2021, 199, 800–811. [Google Scholar] [CrossRef]
- Kharissova, O.V.; Kharisov, B.I.; González, C.M.O.; Méndez, Y.P.; López, I. Greener synthesis of chemical compounds and materials. R. Soc. Open Sci. 2019, 6, 191378. [Google Scholar] [CrossRef] [PubMed]
- Talapko, J.; Matijević, T.; Juzbašić, M.; Antolović-Požgain, A.; Škrlec, I. Antibacterial activity of silver and its application in dentistry, cardiology and dermatology. Microorganisms 2020, 8, 1400. [Google Scholar] [CrossRef]
- Patra, J.K.; Baek, K.H. Antibacterial activity and synergistic antibacterial potential of biosynthesized silver nanoparticles against foodborne pathogenic bacteria along with its anticandidal and antioxidant effects. Front. Microbiol. 2017, 8, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Wypij, M.; Czarnecka, J.; Świecimska, M.; Dahm, H.; Rai, M.; Golinska, P. Synthesis, characterization and evaluation of antimicrobial and cytotoxic activities of biogenic silver nanoparticles synthesized from Streptomyces xinghaiensis OF1 strain. World J. Microbiol. Biotechnol. 2018, 34, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Bakhtiari-Sardari, A.; Mashreghi, M.; Eshghi, H.; Behnam-Rasouli, F.; Lashani, E.; Shahnavaz, B. Comparative evaluation of silver nanoparticles biosynthesis by two cold-tolerant Streptomyces strains and their biological activities. Biotechnol. Lett. 2020, 42, 1985–1999. [Google Scholar] [CrossRef] [PubMed]
- Timková, I.; Sedláková-Kaduková, J.; Pristaš, P. Biosorption and bioaccumulation abilities of actinomycetes/streptomycetes isolated from metal contaminated sites. Separations 2018, 5, 54. [Google Scholar] [CrossRef]
- Aref, M.S.; Salem, S.S. Bio-callus synthesis of silver nanoparticles, characterization, and antibacterial activities via Cinnamomum camphora callus culture. Biocatal. Agric. Biotechnol. 2020, 27, 101689. [Google Scholar] [CrossRef]
- Gu, H.; Chen, X.; Chen, F.; Zhou, X.; Parsaee, Z. Ultrasound-assisted biosynthesis of CuO-NPs using brown alga Cystoseira trinodis: Characterization, photocatalytic AOP, DPPH scavenging and antibacterial investigations. Ultrason. Sonochem. 2018, 41, 109–119. [Google Scholar] [CrossRef]
- Casjens, S. The diverse and dynamic structure of bacterial genomes. Annu. Rev. Genet. 1998, 32, 339–377. Available online: https://pubmed.ncbi.nlm.nih.gov/9928484/ (accessed on 26 February 2021). [CrossRef] [PubMed]
- Cases, I.; De Lorenzo, V.; Ouzounis, C.A. Transcription regulation and environmental adaptation in bacteria. Trends Microbiol. 2013, 11, 248–253. [Google Scholar] [CrossRef]
- Prakasham, R.S.; Kumar, B.S.; Kumar, Y.S.; Shankar, G.G. Characterization of silver nanoparticles synthesized by using marine isolate Streptomyces albidoflavus. J. Microbiol. Biotechnol. 2012, 22, 614–621. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T. Comparison of the Cell-Wall Composition of Morphologically Distinct. J. Bacteriol. 1965, 89, 444–453. [Google Scholar] [CrossRef] [PubMed]
- El-Naggar, N.E.-A.; Abdelwahed, N.A.M. Application of statistical experimental design for optimization of silver nanoparticles biosynthesis by a nanofactory Streptomyces viridochromogenes. J. Microbiol. 2014, 52, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.; Liao, T.-H.; Wang, J.; Ye, Y.; Wei, Y.-C.; Zhou, H.-K.; Xiao, Y.; Zhi, X.-Y.; Shao, Z.-H.; Lyu, L.-D.; et al. A recently evolved diflavin-containing monomeric nitrate reductase is responsible for highly efficient bacterial nitrate assimilation. J. Biol. Chem. 2020, 295, 5051–5066. [Google Scholar] [CrossRef]
- Cuevas, R.; Durán, N.; Diez, M.C.; Tortella, G.R.; Rubilar, O. Extracellular Biosynthesis of Copper and Copper Oxide Nanoparticles byStereum hirsutum, a Native White-Rot Fungus from Chilean Forests. J. Nanomater. 2015, 2015, 789089. [Google Scholar] [CrossRef]
