Discovery and Biological Evaluation of CD147 N-Glycan Inhibitors: A New Direction in the Treatment of Tumor Metastasis
Abstract
1. Introduction
2. Results
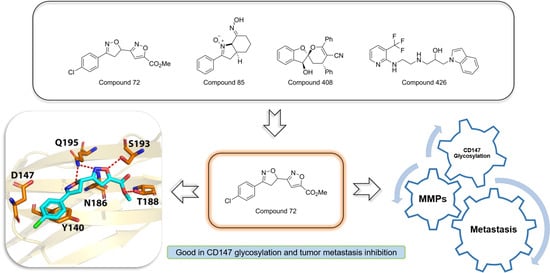
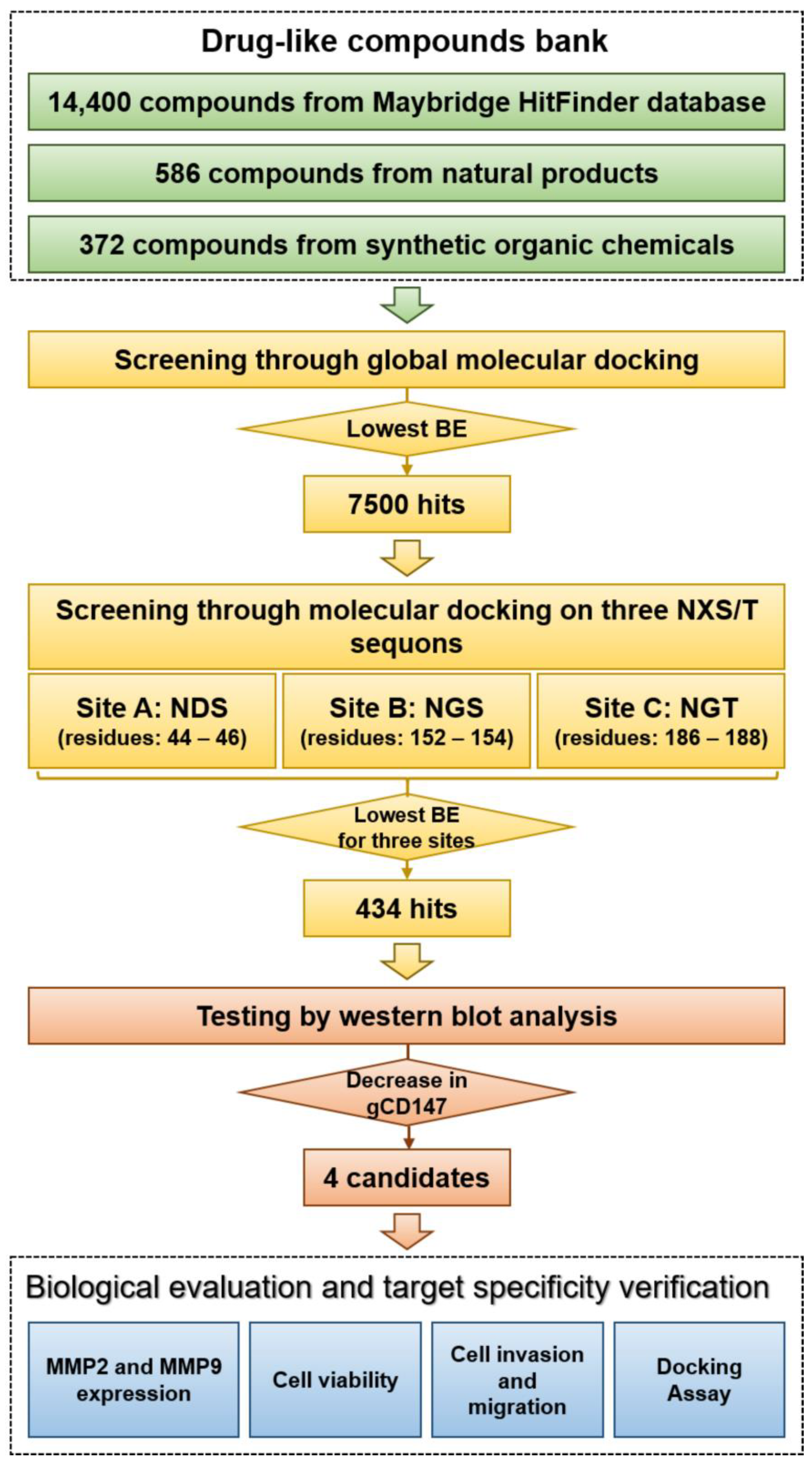
2.1. Virtual Screening and Hit Validation
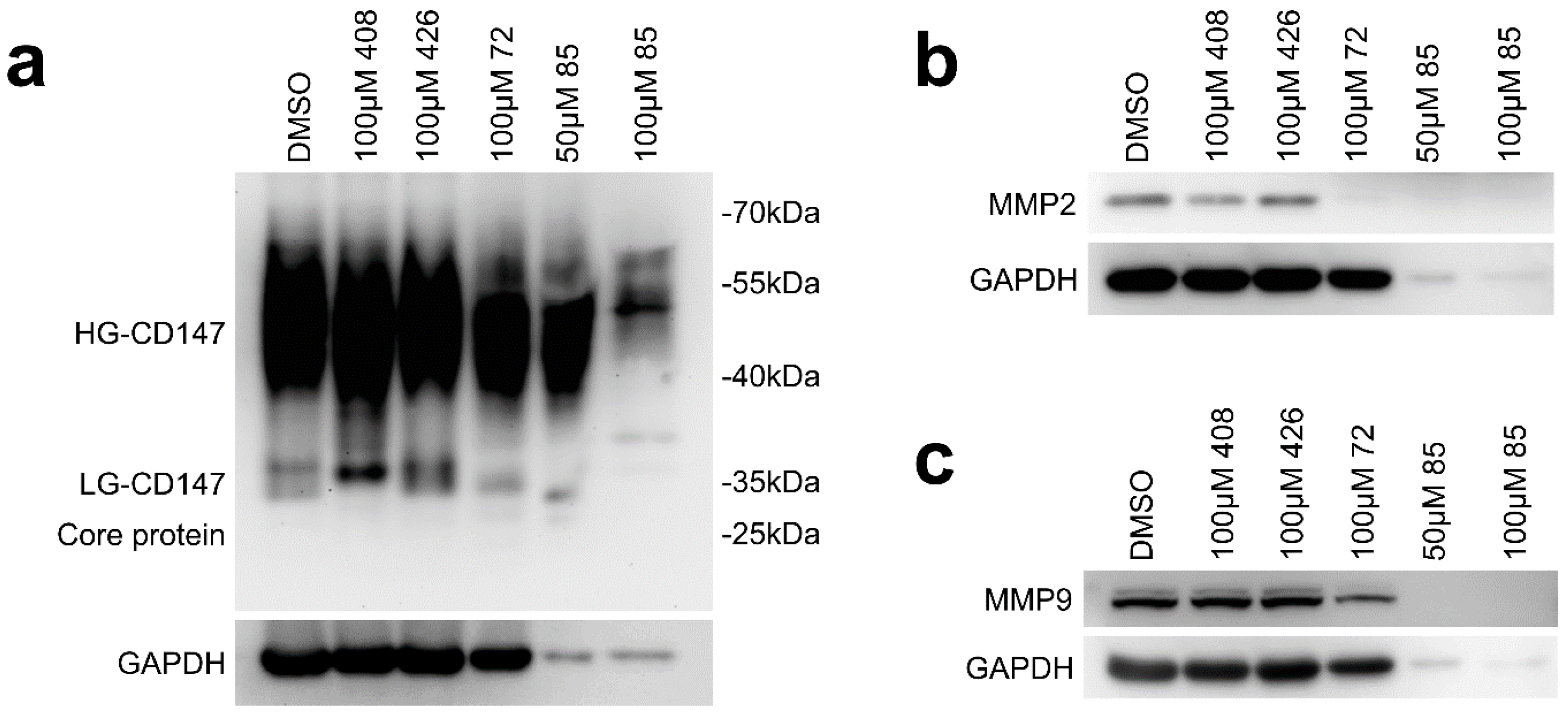
2.2. Effect of Candidate Inhibitors on MMPs Expression
2.3. Cytotoxicity Test of Candidate Inhibitors
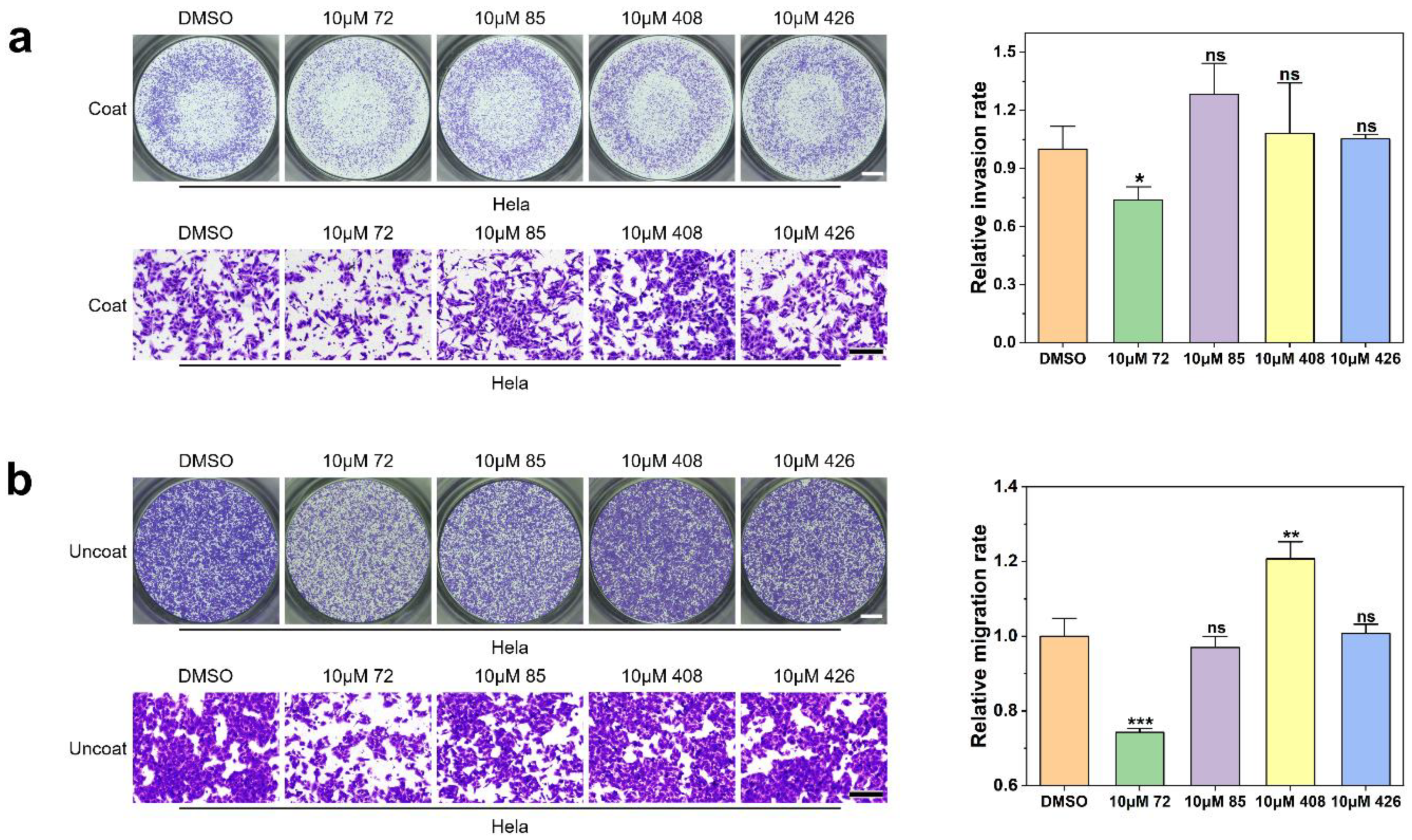
2.4. Effect of Candidate Inhibitors on Tumor Cell Invasion and Migration
2.5. Target Validation of Compound 72
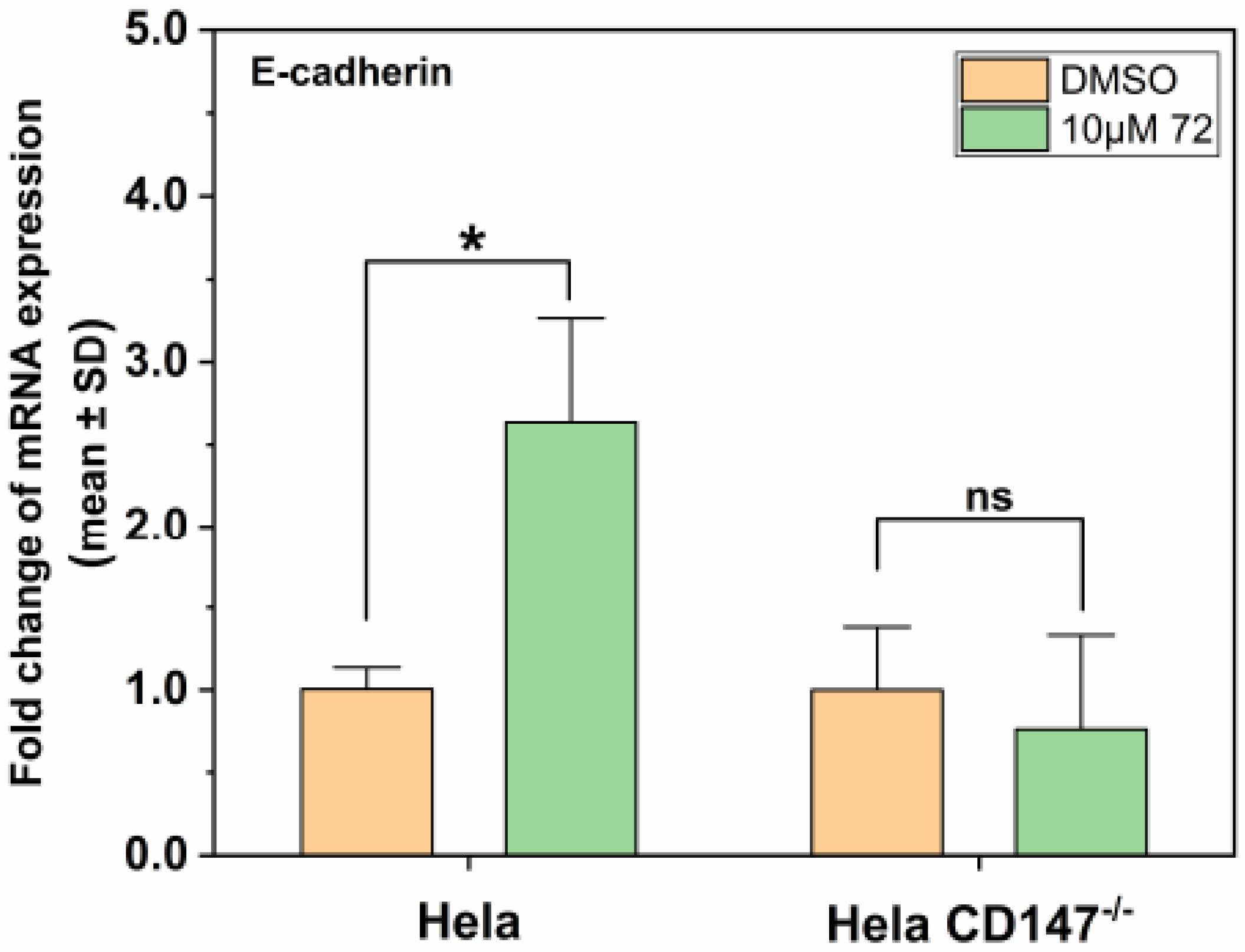
2.6. Effect of Compound 72 on Tumor-Associated Molecules by Targeting CD147
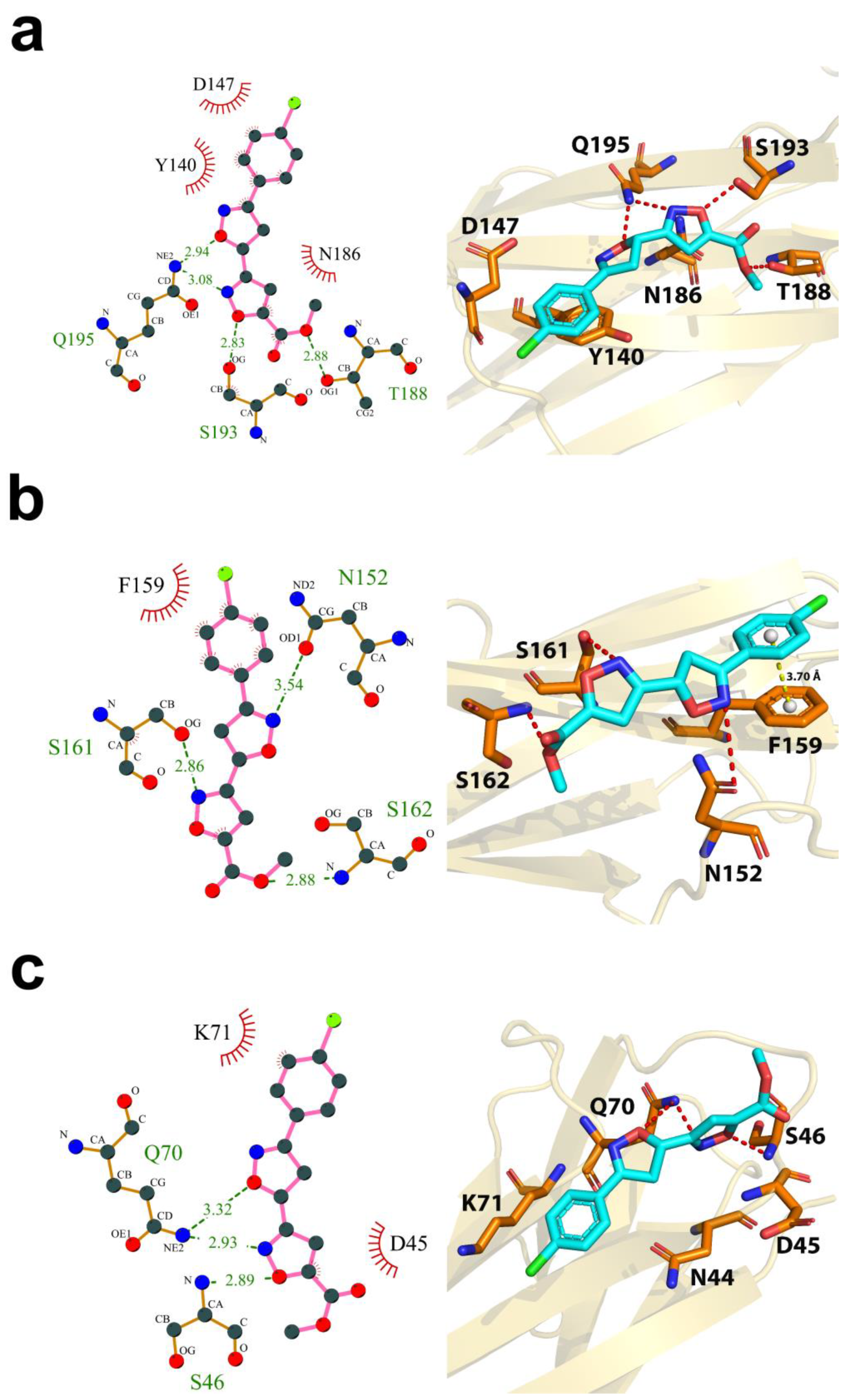
2.7. Computational Characterization of the Binding Modes between Compound 72 and CD147 Glycosylation Sites
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Compounds
4.3. Cell Culture
4.4. CRIPSR–Cas9 Genome Editing
4.5. CCK-8 Cell Cytotoxity Assay
4.6. Transwell Assay
4.7. Virtual Screening and Molecular Docking
4.8. Western Blot Analysis
4.9. RNA Extraction, Reverse Transcription, and Real-Time Quantitative Polymerase Chain Reaction (qPCR)
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Steeg, P.S. Targeting metastasis. Nat. Rev. Cancer 2016, 16, 201–218. [Google Scholar] [CrossRef]
- Guan, X. Cancer metastases: Challenges and opportunities. Acta Pharm. Sin. B 2015, 5, 402–418. [Google Scholar] [CrossRef] [PubMed]
