Studying the Binding Modes of Novel 2-Aminopyridine Derivatives as Effective and Selective c-Met Kinase Type 1 Inhibitors Using Molecular Modeling Approaches
Abstract
1. Introduction
2. Results and Discussion
2.1. The Studied c-Met Inhibitors
2.2. Atom-Based CoMFA and CoMSIA Models
2.2.1. The 3D-QSAR Models
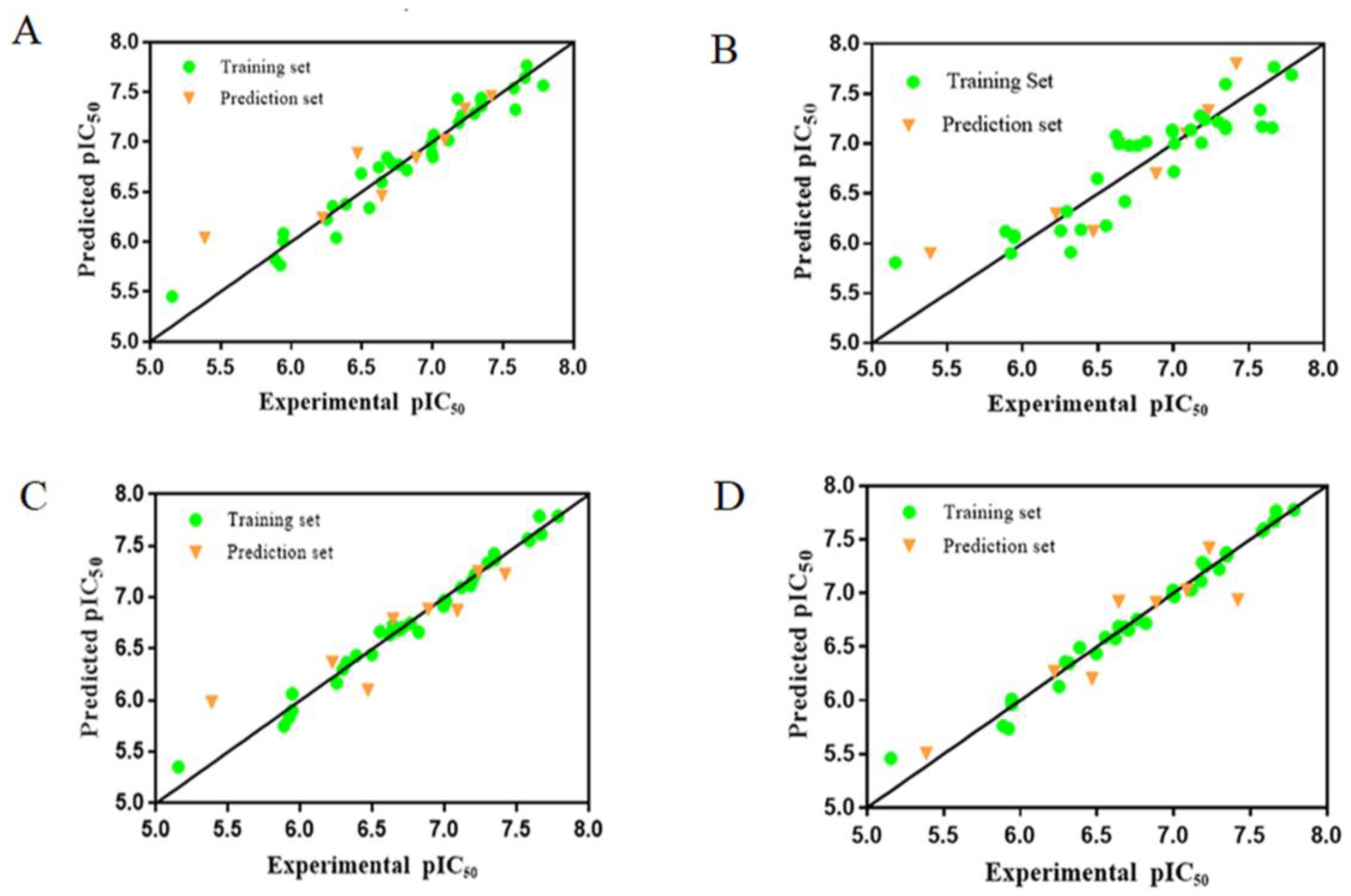
2.2.2. CoMFA Results
2.2.3. CoMSIA Results
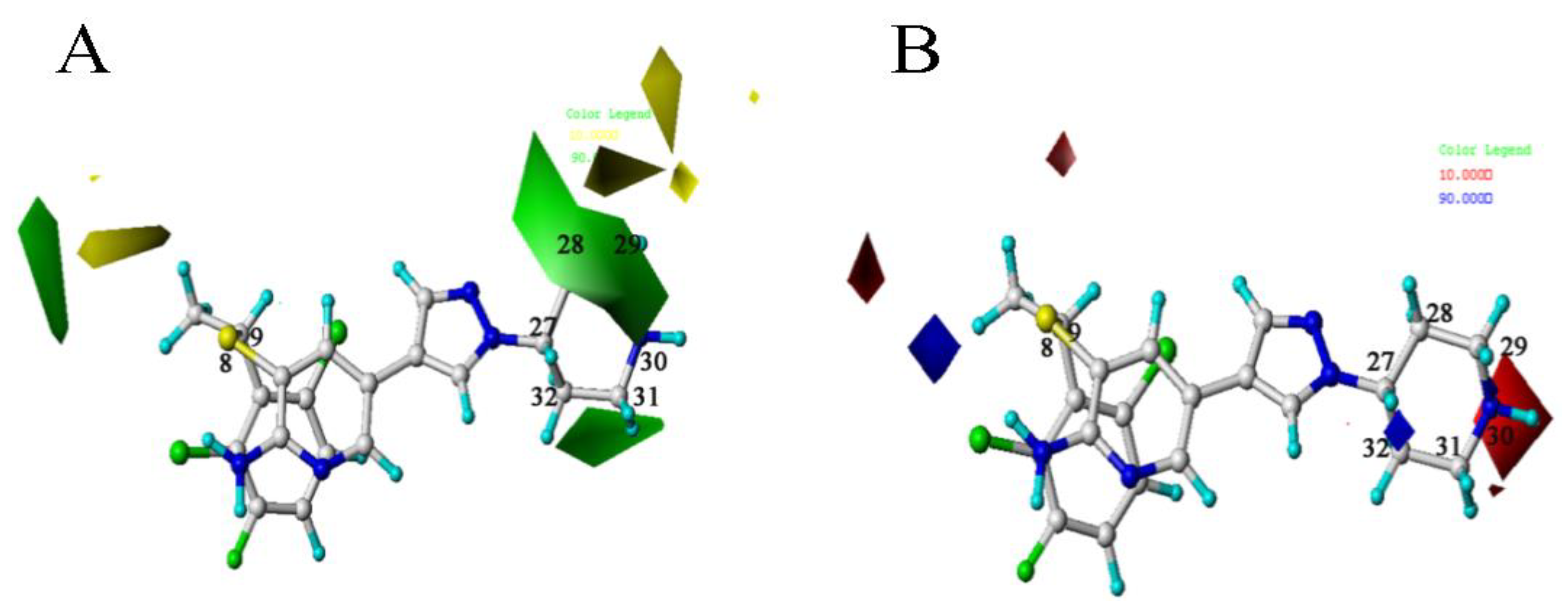
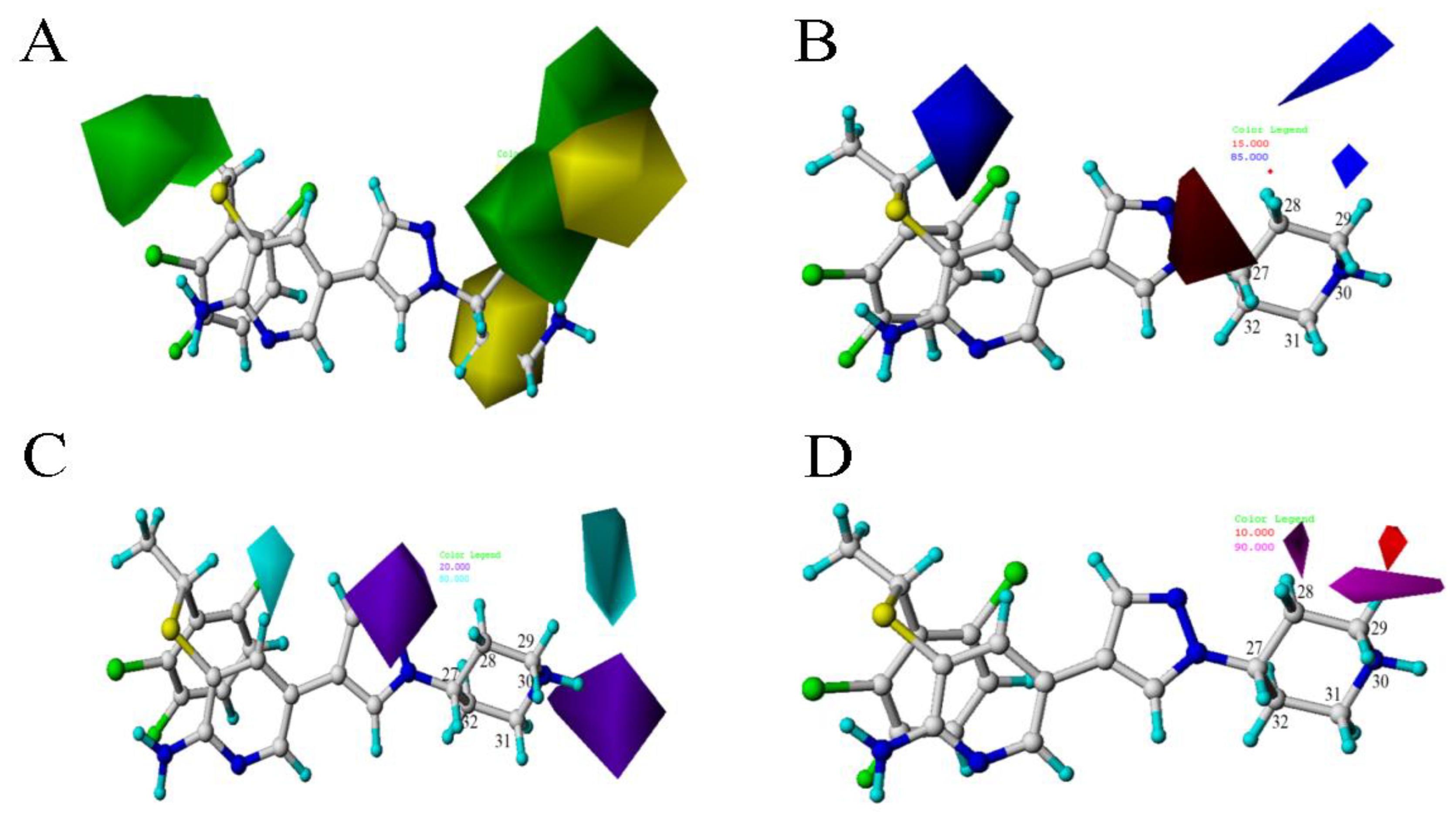
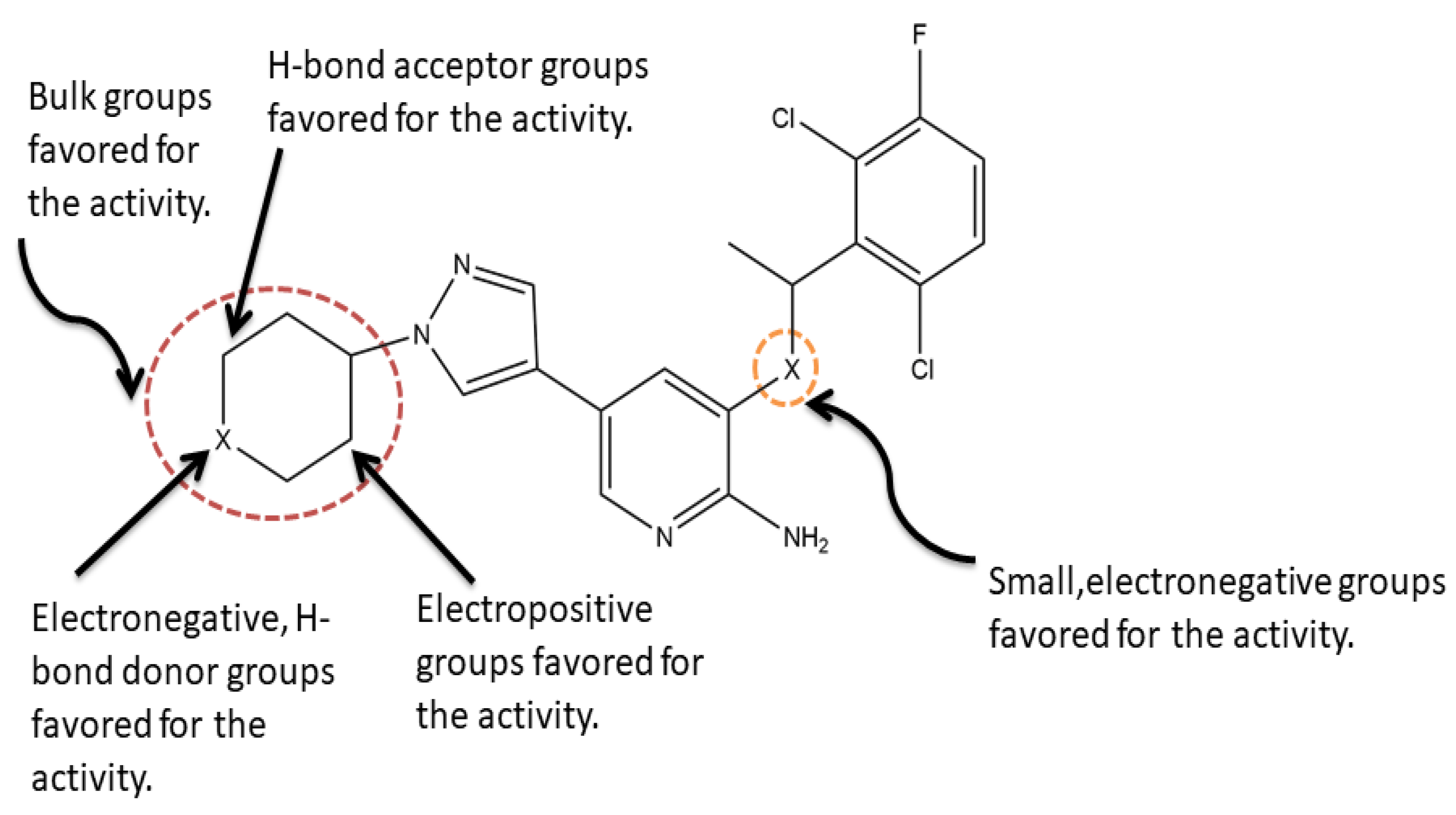
2.2.4. Contour Map Analysis
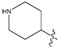 is larger than –CH3, so the activity of compound 4 is higher than compound 6. There is a yellow area near the C8 position. Taking compounds 4 and 1 as well as compounds 5 and 2 as examples, –S at the C8 position is smaller than
is larger than –CH3, so the activity of compound 4 is higher than compound 6. There is a yellow area near the C8 position. Taking compounds 4 and 1 as well as compounds 5 and 2 as examples, –S at the C8 position is smaller than  , so the corresponding compounds have higher activity values.