- Khodadadi, B.; Bordbar, M.; Yeganeh-Faal, A.; Nasrollahzadeh, M. Green synthesis of Ag nanoparticles/clinoptilolite using Vaccinium macrocarpon fruit extract and its excellent catalytic activity for reduction of organic dyes. J. Alloys Compd. 2017, 719, 82–88. [Google Scholar] [CrossRef]
- Nasrollahzadeh, M.; Sajjadi, M.; Tahsili, M.R.; Shokouhimehr, M.; Varma, R.S. Synthesis of 1-Substituted 1H-1,2,3,4-Tetrazoles Using Biosynthesized Ag/Sodium Borosilicate Nanocomposite. Acs Omega 2019, 4, 8985–9000. [Google Scholar] [CrossRef]
- Kanagasubbulakshmi, S.; Kadirvelu, K. Green synthesis of Iron oxide nanoparticles using Lagenaria siceraria and evaluation of its Antimicrobial activity. Def. Life Sci. J. 2017, 2, 422–427. [Google Scholar] [CrossRef]
- Rahman, S.S.U.; Qureshi, M.T.; Sultana, K.; Rehman, W.; Khan, M.Y.; Asif, M.H.; Farooq, M.; Sultana, N. Single step growth of iron oxide nanoparticles and their use as glucose biosensor. Results Phys. 2017, 7, 4451–4456. [Google Scholar] [CrossRef]
- Eid, A.M.; Fouda, A.; Niedbała, G.; Hassan, S.E.-D.; Salem, S.S.; Abdo, A.M.; Hetta, H.F.; Shaheen, T.I. Endophytic Streptomyces laurentii Mediated Green Synthesis of Ag-NPs with Antibacterial and Anticancer Properties for Developing Functional Textile Fabric Properties. Antibiotics 2020, 9, 641. [Google Scholar] [CrossRef]
- Baygar, T.; Ugur, A. Biosynthesis of Silver Nanoparticles by Streptomyces griseorubens isolated from Soil and Their Antioxidant Activity. Iet Nanobiotechnol. 2017, 11, 286–291. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.J.; Trevisan, J.; Bassan, P.; Bhargava, R.; Butler, H.J.; Dorling, K.M.; Fielden, P.R.; Fogarty, S.W.; Fullwood, N.J.; Heys, K.A.; et al. Using Fourier transform IR spectroscopy to analyze biological materials. Nat. Protoc. 2014, 9, 1771–1791. [Google Scholar] [CrossRef]
- Syame, S.M.; Mohamed, W.S.; Mahmoud, R.K.; Omara, S.T. Synthesis of Copper-Chitosan Nanocomposites and its Application in Treatment of Local Pathogenic Isolates Bacteria. Orient. J. Chem. 2017, 33, 2959–2969. [Google Scholar] [CrossRef]
- Hamedi, A.; Trotta, F.; Zarandi, M.B.; Zanetti, M.; Caldera, F.; Anceschi, A.; Nateghi, M.R. In Situ Synthesis of MIL-100(Fe) at the Surface of Fe3O4@AC as Highly Efficient Dye Adsorbing Nanocomposite. Int. J. Mol. Sci. 2019, 20, 5612. [Google Scholar] [CrossRef] [PubMed]
- Banik, M.; Patra, M.; Dutta, D.; Mukherjee, R.; Basu, T. A simple robust method of synthesis of copper–silver core–shell nano-particle: Evaluation of its structural and chemical properties with anticancer potency. Nanotechnology 2018, 29, 325102. [Google Scholar] [CrossRef]
- Tang, M.; Zhang, S.; Li, X.; Pang, X.; Qiu, H. Fabrication of magnetically recyclable Fe 3 O 4 @Cu nanocomposites with high catalytic performance for the reduction of organic dyes and 4-nitrophenol. Mater. Chem. Phys. 2014, 148, 639–647. [Google Scholar] [CrossRef]
- Rudakiya, D.M.; Pawar, K. Bactericidal potential of silver nanoparticles synthesized using cell-free extract of Comamonas acidovorans: In vitro and in silico approaches. 3 Biotech. 2017, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Hassan, S.E.-D.; Fouda, A.; Radwan, A.A.; Salem, S.S.; Barghoth, M.G.; Awad, M.A.; Abdo, A.M.; El-Gamal, M.S. Endophytic actinomycetes Streptomyces spp mediated biosynthesis of copper oxide nanoparticles as a promising tool for biotechnological applications. Jbic J. Biol. Inorg. Chem. 2019, 24, 377–393. [Google Scholar] [CrossRef]