- Lambert, A.W.; Pattabiraman, D.R.; Weinberg, R.A. Emerging Biological Principles of Metastasis. Cell 2017, 168, 670–691. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wu, X.; Cheng, X. Advances in diagnosis and treatment of metastatic cervical cancer. J. Gynecol. Oncol. 2016, 27, e43. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.; Pantel, K.; Kang, Y. Tumor metastasis: Moving new biological insights into the clinic. Nat. Med. 2013, 19, 1450–1464. [Google Scholar] [CrossRef] [PubMed]
- Munkley, J.; Elliott, D.J. Hallmarks of glycosylation in cancer. Oncotarget 2016, 7, 35478–35489. [Google Scholar] [CrossRef]
- Schwarz, F.; Aebi, M. Mechanisms and principles of N-linked protein glycosylation. Curr. Opin. Struct. Biol. 2011, 21, 576–582. [Google Scholar] [CrossRef]
- Helenius, A.; Aebi, M. Intracellular functions of N-linked glycans. Science 2001, 291, 2364–2369. [Google Scholar] [CrossRef]
- Cheung, J.C.; Reithmeier, R.A.F. Scanning N-glycosylation mutagenesis of membrane proteins. Methods 2007, 41, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Biswas, C.; Zhang, Y.; DeCastro, R.; Guo, H.; Nakamura, T.; Kataoka, H.; Nabeshima, K. The Human Tumor Cell-derived Collagenase Stimulatory Factor (Renamed EMMPRIN) Is a Member of the Immunoglobulin Superfamily. Cancer Res. 1995, 55, 434–439. [Google Scholar] [PubMed]
- Miyauchi, T.; Masuzawa, Y.; Muramatsu, T. The basigin group of the immunoglobulin superfamily: Complete conservation of a segment in and around transmembrane domains of human and mouse basigin and chicken HT7 antigen. J. Biochem. 1991, 110, 770–774. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Zhao, S.; Shen, F.; Liang, J.; Chen, J. Overexpression of CD147 is associated with poor prognosis, tumor cell migration and ERK signaling pathway activation in hepatocellular carcinoma. Exp. Ther. Med. 2017, 14, 2637–2642. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Abudumijiti, H.; Xu, L.; Hasim, A. CD147 promotes cervical cancer migration and invasion by up-regulating fatty acid synthase expression. Int. J. Clin. Exp. Pathol. 2019, 12, 4280. [Google Scholar]
- Grass, G.D.; Dai, L.; Qin, Z.Q.; Parsons, C.; Toole, B.P. CD147: Regulator of Hyaluronan Signaling in Invasiveness and Chemoresistance. Hyaluronan Signal. Turnover 2014, 123, 351–373. [Google Scholar] [CrossRef]
- Hibino, T.; Sakaguchi, M.; Miyamoto, S.; Yamamoto, M.; Motoyama, A.; Hosoi, J.; Shimokata, T.; Ito, T.; Tsuboi, R.; Huh, N.H. S100A9 is a novel ligand of EMMPRIN that promotes melanoma metastasis. Cancer Res. 2013, 73, 172–183. [Google Scholar] [CrossRef]
- Iacono, K.T.; Brown, A.L.; Greene, M.I.; Saouaf, S.J. CD147 immunoglobulin superfamily receptor function and role in pathology. Exp. Mol. Pathol. 2007, 83, 283–295. [Google Scholar] [CrossRef]
- Xin, X.; Zeng, X.; Gu, H.; Li, M.; Tan, H.; Jin, Z.; Hua, T.; Shi, R.; Wang, H. CD147/EMMPRIN overexpression and prognosis in cancer: A systematic review and meta-analysis. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef]
- Li, Y.; Xu, J.; Chen, L.; Zhong, W.D.; Zhang, Z.; Mi, L.; Zhang, Y.; Liao, C.G.; Bian, H.J.; Jiang, J.L.; et al. HAb18G (CD147), a cancer-associated biomarker and its role in cancer detection. Histopathology 2009, 54, 677–687. [Google Scholar] [CrossRef]
