Multiple Mechanisms of the Synthesized Antimicrobial Peptide TS against Gram-Negative Bacteria for High Efficacy Antibacterial Action In Vivo
Abstract
1. Introduction
2. Results and Discussion
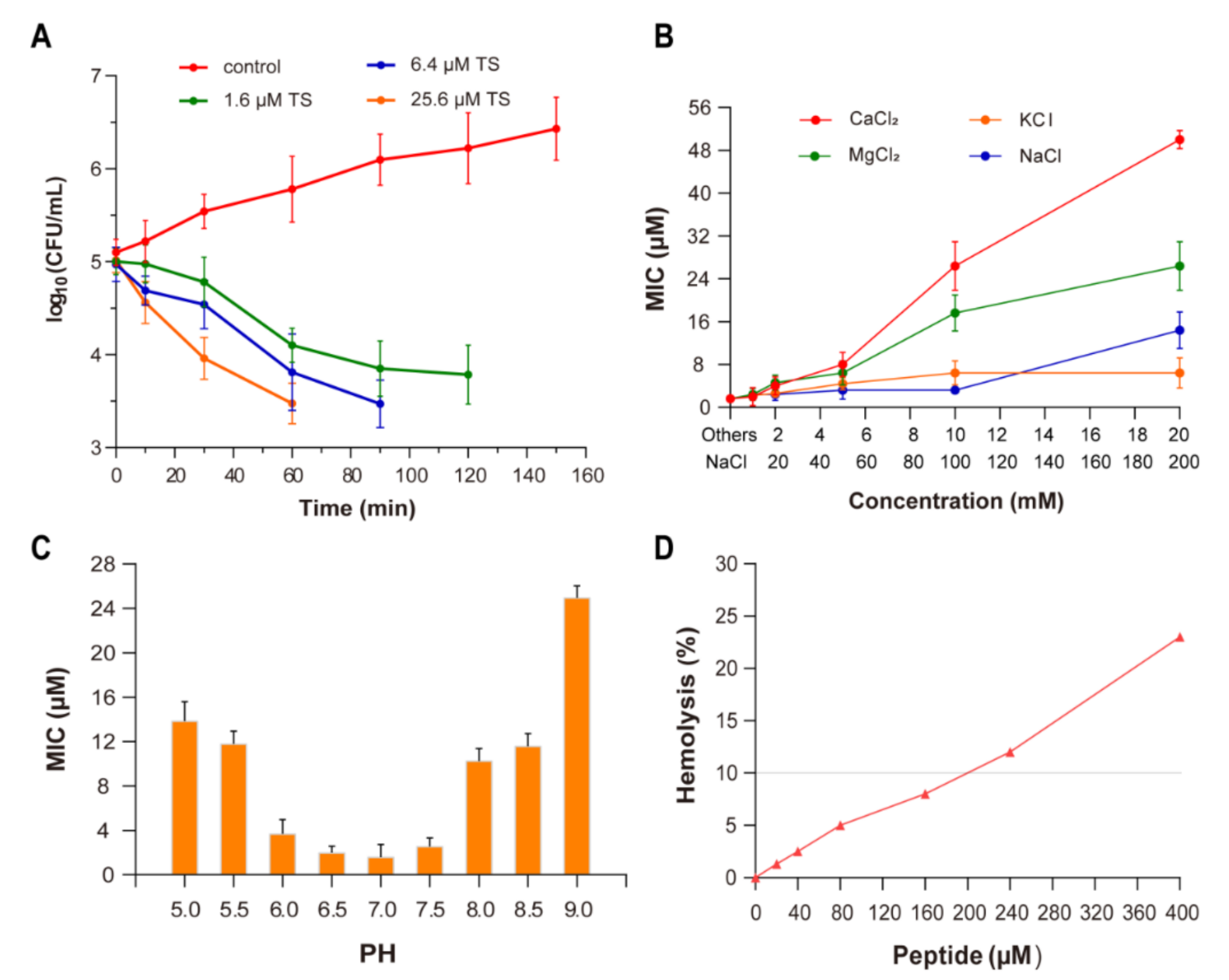
2.1. Antibacterial and Bactericidal Activity
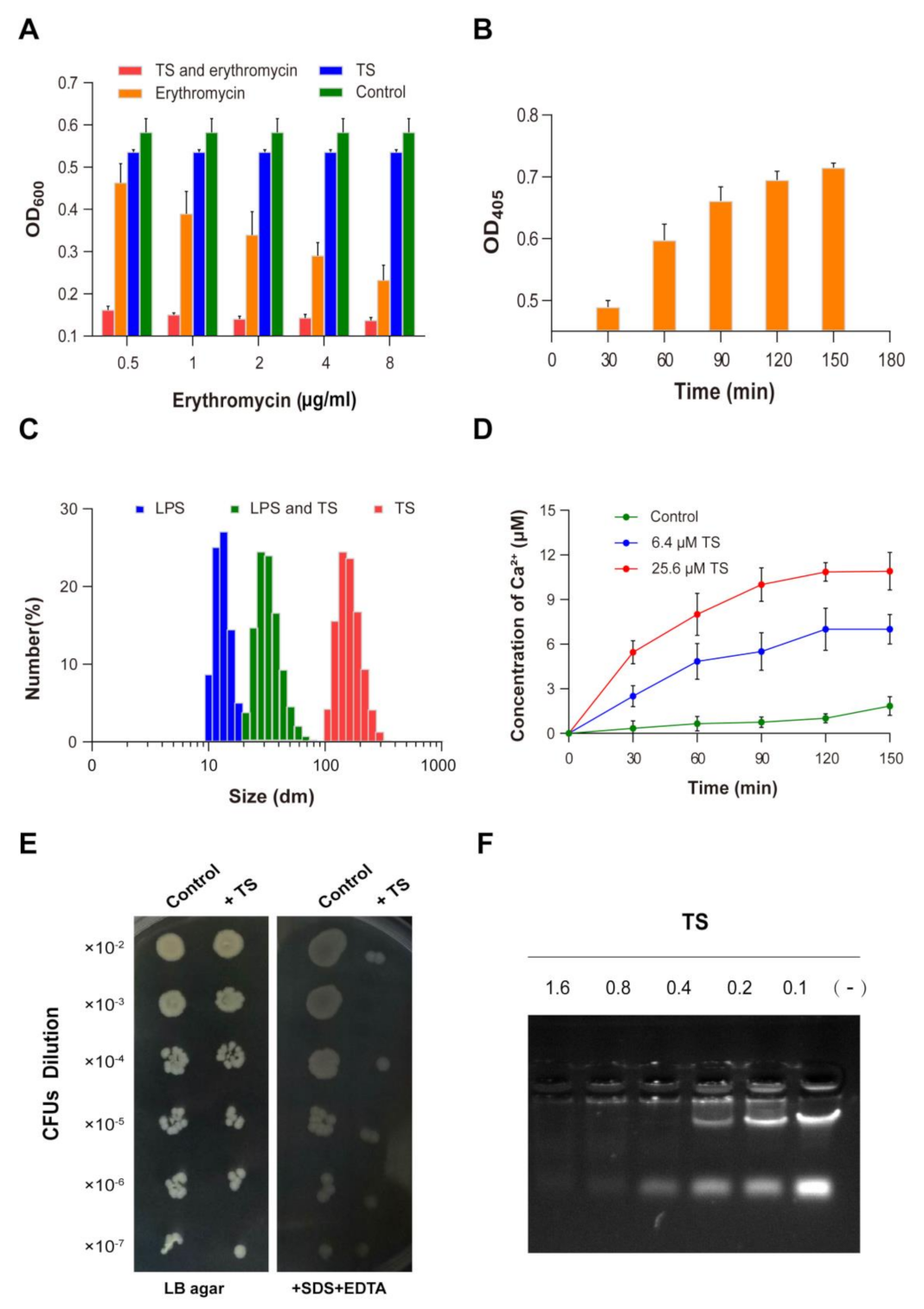
2.2. Effect of Ions
2.3. Effect of pH
2.4. Hemolytic Activity
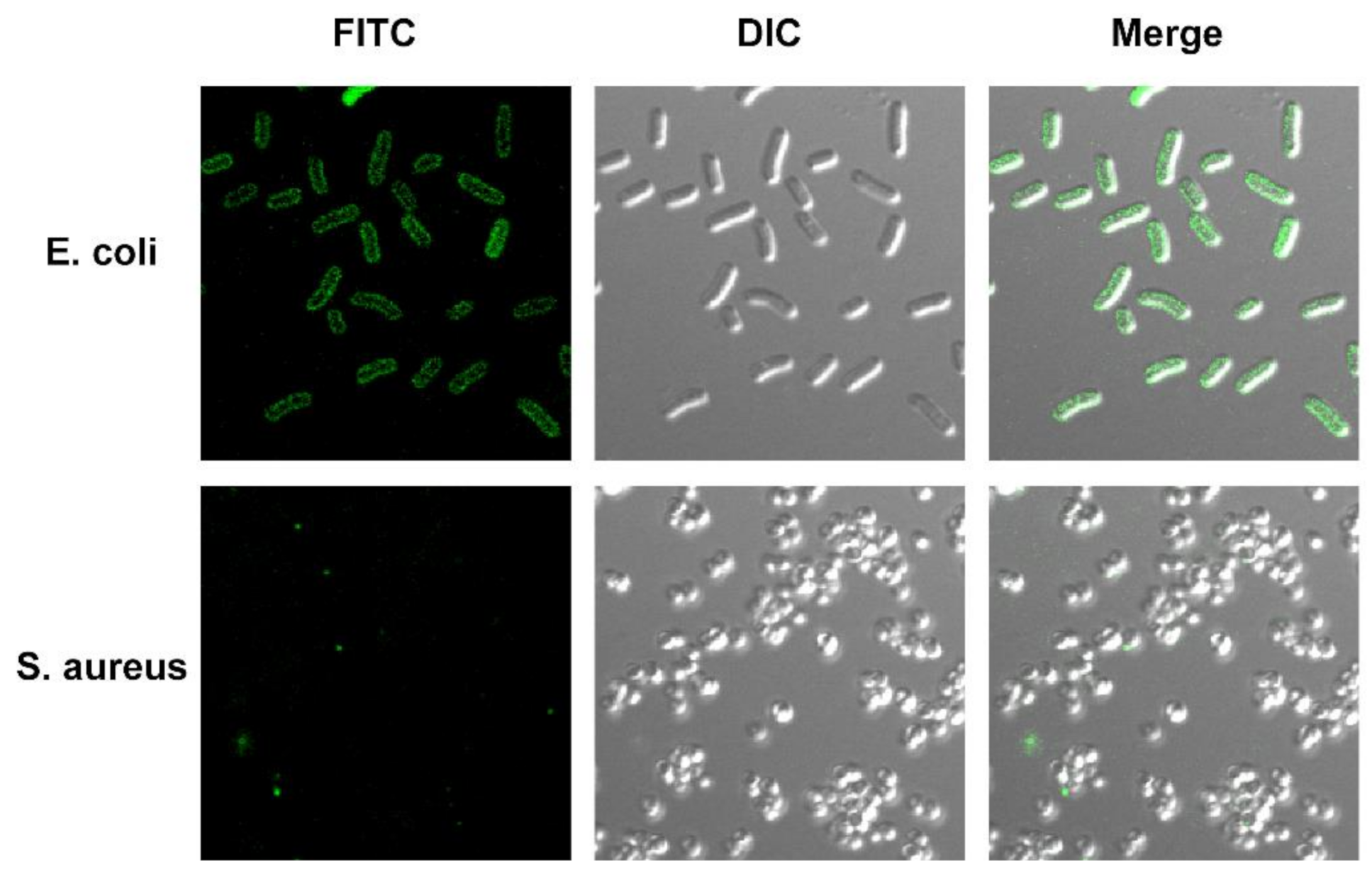
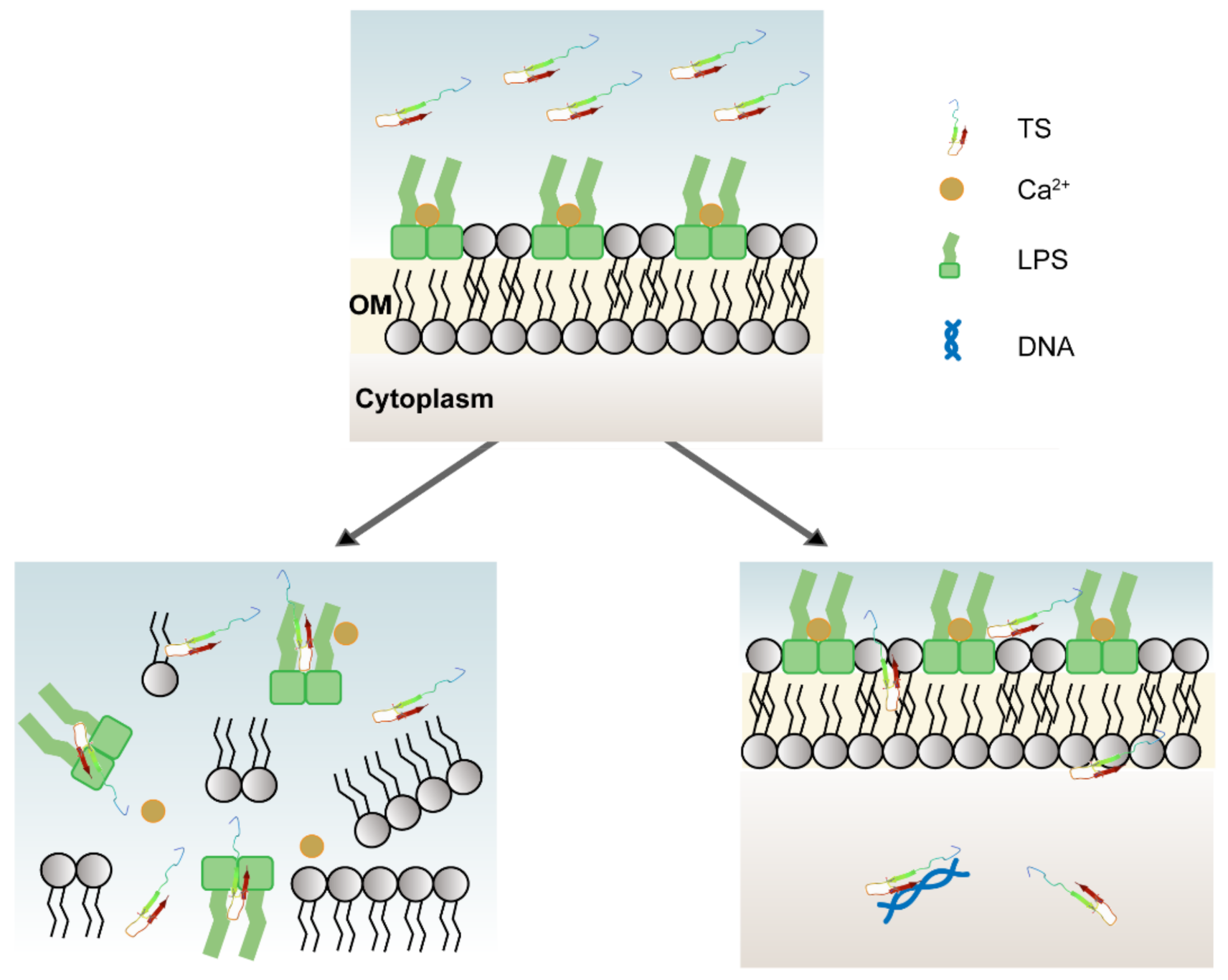
2.5. Localization of Antimicrobial Peptide TS on Bacterial Cells
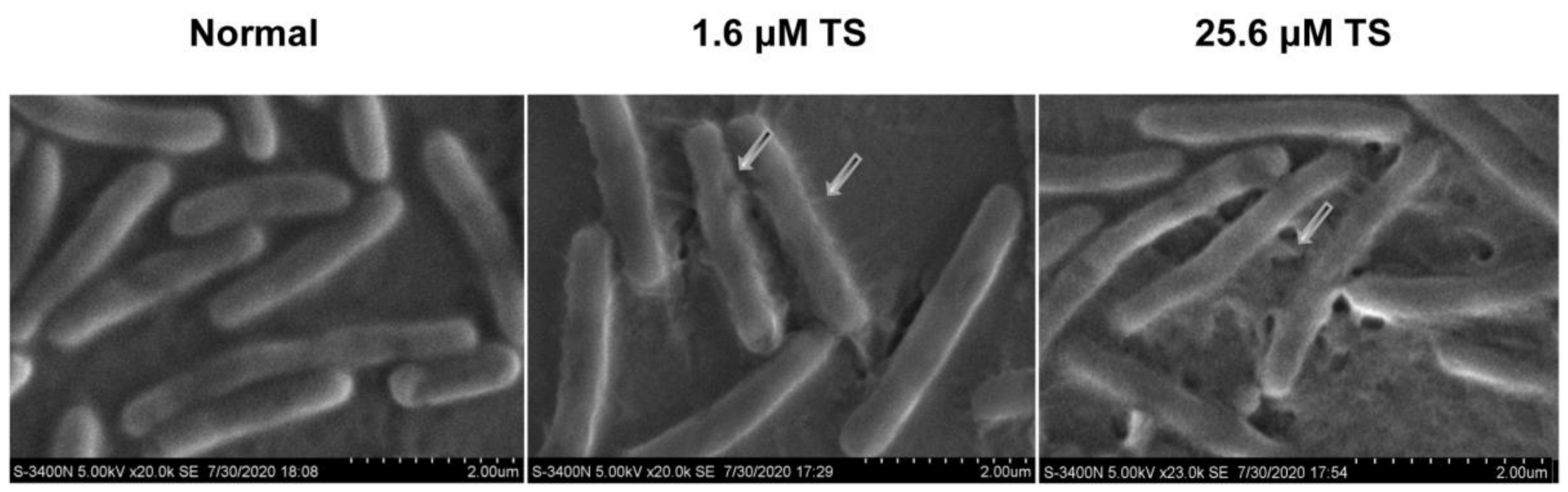
2.6. Morphological Changes of Antimicrobial Peptide TS on Bacterial Cell Membrane
2.7. Permeabilization of the Outer Membrane (OM)
2.8. Permeabilization of the Inner Membrane (IM)
2.9. Interaction of Peptides with Lipopolysaccharides
2.10. Susceptibility to Detergents
2.11. Interaction of Antimicrobial Peptide TS with Cellular DNA
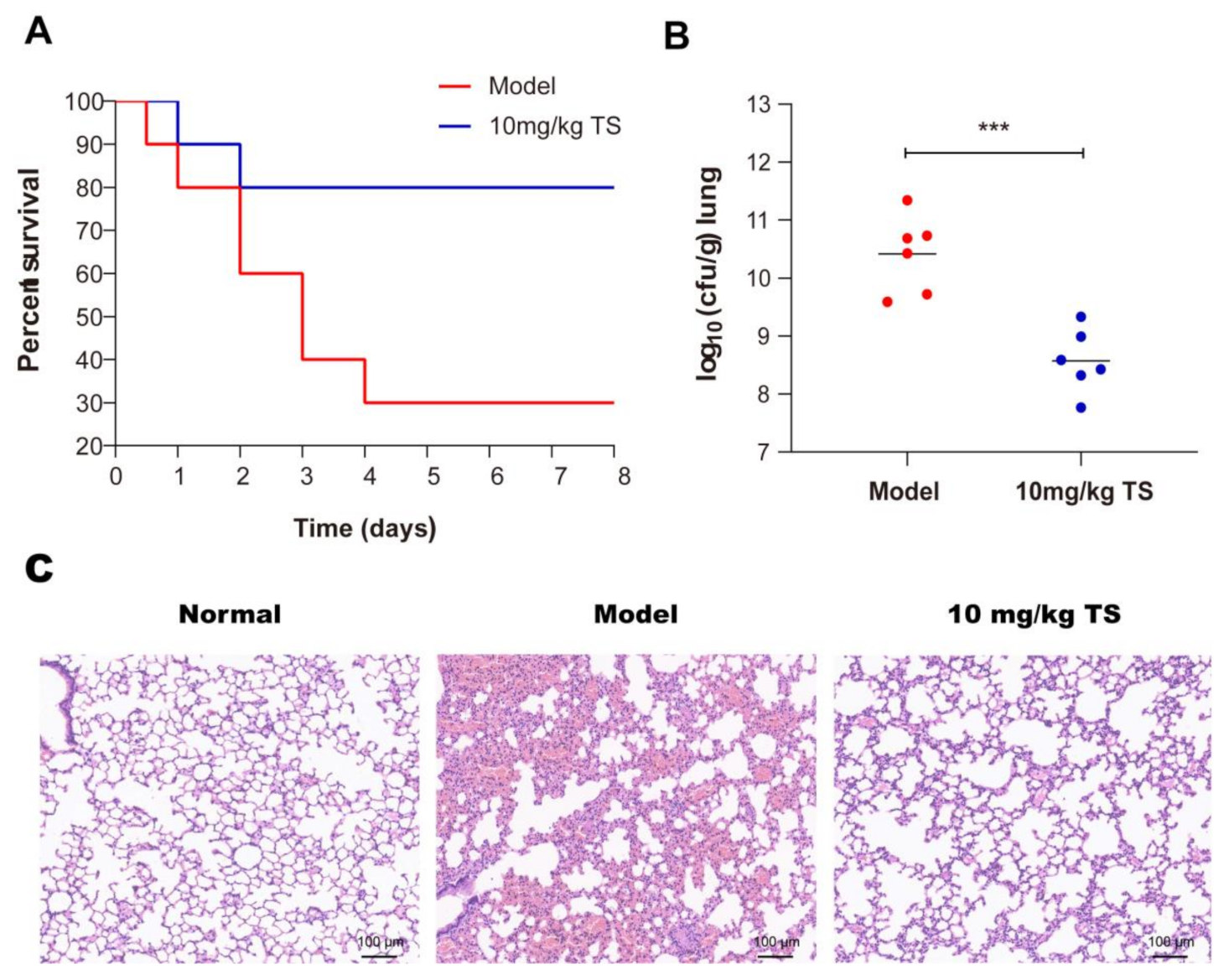
2.12. Antimicrobial Peptide TS Protects Mice Infected with E. coli ATCC 25922
3. Materials and Methods
3.1. Synthesis and Purification of Antimicrobial Peptide TS
3.2. Bacterial Strains
3.3. Minimum Inhibitory Concentration (MIC)
3.4. Time Killing Curve Assay
3.5. Effect of Ions and pH
3.6. Hemolysis Assay
3.7. Confocal Laser Scanning Microscopy
3.8. Scanning Electron Microscopy (SEM)
3.9. OM Permeabilization Assay
3.10. IM Permeabilization Assay
3.11. DLS
3.12. Calcium Ion Release Assay
3.13. Susceptibility to Detergents
3.14. DNA Binding Assay
3.15. Animal
3.16. Mouse Pneumonia Model
3.17. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Eichenberger, E.M.; Thaden, J.T. Epidemiology and mechanisms of resistance of extensively drug resistant gram-negative bacteria. Antibiotics 2019, 8, 37. [Google Scholar] [CrossRef] [PubMed]
