Anticancer Activity of Lesbicoumestan in Jurkat Cells via Inhibition of Oxidative Stress-Mediated Apoptosis and MALT1 Protease
Abstract
1. Introduction
2. Results
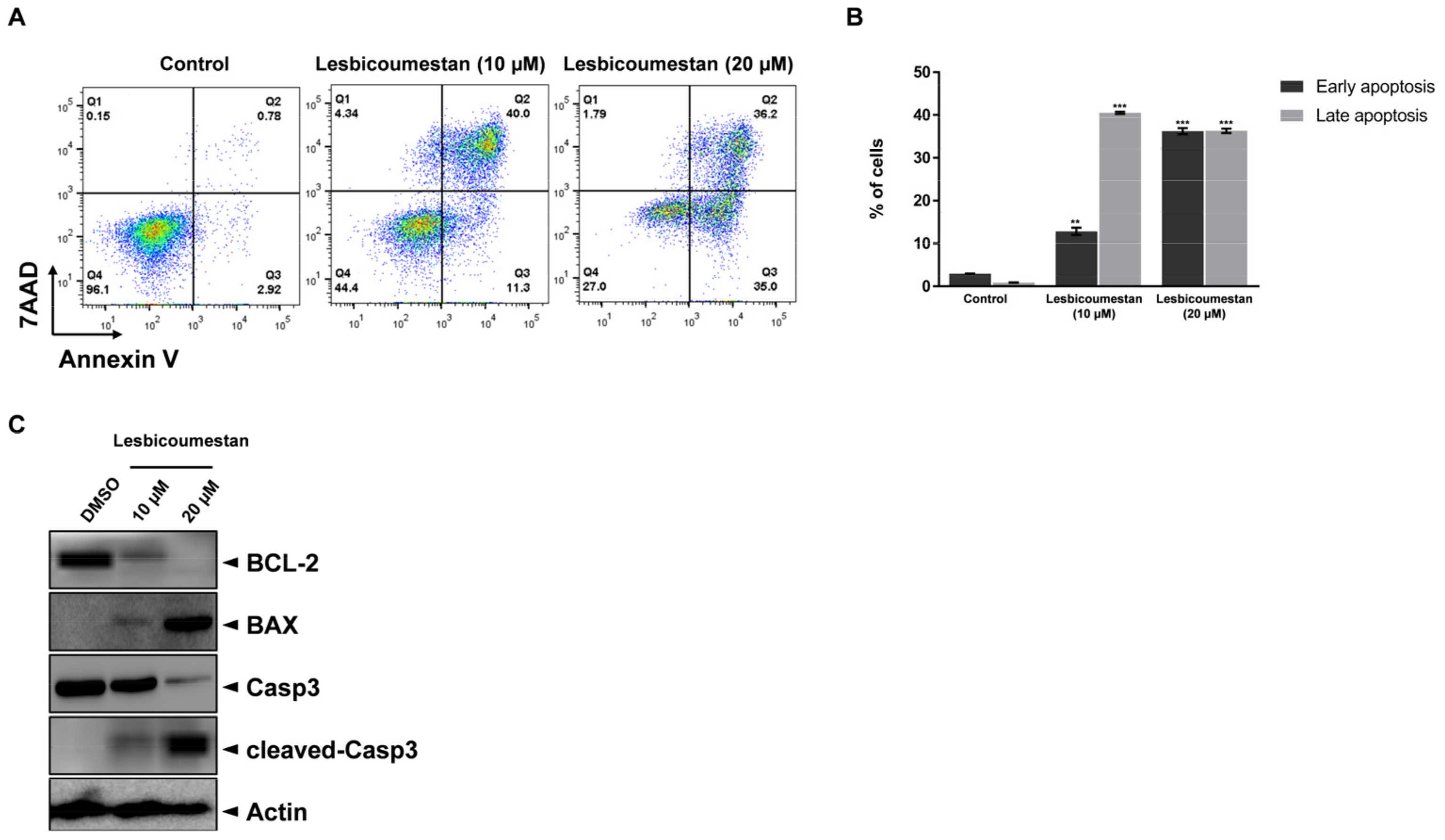
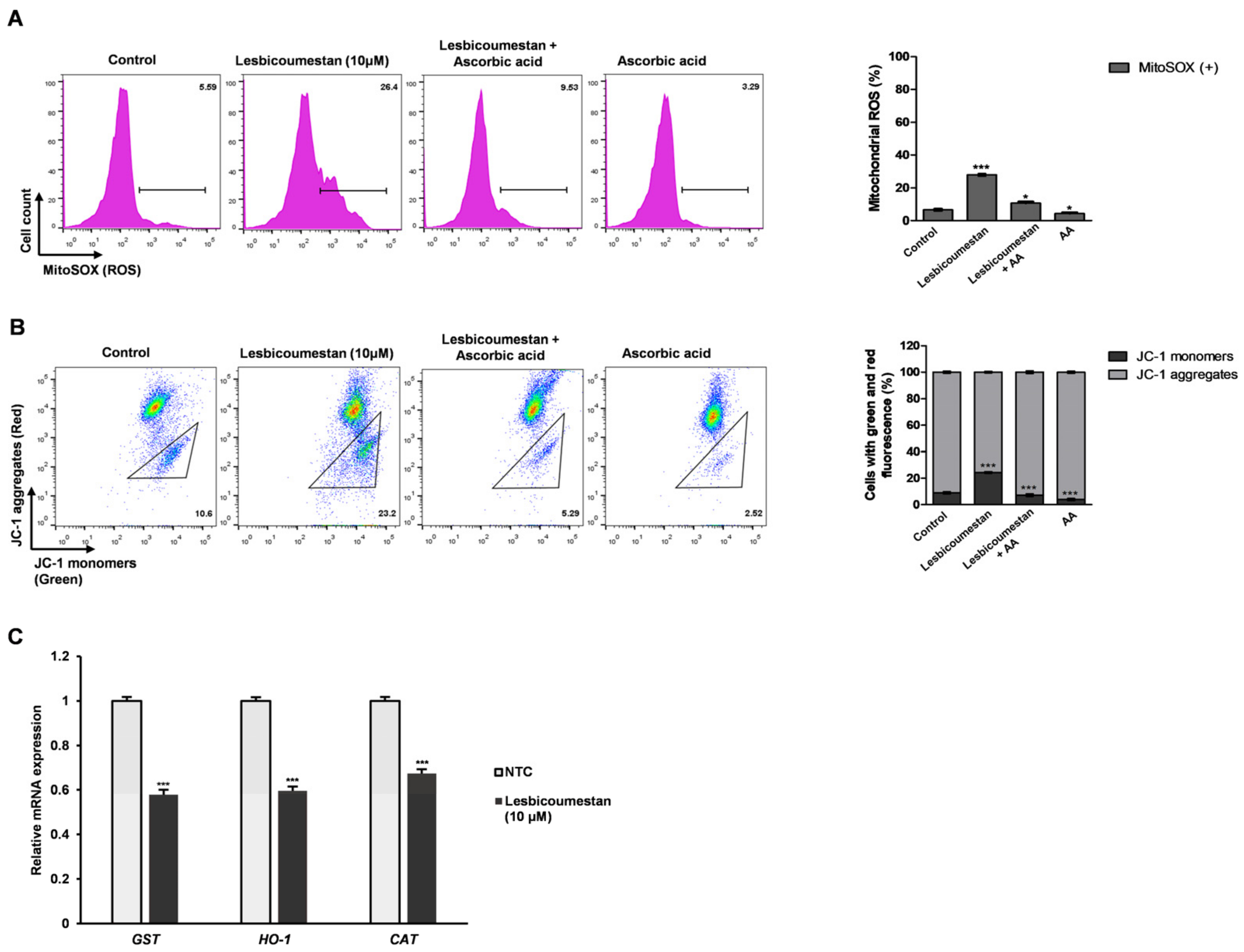
2.1. Lesbicoumestan-Induced Mitochondrial and