- Xiong, L.; Edwards, C.K.; Zhou, L. The biological function and clinical utilization of CD147 in human diseases: A review of the current scientific literature. Int. J. Mol. Sci. 2014, 15, 17411–17441. [Google Scholar] [CrossRef]
- Zhang, F.; Zeng, Y.L.; Zhang, X.G.; Chen, W.J.; Yang, R.; Li, S.J. RNA interference targeting extracellular matrix metalloproteinase inducer (CD147) inhibits growth and increases chemosensitivity in human cervical cancer cells. Eur. J. Gynaecol. Oncol. 2013, 34, 429–435. [Google Scholar]
- Riethdorf, S.; Reimers, N.; Assmann, V.; Kornfeld, J.W.; Terracciano, L.; Sauter, G.; Pantel, K. High incidence of EMMPRIN expression in human tumors. Int. J. Cancer 2006, 119, 1800–1810. [Google Scholar] [CrossRef]
- Landras, A.; de Moura, C.R.; Jouenne, F.; Lebbe, C.; Menashi, S.; Mourah, S. CD147 is a promising target of tumor progression and a prognostic biomarker. Cancers 2019, 11, 1803. [Google Scholar] [CrossRef]
- Fan, X.; Wu, W.; Shi, H.; Han, J. RNA interference targeting CD147 inhibits the invasion of human cervical squamous carcinoma cells by downregulating MMP-9. Cell Biol. Int. 2013, 37, 737–741. [Google Scholar] [CrossRef]
- Cui, J.; Huang, W.; Wu, B.; Jin, J.; Jing, L.; Shi, W.P.; Liu, Z.Y.; Yuan, L.; Luo, D.; Li, L.; et al. N-glycosylation by N-acetylglucosaminyltransferase V enhances the interaction of CD147/basigin with integrin β1 and promotes HCC metastasis. J. Pathol. 2018, 245, 41–52. [Google Scholar] [CrossRef]
- Pinho, S.S.; Reis, C.A. Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer 2015, 15, 540–555. [Google Scholar] [CrossRef]