- Doi, Y.; Bonomo, R.A.; Hooper, D.C.; Kaye, K.S.; Johnson, J.R.; Clancy, C.J.; Thaden, J.T.; Stryjewski, M.E.; van Duin, D.; Gram-Negative Committee of the Antibacterial Resistance Leadership Group. Gram-negative bacterial infections: Research priorities, accomplishments, and future directions of the Antibacterial Resistance Leadership Group. Clin. Infect. Dis. 2017, 64, S30–S35. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States; CDC: Atlanta, GA, USA, 2019. [Google Scholar]
- Nadimpalli, M.; Delarocque-Astagneau, E.; Love, D.C.; Price, L.B.; Huynh, B.-T.; Collard, J.-M.; Lay, K.S.; Borand, L.; Ndir, A.; Walsh, T.R.; et al. Combating global antibiotic resistance: Emerging one health concerns in lower-and middle-income countries. Clin. Infect. Dis. 2018, 66, 963–969. [Google Scholar] [CrossRef] [PubMed]
- Butler, M.S.; Blaskovich, M.A.; Cooper, M.A. Antibiotics in the clinical pipeline. J. Antibiot. 2013, 66, 571–591. [Google Scholar] [CrossRef] [PubMed]
- Bahar, A.A.; Ren, D. Antimicrobial peptides. Pharmaceuticals 2013, 6, 1543–1575. [Google Scholar] [CrossRef]
- Pasupuleti, M.; Schmidtchen, A.; Malmsten, M. Antimicrobial peptides: Key components of the innate immune system. Crit. Rev. Biotechnol. 2012, 32, 143–171. [Google Scholar] [CrossRef]
- Radek, K.; Gallo, R.L. Antimicrobial peptides: Natural effectors of the innate immune system. Semin. Immunopathol. 2007, 29, 27–43. [Google Scholar] [CrossRef]
- Sun, Y.; Dong, W.; Sun, L.; Ma, L.; Shang, D. Insights into the membrane interaction mechanism and antibacterial properties of chensinin-1b. Biomaterials 2015, 37, 299–311. [Google Scholar] [CrossRef]