, so the corresponding compounds have higher activity values. is higher than compound 8 with
is higher than compound 8 with 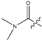 .
. with
with  group at this position can increase the activity.
group at this position can increase the activity.2.3. Docking-Based CoMFA and CoMSIA Models
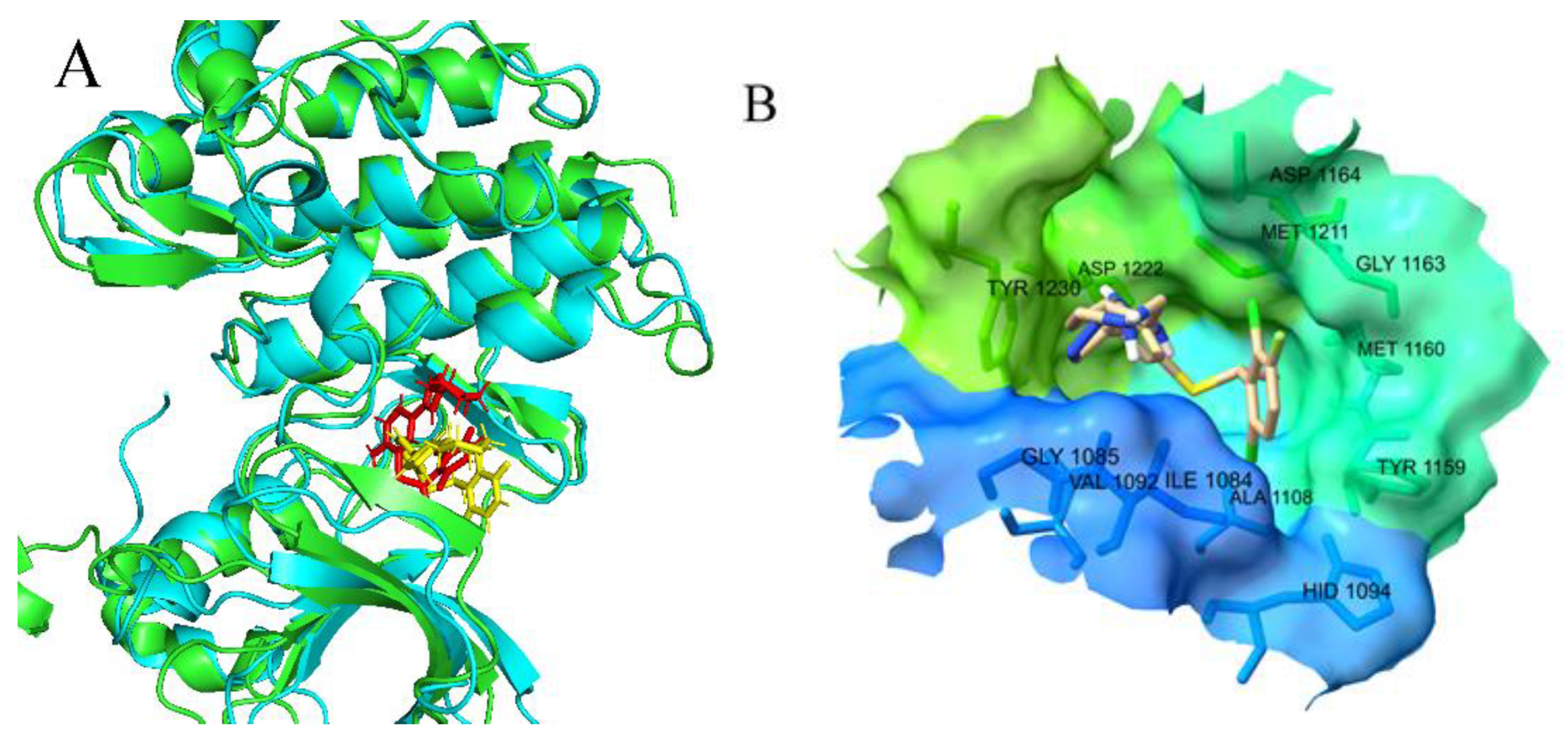
2.3.1. Molecular Docking Analysis
2.3.2. Docking-Based CoMFA and CoMSIA Model Results
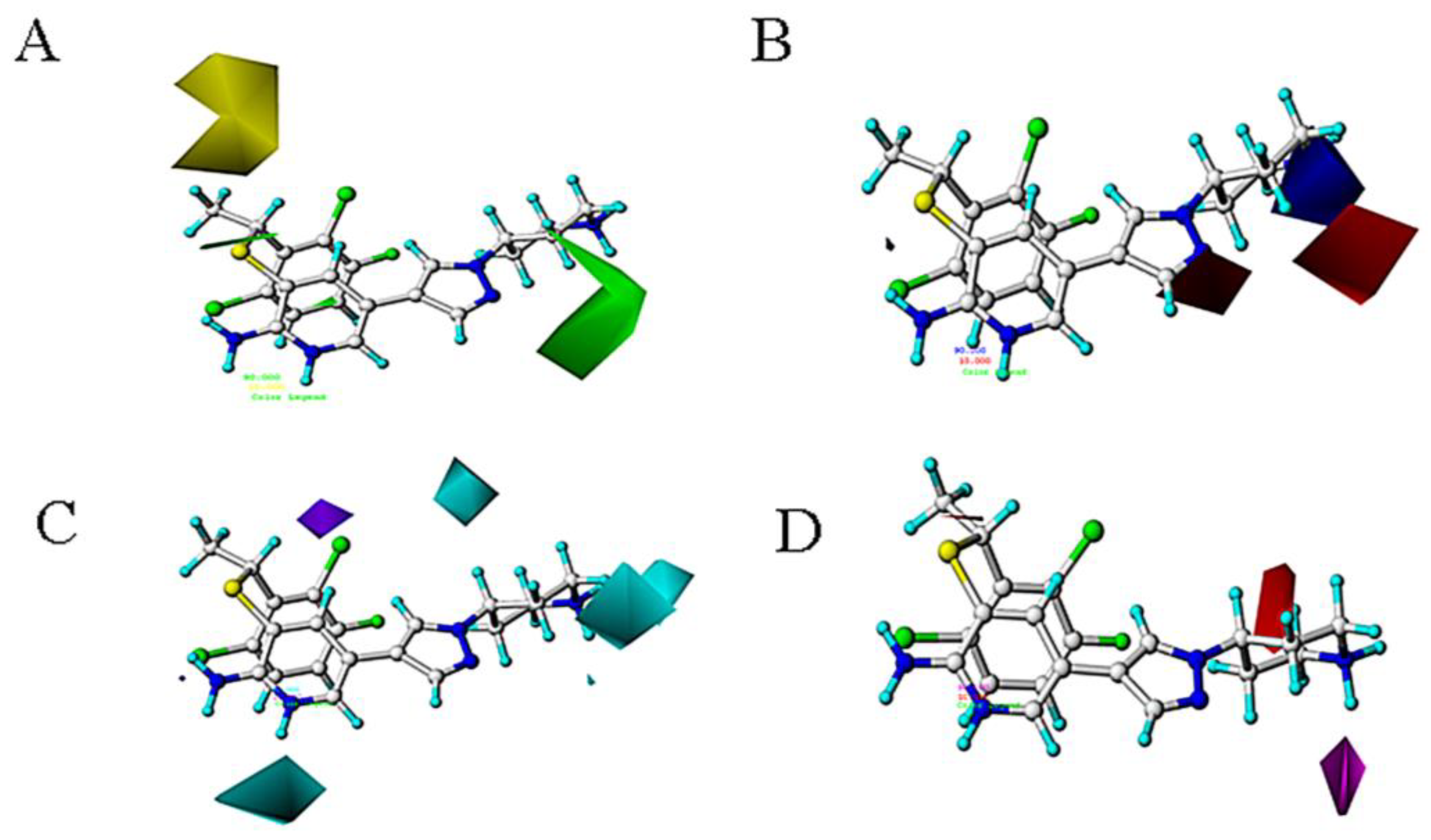
2.3.3. Contour Map Analysis
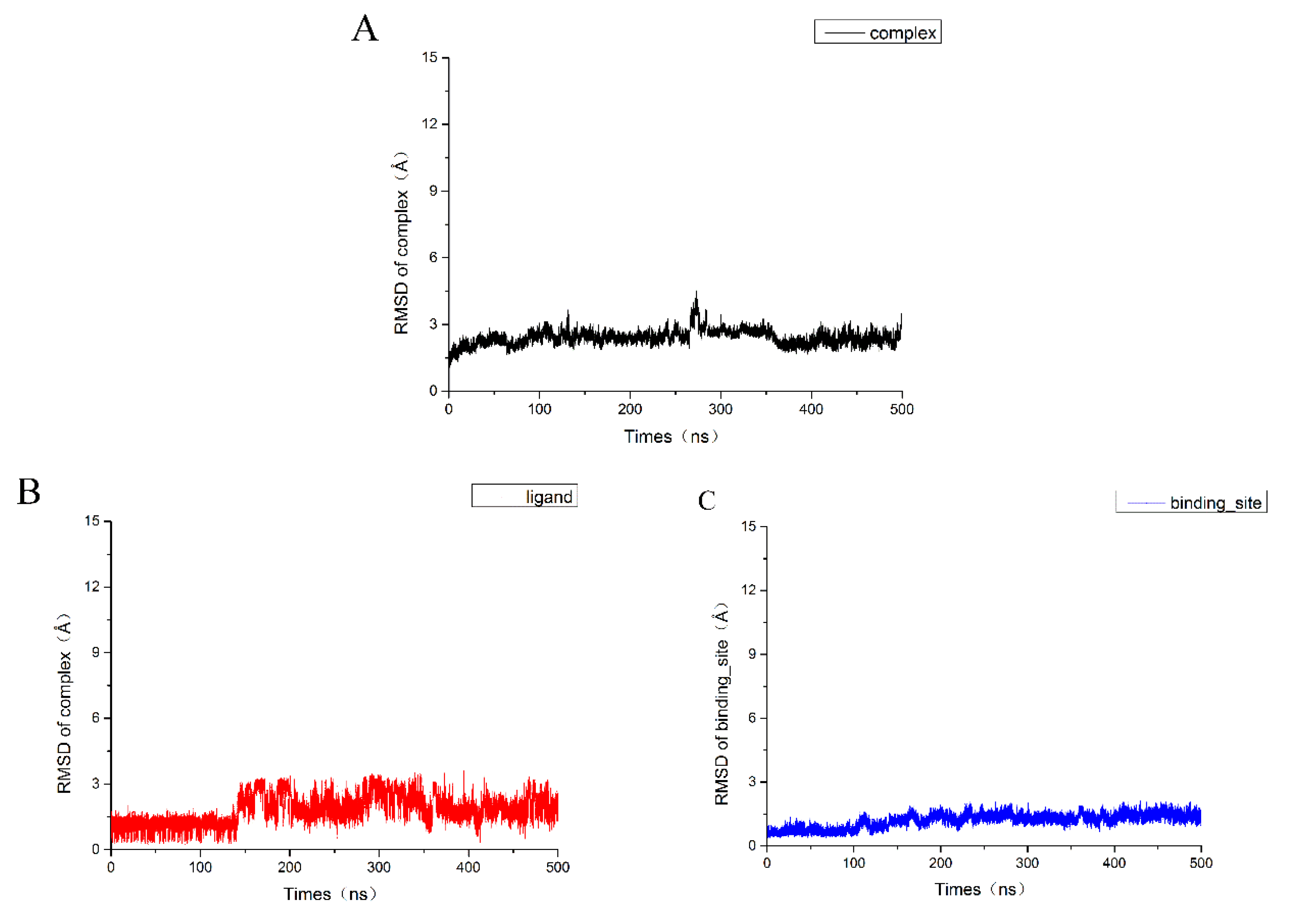
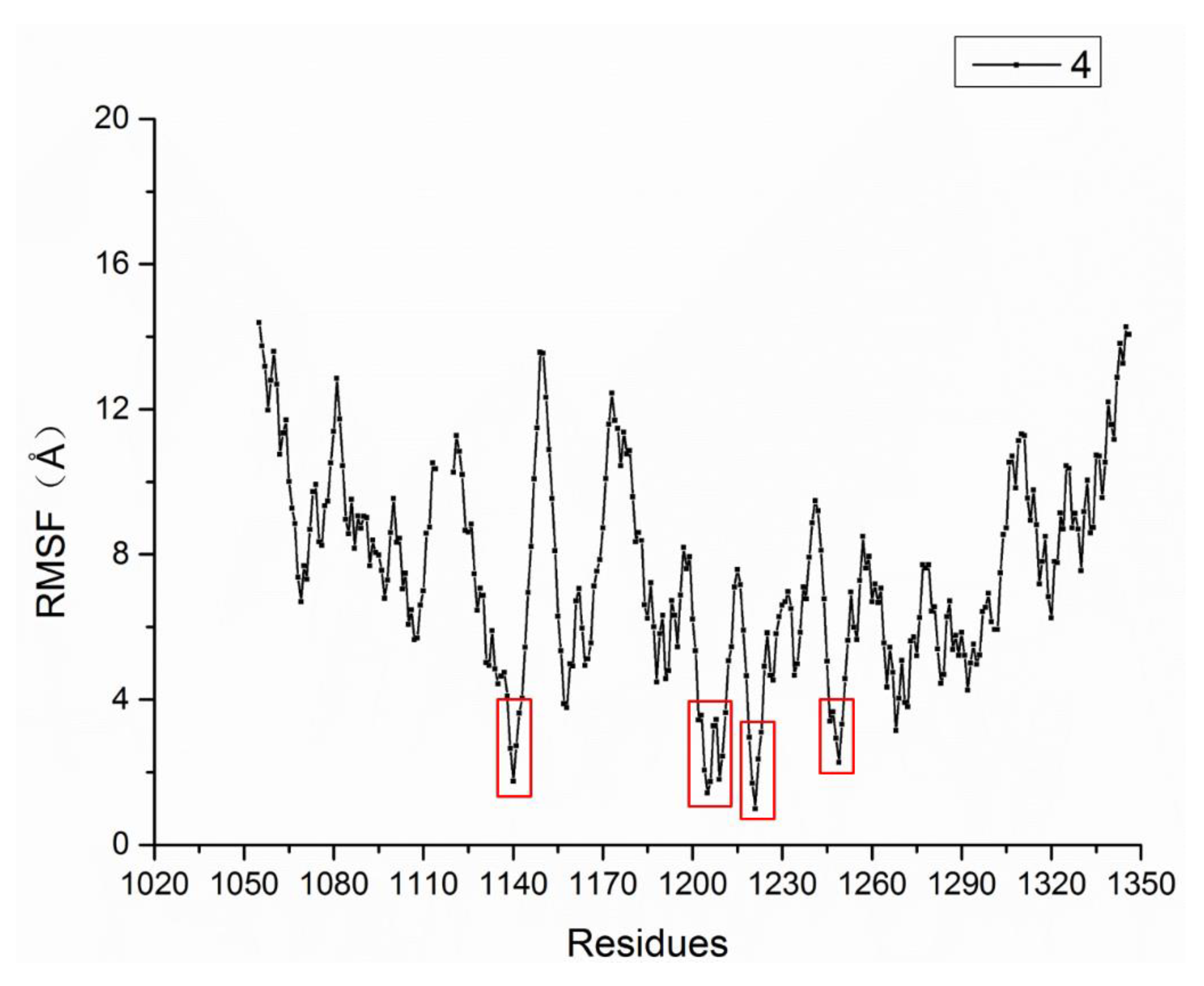
2.4. Molecular Dynamics Simulation Results
3. Materials and Methods
3.1. Molecular Conformation
3.2. Molecular Docking
3.3. Molecular Alignment
3.4. CoMFA and CoMSIA Modeling
3.5. Model Validation
3.6. Molecular Dynamics Simulation
3.7. Binding Free Energy Calculations
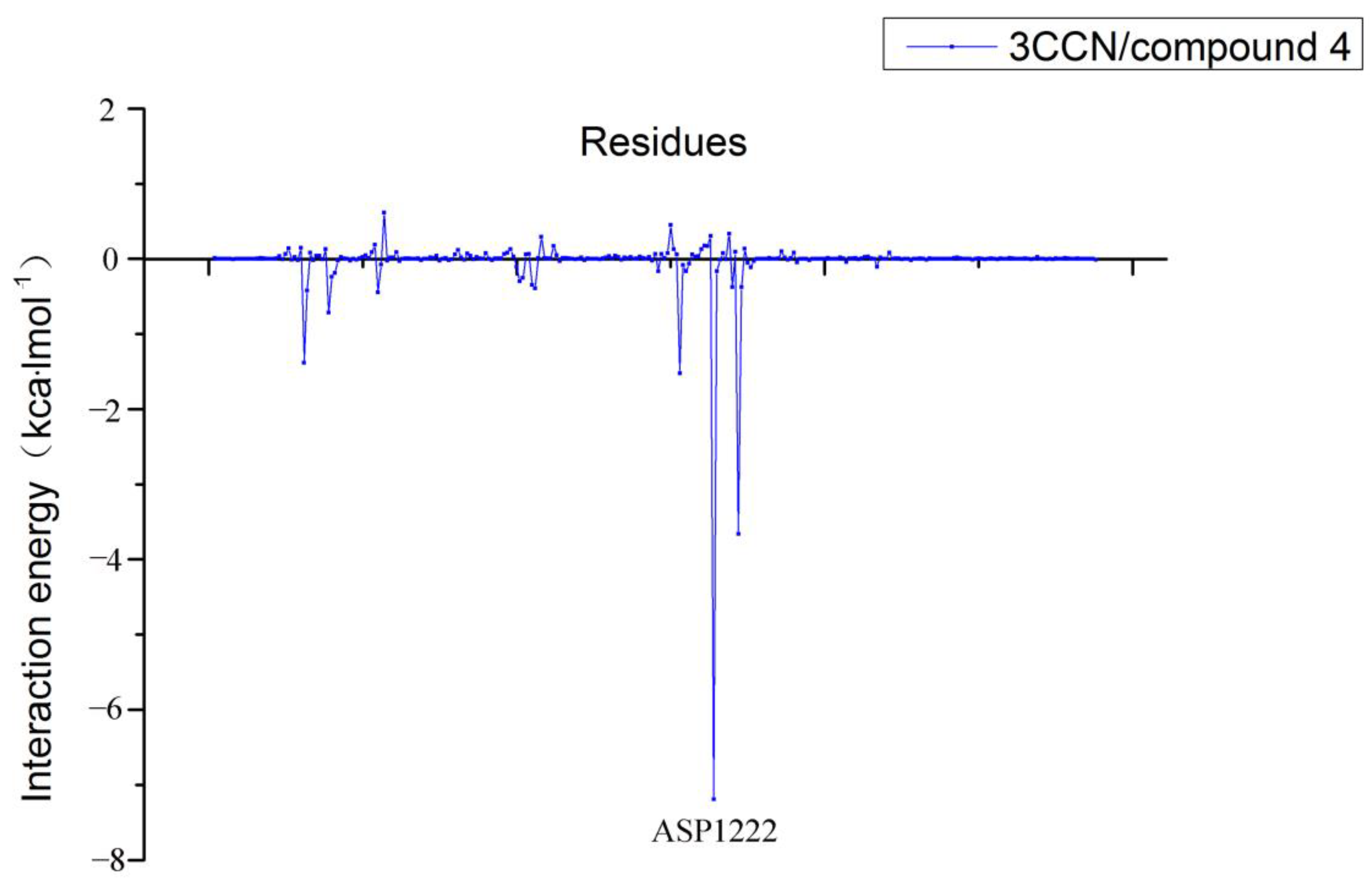
3.8. Per-Residue Free Energy Decomposition Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Vogelzang, N.J.; Benowitz, S.I.; Adams, S.; Aghajanian, C.; Chang, S.M.; Dreyer, Z.E.; Janne, P.A.; Ko, A.H.; Masters, G.A.; Odenike, O.; et al. Clinical cancer advances 2011: Annual Report on Progress Against Cancer from the American Society of Clinical Oncology. J. Clin. Oncol. 2012, 30, 88–109. [Google Scholar] [CrossRef] [PubMed]
- Yamaoka, T.; Kusumoto, S.; Ando, K.; Ohba, M.; Ohmori, T. Receptor Tyrosine Kinase-Targeted Cancer Therapy. Int. J. Mol. Sci. 2018, 19, 3491. [Google Scholar] [CrossRef] [PubMed]
- Blume-Jensen, P.; Hunter, T. Oncogenic kinase signalling. Nature 2001, 411, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Trusolino, L.; Bertotti, A.; Comoglio, P.M. MET signalling: Principles and functions in development, organ regeneration and cancer. Nat. Rev. Mol. Cell Biol. 2010, 11, 834–848. [Google Scholar] [CrossRef]
- Christensen, J.G.; Burrows, J.; Salgia, R. c-Met as a target for human cancer and characterization of inhibitors for therapeutic intervention. Cancer Lett. 2005, 225, 1–26. [Google Scholar] [CrossRef]
- Chen, R.L.; Zhao, J.; Zhang, X.C.; Lou, N.N.; Chen, H.J.; Yang, X.; Su, J.; Xie, Z.; Zhou, Q.; Tu, H.Y.; et al. Crizotinib in advanced non-small-cell lung cancer with concomitant ALK rearrangement and c-Met overexpression. BMC Cancer 2018, 18, 1171. [Google Scholar] [CrossRef]
- Burbridge, M.F.; Bossard, C.J.; Saunier, C.; Fejes, I.; Bruno, A.; Léonce, S.; Ferry, G.; Da Violante, G.; Bouzom, F.; Cattan, V.; et al. S49076 is a novel kinase inhibitor of MET, AXL, and FGFR with strong preclinical activity alone and in association with bevacizumab. Mol. Cancer Ther. 2013, 12, 1749–1762. [Google Scholar] [CrossRef]