- Bai, Y.; Huang, W.; Ma, L.T.; Jiang, J.L.; Chen, Z.N. Importance of N-glycosylation on CD147 for its biological functions. Int. J. Mol. Sci. 2014, 15, 6356–6377. [Google Scholar] [CrossRef]
- Tyler, R.E.; Pearce, M.M.P.; Shaler, T.A.; Olzmann, J.A.; Greenblatt, E.J.; Kopito, R.R. Unassembled CD147 is an endogenous endoplasmic reticulum-associated degradation substrate. Mol. Biol. Cell 2012, 23, 4668–4678. [Google Scholar] [CrossRef]
- Huang, W.; Luo, W.J.; Zhu, P.; Tang, J.; Yu, X.L.; Cui, H.Y.; Wang, B.; Zhang, Y.; Jiang, J.L.; Chen, Z.N. Modulation of CD147-induced matrix metalloproteinase activity: Role of CD147 N-glycosylation. Biochem. J. 2013, 449, 437–448. [Google Scholar] [CrossRef]
- Xu, J.; Shen, Z.Y.; Chen, X.G.; Zhang, Q.; Bian, H.J.; Zhu, P.; Xu, H.Y.; Song, F.; Yang, X.M.; Mi, L.; et al. A randomized controlled trial of licartin for preventing hepatoma recurrence after liver transplantation. Hepatology 2007, 45, 269–276. [Google Scholar] [CrossRef]
- Wang, Y.; Yuan, L.; Yang, X.M.; Wei, D.; Wang, B.; Sun, X.X.; Feng, F.; Nan, G.; Wang, Y.; Chen, Z.N.; et al. A chimeric antibody targeting CD147 inhibits hepatocellular carcinoma cell motility via FAK-PI3K-Akt-Girdin signaling pathway. Clin. Exp. Metastasis 2015, 32, 39–53. [Google Scholar] [CrossRef]
- Fu, Z.G.; Wang, L.; Cuiv, H.Y.; Peng, J.L.; Wangc, S.J.; Gengc, J.J.; Liu, J.D.; Feng, F.; Song, F.; Li, L.; et al. A novel small-molecule compound targeting CD147 inhibits the motility and invasion of hepatocellular carcinoma cells. Oncotarget 2016, 7, 9429–9447. [Google Scholar] [CrossRef]