- Li, Y.; Xiang, Q.; Zhang, Q.; Huang, Y.; Su, Z.-J. Overview on the recent study of antimicrobial peptides: Origins, functions, relative mechanisms and application. Peptides 2012, 37, 207–215. [Google Scholar] [CrossRef]
- Berglund, N.A.; Piggot, T.J.; Jefferies, D.; Sessions, R.B.; Bond, P.J.; Khalid, S. Interaction of the antimicrobial peptide polymyxin B1 with both membranes of E. coli: A molecular dynamics study. PLoS Comput. Biol. 2015, 11, e1004180. [Google Scholar] [CrossRef]
- Haney, E.F.; Petersen, A.P.; Lau, C.K.; Jing, W.; Storey, D.G.; Vogel, H.J. Mechanism of action of puroindoline derived tryp-tophan-rich antimicrobial peptides. Biochim. Biophys. Acta 2013, 1828, 1802–1813. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-C.; Kim, J.-Y.; Jeong, C.; Yoo, S.; Hahm, K.-S.; Park, Y. A plausible mode of action of pseudin-2, an antimicrobial peptide from Pseudis paradoxa. Biochim. Biophys. Acta. 2011, 1808, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Alfred, R.L.; Palombo, E.A.; Panozzo, J.F.; Bhave, M. The antimicrobial domains of wheat puroindolines are cell-penetrating peptides with possible intracellular mechanisms of action. PLoS ONE 2013, 8, e75488. [Google Scholar] [CrossRef] [PubMed]
- Nam, B.H.; Moon, J.Y.; Park, E.H.; Kim, Y.O.; Kim, D.G.; Kong, H.J.; Kim, W.J.; Jee, Y.J.; An, C.M.; Park, N.G.; et al. Antimicrobial activity of peptides derived from olive flounder lipopolysaccharide binding protein/bactericidal permeability-increasing protein (LBP/BPI). Mar. Drugs 2014, 12, 5240–5257. [Google Scholar] [CrossRef]
- Jiang, H.; Du, H.; Wang, L.M.; Wang, P.Z.; Bai, X.F. Hemorrhagic fever with renal syndrome: Pathogenesis and clinical picture. Front. Cell Infect. Microbiol. 2016, 6, 1. [Google Scholar] [CrossRef]