- Boezio, A.A.; Copeland, K.W.; Rex, K.; Albrecht, B.K.; Bauer, D.; Bellon, S.F.; Boezio, C.; Broome, M.A.; Choquette, D.; Coxon, A.; et al. Discovery of (R)-6-(1-(8-Fluoro-6-(1-methyl-1H-pyrazol-4-yl)-[1,2,4]triazolo [4,3-a]pyridin-3-yl)ethyl)-3-(2-methoxyethoxy)-1,6-naphthyridin-5(6H)-one (AMG 337), a Potent and Selective Inhibitor of MET with High Unbound Target Coverage and Robust In Vivo Antitumor Activity. J. Med. Chem. 2016, 59, 2328–2342. [Google Scholar]
- Jia, H.; Dai, G.; Weng, J.; Zhang, Z.; Wang, Q.; Zhou, F.; Jiao, L.; Cui, Y.; Ren, Y.; Fan, S.; et al. Discovery of (S)-1-(1-(Imidazo[1,2-a]pyridin-6-yl)ethyl) -6-(1-methyl-1H-pyrazol-4-yl)-1H-[1,2,3]triazolo[4,5-b]pyrazine (volitinib) as a highly potent and selective mesenchymal-epithelial transition factor (c-Met) inhibitor in clinical development for treatment of cancer. J. Med. Chem. 2014, 57, 7577–7589. [Google Scholar]
- Cui, J.J. Targeting receptor tyrosine kinase MET in cancer: Small molecule inhibitors and clinical progress. J. Med. Chem. 2014, 57, 4427–4453. [Google Scholar] [CrossRef]
- Yakes, F.M.; Chen, J.; Tan, J.; Yamaguchi, K.; Shi, Y.; Yu, P.; Qian, F.; Chu, F.; Bentzien, F.; Cancilla, B.; et al. Cabozantinib (XL184), a novel MET and VEGFR2 inhibitor, simultaneously suppresses metastasis, angiogenesis, and tumor growth. Mol. Cancer Ther. 2011, 10, 2298–2308. [Google Scholar] [CrossRef]
- Qian, F.; Engst, S.; Yamaguchi, K.; Yu, P.; Won, K.A.; Mock, L.; Lou, T.; Tan, J.; Li, C.; Tam, D.; et al. Inhibition of tumor cell growth, invasion, and metastasis by EXEL-2880 (XL880, GSK1363089), a novel inhibitor of HGF and VEGF receptor tyrosine kinases. Cancer Res. 2009, 69, 8009–8016. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.D.; Kaufman, M.D.; Leary, C.B.; Turner, B.A.; Wise, S.C.; Ahn, Y.M.; Booth, R.J.; Caldwell, T.M.; Ensinger, C.L.; Hood, M.M.; et al. Altiratinib Inhibits Tumor Growth, Invasion, Angiogenesis, and Microenvironment-Mediated Drug Resistance via Balanced Inhibition of MET, TIE2, and VEGFR2. Mol. Cancer Ther. 2015, 14, 2023–2034. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.B.; Peek, V.L.; Ajamie, R.; Buchanan, S.G.; Graff, J.R.; Heidler, S.A.; Hui, Y.H.; Huss, K.L.; Konicek, B.W.; Manro, J.R.; et al. LY2801653 is an orally bioavailable multi-kinase inhibitor with potent activity against MET, MST1R, and other oncoproteins, and displays anti-tumor activities in mouse xenograft models. Investig. New Drugs 2013, 31, 833–844. [Google Scholar] [CrossRef] [PubMed]
- Eathiraj, S.; Palma, R.; Volckova, E.; Hirschi, M.; France, D.S.; Ashwell, M.A.; Chan, T.C. Discovery of a novel mode of protein kinase inhibition characterized by the mechanism of inhibition of human mesenchymal-epithelial transition factor (c-Met) protein autophosphorylation by ARQ 197. J. Biol. Chem. 2011, 286, 20666–20676. [Google Scholar] [CrossRef]