- Steet, R.A.; Melançon, P.; Kuchta, R.D. 3’-Azidothymidine potently inhibits the biosynthesis of highly branched N-linked oligosaccharides and poly-N-acetyllactosamine chains in cells. J. Biol. Chem. 2000, 275, 26812–26820. [Google Scholar] [CrossRef]
- Xu, X.; Hu, Y.; Ni, J.; Hu, S.; Jiang, Z.; Xu, L.; Liu, C.; Hua, D.; Wu, S. 3’-Azidothymidine may potently inhibit the biosynthesis of polylactosamine chains on highly glycosylated-CD147 and reduce matrix metalloproteinase-2 expression in SGC-7901 and U251 cells. Mol. Med. Rep. 2015, 11, 4713–4719. [Google Scholar] [CrossRef]
- Tang, W.; Chang, S.B.; Hemler, M.E. Links between CD147 function, glycosylation, and caveolin-1. Mol. Biol. Cell 2004, 15, 4043–4050. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef]
- Cragg, G.M.; Newman, D.J. Natural products: A continuing source of novel drug leads. Biochim. Biophys. Acta Gen. Subj. 2013, 1830, 3670–3695. [Google Scholar] [CrossRef]
- Tian, T.; Li, L.; Xue, J.; Zhang, J.; Li, Y. Enantioselective syntheses of spiroketals via a tandem reaction of Cu(I)-catalyzed cycloetherification and hydrogen-bond-induced [4 + 2] cyclization. J. Org. Chem. 2015, 80, 4189–4200. [Google Scholar] [CrossRef]
- Peng, X.X.; Deng, Y.J.; Yang, X.L.; Zhang, L.; Yu, W.; Han, B. Iminoxyl radical-promoted dichotomous cyclizations: Efficient oxyoximation and aminooximation of alkenes. Org. Lett. 2014, 16, 4650–4653. [Google Scholar] [CrossRef]
- Wu, J.; Hao, Z.W.; Zhao, Y.X.; Yang, X.M.; Tang, H.; Zhang, X.; Song, F.; Sun, X.X.; Wang, B.; Nan, G.; et al. Full-length soluble CD147 promotes MMP-2 expression and is a potential serological marker in detection of hepatocellular carcinoma. J. Transl. Med. 2014, 12, 1–13. [Google Scholar] [CrossRef]