- Fehlbaum, P.; Bulet, P.; Chernysh, S.; Briand, J.P.; Roussel, J.P.; Letellier, L.; Hetru, C.; Hoffmann, J.A. Structure-activity analysis of thanatin, a 21-residue inducible insect defense peptide with sequence homology to frog skin antimicrobial peptides. Proc. Natl. Acad. Sci. USA 1996, 93, 1221–1225. [Google Scholar] [CrossRef]
- Wu, G.; Wu, P.; Xue, X.; Yan, X.; Liu, S.; Zhang, C.; Shen, Z.; Xi, T. Application of S-thanatin, an antimicrobial peptide derived from thanatin, in mouse model of Klebsiella pneumoniae infection. Peptides 2013, 45, 73–77. [Google Scholar] [CrossRef]
- Wu, G.; Ding, J.; Li, H.; Li, L.; Zhao, R.; Shen, Z.; Fan, X.; Xi, T. Effects of cations and pH on antimicrobial activity of thanatin and s-thanatin against Escherichia coli ATCC25922 and B. subtilis ATCC. Curr. Microbiol. 2008, 57, 552–557. [Google Scholar] [CrossRef]
- Wu, G.; Fan, X.; Li, L.; Wang, H.; Ding, J.; Hongbin, W.; Zhao, R.; Gou, L.; Shen, Z.; Xi, T. Interaction of antimicrobial peptide s-thanatin with lipopolysaccharide in vitro and in an experimental mouse model of septic shock caused by a multidrug-resistant clinical isolate of Escherichia coli. Int. J. Antimicrob. Agents 2010, 35, 250–254. [Google Scholar] [CrossRef]
- Hoover, D.M.; Wu, Z.; Tucker, K.; Lu, W.; Lubkowski, J. Antimicrobial characterization of human beta-defensin 3 derivatives. Antimicrob. Agents Chemother. 2003, 47, 2804–2809. [Google Scholar] [CrossRef]
- Cox, D.L.; Sun, Y.; Liu, H.; I Lehrer, R.; Shafer, W.M. Susceptibility of Treponema pallidum to host-derived antimicrobial peptides. Peptides 2003, 24, 1741–1746. [Google Scholar] [CrossRef] [PubMed]