- Ibrahim, S.; Ahmad, Z.; Manzoor, M.Z.; Mujahid, M.; Faheem, Z.; Adnan, A. Optimization for biogenic microbial synthesis of silver nanoparticles through response surface methodology, characterization, their antimicrobial, antioxidant, and catalytic potential. Sci. Rep. 2021, 11, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Fouda, A.; Hassan, S.E.D.; Abdo, A.M.; El-Gamal, M.S. Antimicrobial, Antioxidant and Larvicidal Activities of Spherical Silver Nanoparticles Synthesized by Endophytic Streptomyces spp. Biol. Trace Elem. Res. 2020, 195, 707–724. [Google Scholar] [CrossRef]
- Monowar, T.; Rahman, S.; Bhore, S.J.; Raju, G.; Sathasivam, K.V. Silver Nanoparticles Synthesized by Using the Endophytic Bacterium Pantoea ananatis are Promising Antimicrobial Agents against Multidrug Resistant Bacteria. Molecules 2018, 23, 3220. [Google Scholar] [CrossRef]
- Sharma, D.; Misba, L.; Khan, A.U. Antibiotics versus biofilm: An emerging battleground in microbial communities. Antimicrob. Resist. Infect. Control. 2019, 8, 1–10. [Google Scholar] [CrossRef]
- Mah, T.-F.C.; O’Toole, G.A. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001, 9, 34–39. [Google Scholar] [CrossRef]
- Majumdar, M.K.; Majumdar, S.K. Effects of Minerals on Neomycin Production by Streptomyces fradiae. Appl. Microbiol. 1965, 13, 190–193. [Google Scholar] [CrossRef]
- Singh, D.; Rathod, V.; Ninganagouda, S.; Hiremath, J.; Singh, A.K.; Mathew, J. Optimization and characterization of silver nanoparticle by endophytic fungi penicillium sp. isolated from curcuma longa (Turmeric) and application studies against MDR E. coli and S. aureus. Bioinorg. Chem. Appl. 2014, 5251, 408021. [Google Scholar]
- Remya, M.; Vijayakumar, R. Isolation and characterization of marine antagonistic actinomycetes from West Coast of India. Med. Biol. 2008, 15, 13–19. [Google Scholar]
- Suthindhiran, K.; Jayasri, M.A.; Dipali, D.; Prasar, A. Screening and characterization of protease producing actinomycetes from marine saltern. J. Basic Microbiol. 2013, 54, 1098–1109. [Google Scholar] [CrossRef]
- Avilala, J.; Golla, N. Antibacterial and Antiviral properties of silver nanoparticles synthesized by marine actinomycetes. Int. J. Pharm. Sci. Res. 2019, 10, 1223. [Google Scholar] [CrossRef]
- Mukherjee, K.; Gupta, R.; Kumar, G.; Kumari, S.; Biswas, S.; Padmanabhan, P. Synthesis of silver nanoparticles by Bacillus clausii and computational profiling of nitrate reductase enzyme involved in production. J. Genet. Eng. Biotechnol. 2018, 16, 527–536. [Google Scholar] [CrossRef] [PubMed]
- Philippot, L.; Piutti, S.; Martin-Laurent, F.; Hallet, S.; Germon, J.C. Molecular analysis of the nitrate-reducing community from unplanted and maize-planted soils. Appl. Environ. Microbiol. 2002, 68, 6121–6128. [Google Scholar] [CrossRef] [PubMed]
- Mintmier, B.; McGarry, J.M.; Sparacino-Watkins, C.E.; Sallmen, J.; Fischer-Schrader, K.; Magalon, A.; McCormick, J.R.; Stolz, J.F.; Schwarz, G.; Bain, D.J.; et al. Molecular cloning, expression and biochemical characterization of periplasmic nitrate reductase from Campylobacter jejuni. Fems Microbiol. Lett. 2018, 365, 151. [Google Scholar] [CrossRef] [PubMed]
- Olaya-Abril, A.; Hidalgo-Carrillo, J.; Luque-Almagro, V.M.; Fuentes-Almagro, C.; Urbano, F.J.; Moreno-Vivián, C.; Richardson, D.J.; Roldán, M.D. Exploring the Denitrification Proteome of Paracoccus denitrificans PD1222. Front. Microbiol. 2018, 9, 1137. [Google Scholar] [CrossRef] [PubMed]
- Hartsock, A.; Shapleigh, J.P. Physiological roles for two periplasmic nitrate reductases in Rhodobacter sphaeroides 2.4.3 (ATCC 17025). J. Bacteriol. 2011, 193, 6483–6489. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, F. Insights on nitrate respiration by Shewanella. Front. Mar. Sci. 2015, 1. [Google Scholar] [CrossRef]
- Farjadian, F.; Akbarizadeh, A.R.; Tayebi, L. Synthesis of novel reducing agent for formation of metronidazole-capped silver nanoparticle and evaluating antibacterial efficiency in gram-positive and gram-negative bacteria. Heliyon 2020, 6, e04747. [Google Scholar] [CrossRef]