- Yan, L.; Zucker, S.; Toole, B.P. Roles of the multifunctional glycoprotein, emmprin (basigin; CD147), in tumour progression. Thromb. Haemost. 2005, 93, 199–204. [Google Scholar] [CrossRef]
- Scherließ, R. The MTT assay as tool to evaluate and compare excipient toxicity in vitro on respiratory epithelial cells. Int. J. Pharm. 2011, 411, 98–105. [Google Scholar] [CrossRef]
- Li, J.H.; Huang, W.; Lin, P.; Wu, B.; Fu, Z.G.; Shen, H.M.; Jing, L.; Liu, Z.Y.; Zhou, Y.; Meng, Y.; et al. N-linked glycosylation at Asn152 on CD147 affects protein folding and stability: Promoting tumour metastasis in hepatocellular carcinoma. Sci. Rep. 2016, 6, 35210. [Google Scholar] [CrossRef]
- Brabletz, T.; Kalluri, R.; Nieto, M.A.; Weinberg, R.A. EMT in cancer. Nat. Rev. Cancer 2018, 18, 128–134. [Google Scholar] [CrossRef]
- Banyard, J.; Bielenberg, D.R. The role of EMT and MET in cancer dissemination. Connect. Tissue Res. 2015, 56, 403–413. [Google Scholar] [CrossRef]
- Weber, G.F. Why does cancer therapy lack effective anti-metastasis drugs? Cancer Lett. 2013, 328, 207–211. [Google Scholar] [CrossRef]
- Shakineshleman, S.H.; Spitalnik, S.L.; Kasturi, L. The Amino Acid at the X Position of an Asn-X-Ser Sequon Is an Important Determinant of N-Linked Core-glycosylation Efficiency. J. Biol. Chem. 1996, 271, 6363–6366. [Google Scholar] [CrossRef]
- Louis, K.S.K.S.; Siegel, A.C.A.C. Mammalian Cell Viability. Methods Mol. Biol. 2011, 740, 7–12. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2009, 31, 455–461. [Google Scholar] [CrossRef]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. Ligplot: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. Des. Sel. 1995, 8, 127–134. [Google Scholar] [CrossRef] [PubMed]