- Holdbrook, D.A.; Singh, S.; Choong, Y.K.; Petrlova, J.; Malmsten, M.; Bond, P.J.; Verma, N.K.; Schmidtchen, A.; Saravanan, R. Influence of pH on the activity of thrombin-derived antimicrobial peptides. Biochim. Biophys. Acta Biomembr. 2018, 1860, 2374–2384. [Google Scholar] [CrossRef] [PubMed]
- Hitchner, M.A.; Santiago-Ortiz, L.E.; Necelis, M.R.; Shirley, D.J.; Palmer, T.J.; Tarnawsky, K.E.; Vaden, T.D.; Caputo, G.A. Activity and characterization of a pH-sensitive antimicrobial peptide. Biochim. Biophys. Acta Biomembr. 2019, 1861, 182984. [Google Scholar] [CrossRef] [PubMed]
- Marcellini, L.; Giammatteo, M.; Aimola, P.; Mangoni, M.L. Fluorescence and electron microscopy methods for exploring an-timicrobial peptides mode(s) of action. Methods Mol. Biol. 2010, 618, 249–266. [Google Scholar]
- Liao, H.; Zhang, F.; Liao, X.; Hu, X.; Chen, Y.; Deng, L. Analysis of Escherichia coli cell damage induced by HPCD using mi-croscopies and fluorescent staining. Int. J. Food Microbiol. 2010, 144, 169–176. [Google Scholar] [CrossRef]
- Vaara, M.; Porro, M. Group of peptides that act synergistically with hydrophobic antibiotics against gram-negative enteric bacteria. Antimicrob. Agents Chemother. 1996, 40, 1801–1805. [Google Scholar] [CrossRef]
- Tang, W.; Zhang, H.; Wang, L.; Qian, H. New cationic antimicrobial peptide screened from boiled-dried anchovies by immo-bilized bacterial membrane liposome chromatography. J. Agric. Food Chem. 2014, 62, 1564–1571. [Google Scholar] [CrossRef]
- Hawrani, A.; Howe, R.A.; Walsh, T.R.; Dempsey, C.E. Thermodynamics of RTA3 peptide binding to membranes and conse-quences for antimicrobial activity. Biochim. Biophys. Acta 2010, 1798, 1254–1262. [Google Scholar] [CrossRef]
- Sutterlin, H.A.; Zhang, S.; Silhavy, T.J. Accumulation of phosphatidic acid increases vancomycin resistance in Escherichia coli. J. Bacteriol. 2014, 196, 3214–3220. [Google Scholar] [CrossRef]