- Gupta, A.; Singh, D.; Singh, S.K.; Singh, V.K.; Singh, A.V.; Kumar, A. Role of actinomycetes in bioactive and nanoparticle synthesis. In Role of Plant Growth Promoting Microorganisms in Sustainable Agriculture and Nanotechnology; Elsevier BV: Amsterdam, The Netherlands, 2019; pp. 163–182. [Google Scholar]
- Kumar, S.V.; Bafana, A.P.; Pawar, P.; Rahman, A.; Dahoumane, S.A.; Jeffryes, C.S. High conversion synthesis of <10 nm starch-stabilized silver nanoparticles using microwave technology. Sci. Rep. 2018, 8, 1–10. [Google Scholar]
- Alahmadi, N.S.; Betts, J.W.; Cheng, F.; Francesconi, M.G.; Kelly, S.M.; Kornherr, A.; Prior, T.J.; Wadhawan, J.D. Synthesis and antibacterial effects of cobalt-cellulose magnetic nanocomposites. Rsc Adv. 2017, 7, 20020–20026. [Google Scholar] [CrossRef]
- Adiguzel, A.O.; Adigüzel, S.K.; Mazmancı, B.; Tuncer, M.; Mazmanci, M.A.; Mazmanci, B. Silver nanoparticle biosynthesis from newly isolated streptomyces genus from soil. Mater. Res. Express 2018, 5, 045402. [Google Scholar] [CrossRef]
- Prabhu, Y.; Rao, K.V.; Sai, V.S.; Pavani, T. A facile biosynthesis of copper nanoparticles: A micro-structural and antibacterial activity investigation. J. Saudi Chem. Soc. 2017, 21, 180–185. [Google Scholar] [CrossRef]
- Chaudhary, R.G.; Tanna, J.A.; Gandhare, N.V.; Rai, A.R.; Juneja, H.D. Synthesis of nickel nanoparticles: Microscopic investigation, an efficient catalyst and effective antibacterial activity. Adv. Mater. Lett. 2015, 6, 990–998. [Google Scholar] [CrossRef]
- Franci, G.; Falanga, A.; Galdiero, S.; Palomba, L.; Rai, M.; Morelli, G.; Galdiero, M. Silver Nanoparticles as Potential Antibacterial Agents. Molecules 2015, 20, 8856–8874. [Google Scholar] [CrossRef]
- Siddique, M.H.; Aslam, B.; Imran, M.; Ashraf, A.; Nadeem, H.; Hayat, S.; Khurshid, M.; Afzal, M.; Malik, I.R.; Shahzad, M.; et al. Research article effect of silver nanoparticles on biofilm formation and EPS production of multidrug-resistant Klebsiella pneumonia. Hindawi BioMed Res. Int. 2020, 2020, 6398165. [Google Scholar]
- Hamed, A.A.; Kabary, H.; Khedr, M.; Emam, A.N. Antibiofilm, antimicrobial and cytotoxic activity of extracellular green-synthesized silver nanoparticles by two marine-derived actinomycete. Rsc Adv. 2020, 10, 10361–10367. [Google Scholar] [CrossRef]
- Rajan, B.M.; Kannabiran, K. Extraction and Identification of Antibacterial Secondary Metabolites from Marine Streptomyces sp. VITBRK2. Int. J. Mol. Cell. Med. 2014, 3, 130–137. [Google Scholar] [PubMed]
- Rashad, F.M.; Fathy, H.M.; El-Zayat, A.S.; Elghonaimy, A.M. Isolation and characterization of multifunctional Streptomyces species with antimicrobial, nematicidal and phytohormone activities from marine environments in Egypt. Microbiol. Res. 2015, 175, 34–47. [Google Scholar] [CrossRef]
- Seong, C.N.; Choi, J.H.; Baik, K.S. An Improved Selective Isolation of Rare Actinomycetes from Forest Soil. J. Microbiol. 2001, 39, 17–23. [Google Scholar]
- Duraipandiyan, V.; Sasi, A.; Islam, V.; Arasu, M.V.; Ignacimuthu, S. Antimicrobial properties of actinomycetes from the soil of Himalaya. J. Mycol. Méd. 2010, 20, 15–20. [Google Scholar] [CrossRef]
- Karthik, L.; Kumar, G.; Kirthi, A.V.; Rahuman, A.A.; Rao, K.V.B. Streptomyces sp. LK3 mediated synthesis of silver nanoparticles and its biomedical application. Bioprocess. Biosyst. Eng. 2014, 37, 261–267. [Google Scholar] [CrossRef]
- Składanowski, M.; Wypij, M.; Laskowski, D.; Golińska, P.; Dahm, H.; Rai, M. Silver and gold nanoparticles synthesized from Streptomyces sp. isolated from acid forest soil with special reference to its antibacterial activity against pathogens. J. Clust. Sci. 2017, 28, 59–79. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).