- Madapa, S.; Tusi, Z.; Batra, S. Advances in the Syntheses of Quinoline and Quinoline-Annulated Ring Systems. Curr. Org. Chem. 2008, 12, 1116–1183. [Google Scholar] [CrossRef]
- Hopfinger, A.J. A QSAR investigation of dihydrofolate reductase inhibition by baker trizaines based upon molecular shape analysis. J. Am. Chem. Soc. 1980, 102, 7196–7206. [Google Scholar] [CrossRef]
- Cramer, R.D.; Patterson, D.E.; Bunce, J.D. Comparative molecular field analysis (CoMFA). 1. Effect of shape on binding of steroids to carrier proteins. J. Am. Chem. Soc. 1988, 110, 5959–5967. [Google Scholar] [CrossRef]
- Klebe, G.; Abraham, U.; Mietzner, T. Molecular similarity indices in a comparative analysis (CoMSIA) of drug molecules to correlate and predict their biological activity. J. Med. Chem. 1994, 37, 4130–4146. [Google Scholar] [CrossRef]
- Zhang, D.; Ai, J.; Liang, Z.; Li, C.; Peng, X.; Ji, Y.; Jiang, H.; Geng, M.; Luo, C.; Liu, H. Discovery of novel 2-aminopyridine-3-carboxamides as c-Met kinase inhibitors. Bioorg. Med. Chem. 2012, 20, 5169–5180. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, X.; Ai, J.; Zhai, Y.; Liang, Z.; Wang, Y.; Chen, Y.; Li, C.; Zhao, F.; Jiang, H.; et al. Synthesis and biological evaluation of 2-amino-5-aryl-3-benzylthiopyridine scaffold based potent c-Met inhibitors. Bioorg. Med. Chem. 2013, 21, 6804–6820. [Google Scholar] [CrossRef] [PubMed]
- Roy, P.P.; Leonard, J.T.; Roy, K. Exploring the impact of size of training sets for the development of predictive QSAR models. Chemom. Intell. Lab. Syst. 2008, 90, 31–42. [Google Scholar] [CrossRef]
- Accelrys Discovery Studio, Version 2.5; Accelrys: San Diego, CA, USA, 2010.
- Fu, T.; Wu, X.; Xiu, Z.; Wang, J.; Yin, L.; Li, G. Understanding the Molecular Mechanism of Binding Modes of Aurora a Inhibitors by Long Time Scale Gpu Dynamics. J. Theor. Comput. Chem. 2013, 12, 1341003. [Google Scholar] [CrossRef]
- Zhong, H.; Bowen, J.P. GALAHAD Tripos, Inc., 1699 South Hanley Road, St. Louis, MO 63144-2319. www.tripos.com. Contact company for pricing information. J. Am. Chem. Soc. 2007, 129, 5780. [Google Scholar]
- Gasteiger, J.; Marsili, M. Iterative partial equalization of orbital electronegativity—A rapid access to atomic charges. Tetrahedron 1980, 36, 3219–3228. [Google Scholar] [CrossRef]
- Gao, C.Z.; Dong, W.; Cui, Z.W.; Yuan, Q.; Hu, X.M.; Wu, Q.M.; Han, X.; Xu, Y.; Min, Z.L. Synthesis, preliminarily biological evaluation and molecular docking study of new Olaparib analogues as multifunctional PARP-1 and cholinesterase inhibitors. J. Enzym. Inhib. Med. Chem. 2019, 34, 150–162. [Google Scholar] [CrossRef] [PubMed]