- Snyder, D.S.; McIntosh, T.J. The lipopolysaccharide barrier: Correlation of antibiotic susceptibility with antibiotic permeability and fluorescent probe binding kinetics. Biochemistry 2000, 39, 11777–11787. [Google Scholar] [CrossRef]
- Anunthawan, T.; De La Fuente-Nunez, C.; Hancock, R.E.; Klaynongsruang, S. Cationic amphipathic peptides KT2 and RT2 are taken up into bacterial cells and kill planktonic and biofilm bacteria. Biochim. Biophys. Acta 2015, 1848, 1352–1358. [Google Scholar] [CrossRef] [PubMed]
- Rautenbach, M.; Vlok, N.M.; Eyeghe-Bickong, H.A.; van der Merwe, M.J.; Stander, M.A. An electrospray ionization mass spectrometry study on the "in vacuo" hetero-oligomers formed by the antimicrobial peptides, surfactin and gramicidin S. J. Am. Soc. Mass Spectrom 2017, 28, 1623–1637. [Google Scholar] [CrossRef] [PubMed]
- Smart, M.; Rajagopal, A.; Liu, W.-K.; Ha, B.-Y. Opposing effects of cationic antimicrobial peptides and divalent cations on bacterial lipopolysaccharides. Phys. Rev. E 2017, 96, 042405. [Google Scholar] [CrossRef] [PubMed]
- Clifton, L.A.; Skoda, M.W.A.; Le Brun, A.P.; Ciesielski, F.; Kuzmenko, I.; Holt, S.A.; Lakey, J.H. Effect of divalent cation removal on the structure of gram-negative bacterial outer membrane models. Langmuir 2015, 31, 404–412. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Wang, Y.; Wang, M.; Tian, Y.; Kang, W.; Liu, H.; Wang, H.; Dou, J.; Zhou, C. Effective antimicrobial activity of Cbf-14, derived from a cathelin-like domain, against penicillin-resistant bacteria. Biomaterials 2016, 87, 32–45. [Google Scholar] [CrossRef] [PubMed]