| Reference Number | Chemical Structure | MW (Da) |
|---|---|---|
| 72 |  | 306.70 |
| 85 | 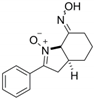 | 244.29 |
| 408 | 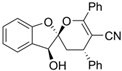 | 381.43 |
| 426 | 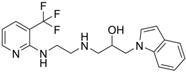 | 378.40 |
| No. | Sequences(5′ to 3′) |
|---|---|
| 1 | CATCTCCATCGACACGCTCG |
| 2 | TCATGAACGGCTCCGAGAGC |
| 3 | CGTCAGAACACATCAACGAG |
| 4 | GTCGTCAGAACACATCAACG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Wang, D.; Ge, Y.; Zhang, L.; Wu, J.; Liu, D. Discovery and Biological Evaluation of CD147 N-Glycan Inhibitors: A New Direction in the Treatment of Tumor Metastasis. Molecules 2021, 26, 33. https://doi.org/10.3390/molecules26010033
Li W, Wang D, Ge Y, Zhang L, Wu J, Liu D. Discovery and Biological Evaluation of CD147 N-Glycan Inhibitors: A New Direction in the Treatment of Tumor Metastasis. Molecules. 2021; 26(1):33. https://doi.org/10.3390/molecules26010033
Chicago/Turabian StyleLi, Wenqian, Daojiong Wang, Yushu Ge, Lei Zhang, Jiang Wu, and Dan Liu. 2021. "Discovery and Biological Evaluation of CD147 N-Glycan Inhibitors: A New Direction in the Treatment of Tumor Metastasis" Molecules 26, no. 1: 33. https://doi.org/10.3390/molecules26010033
APA StyleLi, W., Wang, D., Ge, Y., Zhang, L., Wu, J., & Liu, D. (2021). Discovery and Biological Evaluation of CD147 N-Glycan Inhibitors: A New Direction in the Treatment of Tumor Metastasis. Molecules, 26(1), 33. https://doi.org/10.3390/molecules26010033