- Böhm, M.; Stürzebecher, J.; Klebe, G. Three-Dimensional Quantitative Structure-Activity Relationship Analyses Using Comparative Molecular Field Analysis and Comparative Molecular Similarity Indices Analysis to Elucidate Selectivity Differences of Inhibitors Binding to Trypsin, Thrombin, and Factor Xa. Med. Chem. 1999, 42, 458–477. [Google Scholar]
- Cruz, J.N.; Costa, J.F.S.; Khayat, A.S.; Kuca, K.; Barros, C.A.L.; Neto, A.M.J.C. Molecular dynamics simulation and binding free energy studies of novel leads belonging to the benzofuran class inhibitors of Mycobacterium tuberculosis Polyketide Synthase 13. J. Biomol. Struct. Dyn. 2019, 37, 1616–1627. [Google Scholar] [CrossRef]
- Wu, X.; Wang, Y.; Wan, S.; Zhang, J.J. Investigation on the binding mechanism of loratinib with the c-ros oncogene 1 (ROS1) receptor tyrosine kinase via molecular dynamics simulation and binding free energy calculations. J. Biomol. Struct. Dyn. 2018, 36, 3106–3113. [Google Scholar] [CrossRef]
- Du, J.; Qiu, M.; Guo, L.; Yao, X. Computational study of the binding mechanism between farnesoid X receptor α and antagonist N-benzyl-N-(3-(tertbutyl)-4-hydroxyphenyl)-2,6-dichloro-4-(dimethylamino) benzamide. J. Biomol. Struct. Dyn. 2019, 37, 1628–1640. [Google Scholar] [CrossRef]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef] [PubMed]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
- Case, D.; Betz, R.; Botello-Smith, W.; Cerutti, D.; Cheatham, T.E., III; Darden, T.; Duke, R.; Giese, T.; Gohlke, H.; Goetz, A.; et al. Amber 2014; University of California: San Francisco, CA, USA, 2014. [Google Scholar]
- Van der Spoel, D.; van Maaren, P.J. The Origin of Layer Structure Artifacts in Simulations of Liquid Water. J. Chem. Theory Comput. 2006, 1, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald:An N·Log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Ryckaert, J.P.; Ciccotti, G.; Berendsen, H.J.C. Numerical integration of Cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef]
- Hou, T.; Wang, J.; Li, Y.; Wang, W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J. Chem. Inf. Model. 2011, 51, 69–82. [Google Scholar] [CrossRef]
- Xu, L.; Sun, H.; Li, Y.; Wang, J.; Hou, T. Assessing the performance of MM/PBSA and MM/GBSA methods. 3. The impact of force fields and ligand charge models. J. Phys. Chem. B 2013, 117, 8408–8421. [Google Scholar] [CrossRef]
- Sun, H.; Li, Y.; Tian, S.; Xu, L.; Hou, T. Assessing the performance of MM/PBSA and MM/GBSA methods. 4. Accuracies of MM/PBSA and MM/GBSA methodologies evaluated by various simulation protocols using PDBbind data set. Phys. Chem. Chem. Phys. 2014, 16, 16719–16729. [Google Scholar] [CrossRef]
- Sun, H.; Li, Y.; Shen, M.; Tian, S.; Xu, L.; Pan, P.; Guan, Y.; Hou, T. Assessing the performance of MM/PBSA and MM/GBSA methods. 5. Improved docking performance using high solute dielectric constant MM/GBSA and MM/PBSA rescoring. Phys. Chem. Chem. Phys. 2014, 16, 22035–22045. [Google Scholar] [CrossRef]