- Ricard, J.-D.; Conti, G.; Boucherie, M.; Hörmann, C.; Poelaert, J.; Quintel, M.; Rubertsson, S.; Torres, A. A European survey of nosocomial infection control and hospital-acquired pneumonia prevention practices. J. Infect. 2012, 65, 285–291. [Google Scholar] [CrossRef]
- Russo, T.A.; Wang, Z.; Davidson, B.A.; Genagon, S.A.; Beanan, J.M.; Olson, R.; Holm, B.A.; Knight, P.R., 3rd; Chess, P.R.; Notter, R.H. Surfactant dysfunction and lung injury due to the E. coli virulence factor hemolysin in a rat pneumonia model. Am. J. Physiol. Lung Cell Mol. Physiol. 2007, 292, L632–L643. [Google Scholar] [CrossRef][Green Version]
- Fihman, V.; Messika, J.; Hajage, D.; Tournier, V.; Gaudry, S.; Magdoud, F.; Barnaud, G.; Billard-Pomares, T.; Branger, C.; Dreyfuss, D.; et al. Five-year trends for ventilator-associated pneumonia: Correlation between microbiological findings and antimicrobial drug consumption. Int. J. Antimicrob. Agents 2015, 46, 518–525. [Google Scholar] [CrossRef]
- Kollef, M.H.; Ricard, J.-D.; Roux, D.; François, B.; Ischaki, E.; Rozgonyi, Z.; Boulain, T.; Ivanyi, Z.; János, G.; Garot, D.; et al. A randomized trial of the amikacin fosfomycin inhalation system for the adjunctive therapy of gram-negative ventilator-associated Pneumonia. Chest 2017, 151, 1239–1246. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. M07: Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically; CLSI: Wayne, PA, USA, 2012. [Google Scholar]
- Skerlavaj, B.; Romeo, D.; Gennaro, R. Rapid membrane permeabilization and inhibition of vital functions of gram-negative bacteria by bactenecins. Infect. Immun. 1990, 58, 3724–3730. [Google Scholar] [CrossRef]
| Bacteria | MIC | ||||
|---|---|---|---|---|---|
| TS (μg/mL)/(μM) | Ampicillin (μg/mL) | Erythromycin (μg/mL) | TrimeThoprim (μg/mL) | AzithromyCin (μg/mL) | |
| E. coli (ATCC 25922) | 4/1.6 | 8 | >64 | <0.5 | 32 |
| K. pneumonia (ATCC 13883) | 4/1.6 | 8 | >64 | 15.6 | 32 |
| P. aeruginosa (ATCC 27853) | 32/12.8 | 8 | >64 | 8 | >64 |
| E. faecium (ATCC 29212) | 64.0/25.6 | 2 | 4 | 2 | 8 |
| S. aureus (ATCC 29213) | >64 | 2 | 1 | 2 | 1 |
| Sequence | Molecular Weight | Extinction Coefficient | GRAVY | Isoelectric Point | |
|---|---|---|---|---|---|
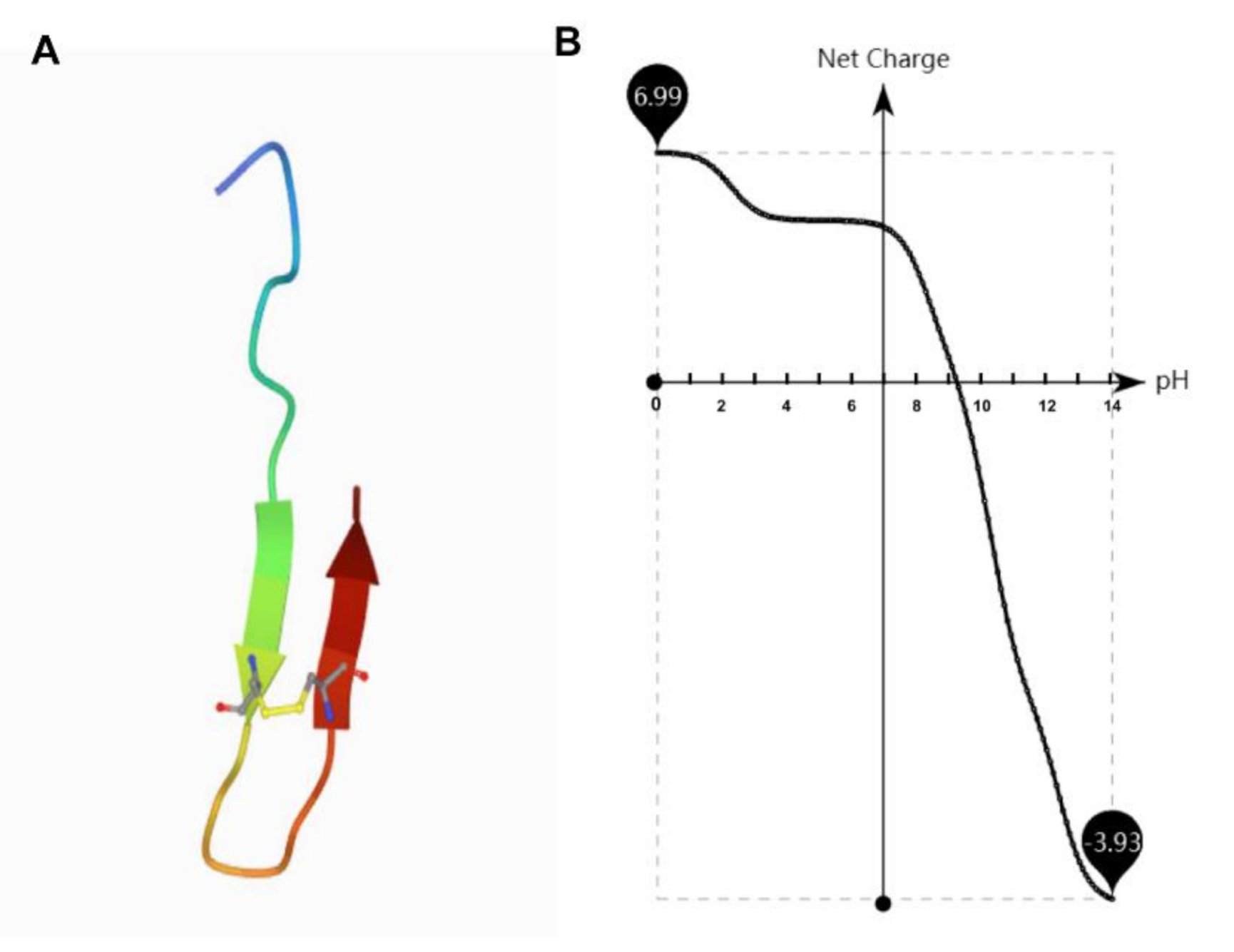
| TS | GSKKPVPIIYCNRRSGKCQRM | 2419.88 | 1400 M−1cm−1 | −0.9 | 10.87 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, R.; Fan, X.; Jiang, X.; Zou, M.; Xiao, H.; Wu, G. Multiple Mechanisms of the Synthesized Antimicrobial Peptide TS against Gram-Negative Bacteria for High Efficacy Antibacterial Action In Vivo. Molecules 2021, 26, 60. https://doi.org/10.3390/molecules26010060
Zhang R, Fan X, Jiang X, Zou M, Xiao H, Wu G. Multiple Mechanisms of the Synthesized Antimicrobial Peptide TS against Gram-Negative Bacteria for High Efficacy Antibacterial Action In Vivo. Molecules. 2021; 26(1):60. https://doi.org/10.3390/molecules26010060
Chicago/Turabian StyleZhang, Rui, Xiaobo Fan, Xinglu Jiang, Mingyuan Zou, Han Xiao, and Guoqiu Wu. 2021. "Multiple Mechanisms of the Synthesized Antimicrobial Peptide TS against Gram-Negative Bacteria for High Efficacy Antibacterial Action In Vivo" Molecules 26, no. 1: 60. https://doi.org/10.3390/molecules26010060
APA StyleZhang, R., Fan, X., Jiang, X., Zou, M., Xiao, H., & Wu, G. (2021). Multiple Mechanisms of the Synthesized Antimicrobial Peptide TS against Gram-Negative Bacteria for High Efficacy Antibacterial Action In Vivo. Molecules, 26(1), 60. https://doi.org/10.3390/molecules26010060