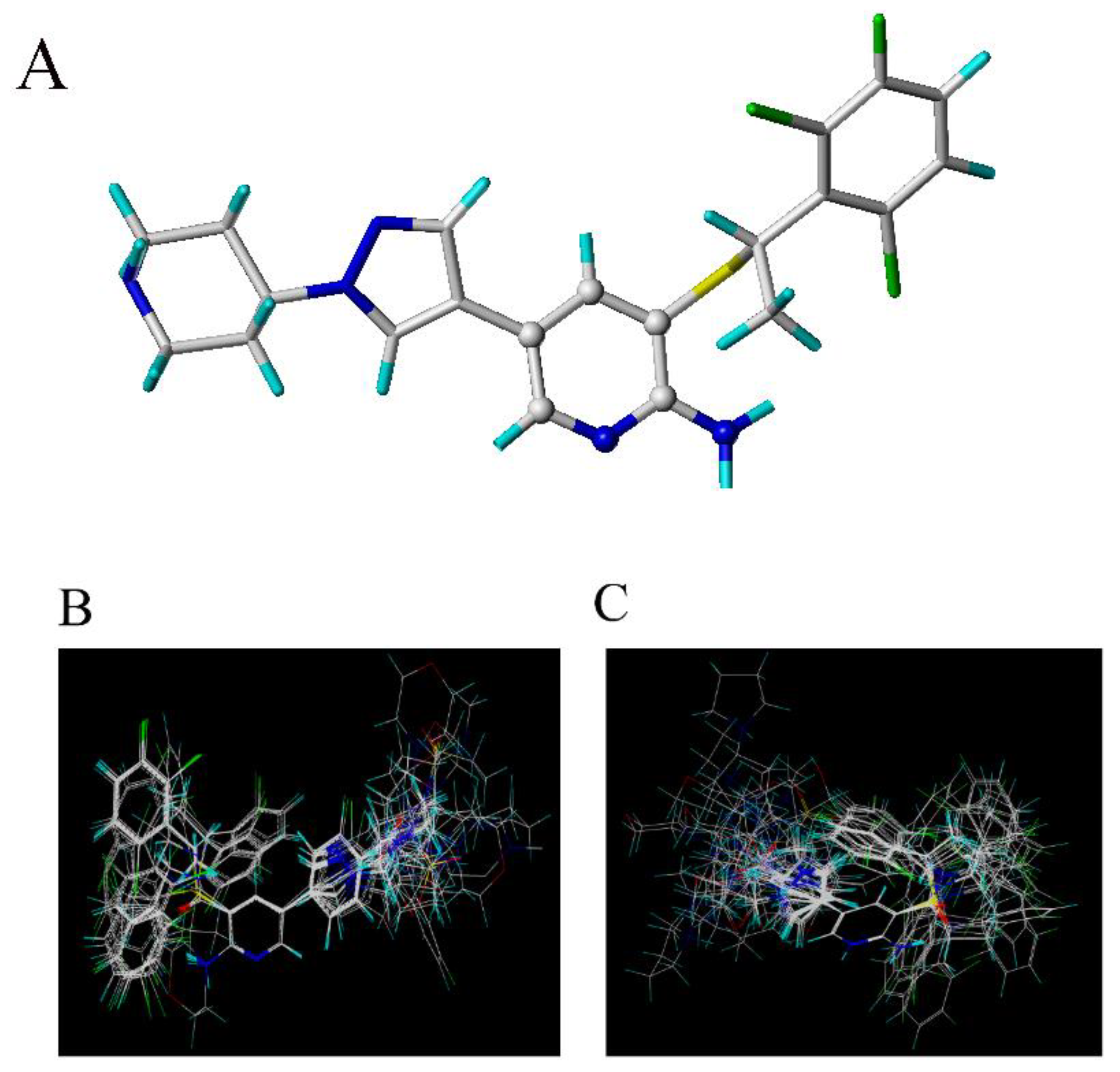



| No. | Activity (pIC50) | ||||
|---|---|---|---|---|---|
| Exp. | Alignment A | Alignment B | |||
| CoMFA | CoMSIA | CoMFA | CoMSIA | ||
| 1 | 6.494 | 6.684 | 6.634 | 6.447 | 6.437 |
| 2 | 7.004 | 6.848 | 6.958 | 6.966 | 6.97 |
| 3 | 6.553 | 6.338 | 6.464 | 6.669 | 6.587 |
| 4 | 7.785 | 7.567 | 7.687 | 7.789 | 7.775 |
| 5 | 7.670 | 7.766 | 7.584 | 7.615 | 7.764 |
| 6 | 6.993 | 6.988 | 7.108 | 6.914 | 6.987 |
| 7 | 7.210 | 7.267 | 7.155 | 7.218 | 7.248 |
| 8 | 6.640 | 6.597 | 6.632 | 6.726 | 6.687 |
| 9 | 7.347 | 7.440 | 7.474 | 7.367 | 7.349 |
| 10 * | 7.231 | 7.332 | 7.333 | 7.248 | 7.416 |
| 11 * | 6.641 | 6.459 | 6.636 | 6.791 | 6.918 |
| 12 * | 7.419 | 7.457 | 7.310 | 7.223 | 6.934 |
| 13 | 6.819 | 6.718 | 6.790 | 6.663 | 6.718 |
| 14 | 7.577 | 7.541 | 7.547 | 7.569 | 7.579 |
| 15 | 7.179 | 7.432 | 7.372 | 7.116 | 7.11 |
| 16* | 6.885 | 6.841 | 6.917 | 6.884 | 6.909 |
| 17 | 7.297 | 7.284 | 7.288 | 7.333 | 7.223 |
| 18 | 7.588 | 7.324 | 7.259 | 7.553 | 7.6 |
| 19 | 7.343 | 7.426 | 7.301 | 7.425 | 7.373 |
| 20 | 7.348 | 7.358 | 7.332 | 7.365 | 7.362 |
| 21 | 6.619 | 6.745 | 7.018 | 6.64 | 6.577 |
| 22 | 7.115 | 7.019 | 7.084 | 7.096 | 7.031 |
| 23 * | 7.088 | 7.021 | 7.145 | 6.87 | 7.024 |
| 24 | 7.656 | 7.647 | 7.685 | 7.789 | 7.67 |
| 25 | 6.996 | 6.892 | 6.786 | 6.97 | 7.026 |
| 26 | 7.188 | 7.193 | 7.239 | 7.158 | 7.283 |
| 27 | 7.009 | 7.070 | 7.000 | 6.965 | 6.968 |
| 28 | 6.707 | 6.791 | 6.750 | 6.707 | 6.655 |
| 29 | 6.761 | 6.771 | 6.784 | 6.747 | 6.755 |
| 30 | 5.920 | 5.766 | 5.797 | 5.833 | 5.736 |
| 31 | 5.155 | 5.450 | 5.600 | 5.354 | 5.46 |
| 32 * | 6.222 | 6.237 | 6.356 | 6.369 | 6.263 |
| 33 * | 5.386 | 6.036 | 6.075 | 5.984 | 5.505 |
| 34 | 6.319 | 6.040 | 5.980 | 6.366 | 6.347 |
| 35 | 5.886 | 5.821 | 5.833 | 5.75 | 5.762 |
| 36 | 5.943 | 6.084 | 6.026 | 6.063 | 6.009 |
| 37 | 6.387 | 6.375 | 6.269 | 6.435 | 6.491 |
| 38 | 6.251 | 6.223 | 6.193 | 6.171 | 6.13 |
| 39 | 6.292 | 6.356 | 6.178 | 6.304 | 6.357 |
| 40 | 6.678 | 6.843 | 6.892 | 6.683 | 6.681 |
| 41 | 5.943 | 6.006 | 5.969 | 5.9 | 5.963 |
| 42 * | 6.468 | 6.887 | 6.863 | 6.102 | 6.202 |
| PLS Statistic | Alignment-A | Alignment-B | ||
|---|---|---|---|---|
| CoMFA | CoMSIA | CoMFA | CoMSIA | |
| Q2 | 0.596 | 0.646 | 0.563 | 0.568 |
| ONC | 2 | 2 | 6 | 2 |
| R2 | 0.950 | 0.931 | 0.985 | 0.983 |
| SEE | 0.150 | 0.175 | 0.085 | 0.089 |
| R2pred | 0.839 | 0.840 | 0.821 | 0.854 |
| F | 160.303 | 76.047 | 286.459 | 260.420 |
| Field distribution | - | - | - | - |
| S (Field distribution) | 47.2% | 19.7% | 54.00% | 14.30% |
| E (Field distribution) | 52.8% | 33.2% | 46.00% | 33.20% |
| A (Field distribution) | - | 19.3% | - | 30.00% |
| D (Field distribution) | - | 27.8% | - | 22.5% |
| Field | ONC | Q2 | R2 | SEE | F | Rate of Contribution |
|---|---|---|---|---|---|---|
| SED | 2 | 0.628 | 0.934 | 0.172 | 79.479 | 0.268:0.525:0.207 |
| SEA | 2 | 0.643 | 0.943 | 0.160 | 92.376 | 0.277:0.392:0.331 |
| SEH | 2 | 0.603 | 0.923 | 0.185 | 67.394 | 0.235:0.440:0.325 |
| SEHA | 2 | 0.620 | 0.929 | 0.179 | 73.004 | 0.191:0.292:0.255:0.262 |
| SEHD | 2 | 0.602 | 0.922 | 0.186 | 66.626 | 0.184:0.375:0.273:0.168 |
| SEAD | 2 | 0.646 | 0.931 | 0.175 | 76.047 | 0.197:0.332:0.193:0.278 |
| SEHAD | 2 | 0.622 | 0.922 | 0.187 | 66.342 | 0.148:0.253:0.160:0.227:0.212 |
| Field | ONC | Q2 | R2 | SEE | F | Rate of Contribution |
|---|---|---|---|---|---|---|
| SED | 3 | 0.556 | 0.981 | 0.094 | 229.485 | 0.255:0.515:0.230 |
| SEA | 3 | 0.55 | 0.976 | 0.105 | 184.036 | 0.219:0.477:0.304 |
| SEH | 2 | 0.52 | 0.984 | 0.087 | 272.649 | 0.182:0.446:0.372 |
| SEHA | 2 | 0.53 | 0.985 | 0.082 | 303.786 | 0.136:0.339:0.306:0.219 |
| SEHD | 2 | 0.526 | 0.991 | 0.065 | 491.572 | 0.154:0.358:0.163:0.325 |
| SEAD | 3 | 0.561 | 0.981 | 0.095 | 228.254 | 0.178:0.378:0.264:0.180 |
| SEHDA | 2 | 0.536 | 0.988 | 0.073 | 385.686 | 0.117:0.286:0.197:0.138:0.262 |
| Interaction | Contribution (kcal/mol) | Standard Deviation |
|---|---|---|
| ∆EvdW | −32.99 | 3.62 |
| ∆Eele | 36.18 | 24.78 |
| ∆GGB | −25.82 | 23.74 |
| ∆GSA | −4.45 | 0.38 |
| ∆Egas | 3.19 | 24.72 |
| ∆Esolv | −30.27 | 23.64 |
| ∆Gbind | −27.08 | 3.80 |
| Acceptor | DonorH | Donor | Frac |
|---|---|---|---|
| ASP_1222@OD2 | MOL@H22 | MOL@N4 | 0.6651 |
| ASP_1222@OD2 | MOL@H20 | MOL@N3 | 0.6569 |
| ASP_1222@OD1 | MOL@H20 | MOL@N3 | 0.6431 |
| ASP_1222@OD1 | MOL@H22 | MOL@N4 | 0.6178 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, Q.; Fu, C.; Li, J. Studying the Binding Modes of Novel 2-Aminopyridine Derivatives as Effective and Selective c-Met Kinase Type 1 Inhibitors Using Molecular Modeling Approaches. Molecules 2021, 26, 52. https://doi.org/10.3390/molecules26010052
Ye Q, Fu C, Li J. Studying the Binding Modes of Novel 2-Aminopyridine Derivatives as Effective and Selective c-Met Kinase Type 1 Inhibitors Using Molecular Modeling Approaches. Molecules. 2021; 26(1):52. https://doi.org/10.3390/molecules26010052
Chicago/Turabian StyleYe, Qianwen, Chenggong Fu, and Jiazhong Li. 2021. "Studying the Binding Modes of Novel 2-Aminopyridine Derivatives as Effective and Selective c-Met Kinase Type 1 Inhibitors Using Molecular Modeling Approaches" Molecules 26, no. 1: 52. https://doi.org/10.3390/molecules26010052
APA StyleYe, Q., Fu, C., & Li, J. (2021). Studying the Binding Modes of Novel 2-Aminopyridine Derivatives as Effective and Selective c-Met Kinase Type 1 Inhibitors Using Molecular Modeling Approaches. Molecules, 26(1), 52. https://doi.org/10.3390/molecules26010052