Caspase-Dependent Apoptosis in Jurkat Cells
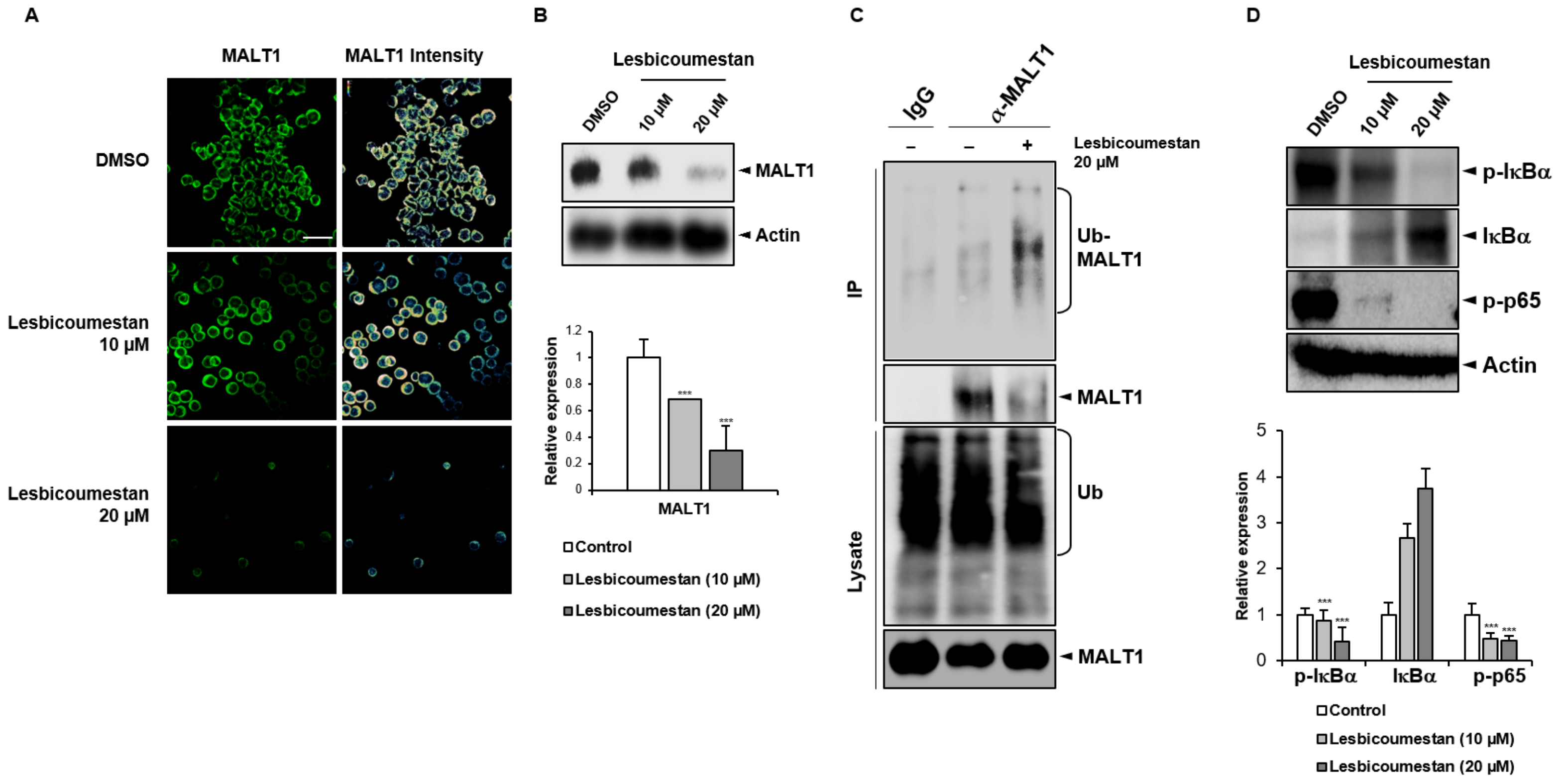
2.2. Lesbicoumestan-Induced Degradation of MALT1
2.3. Molecular Docking
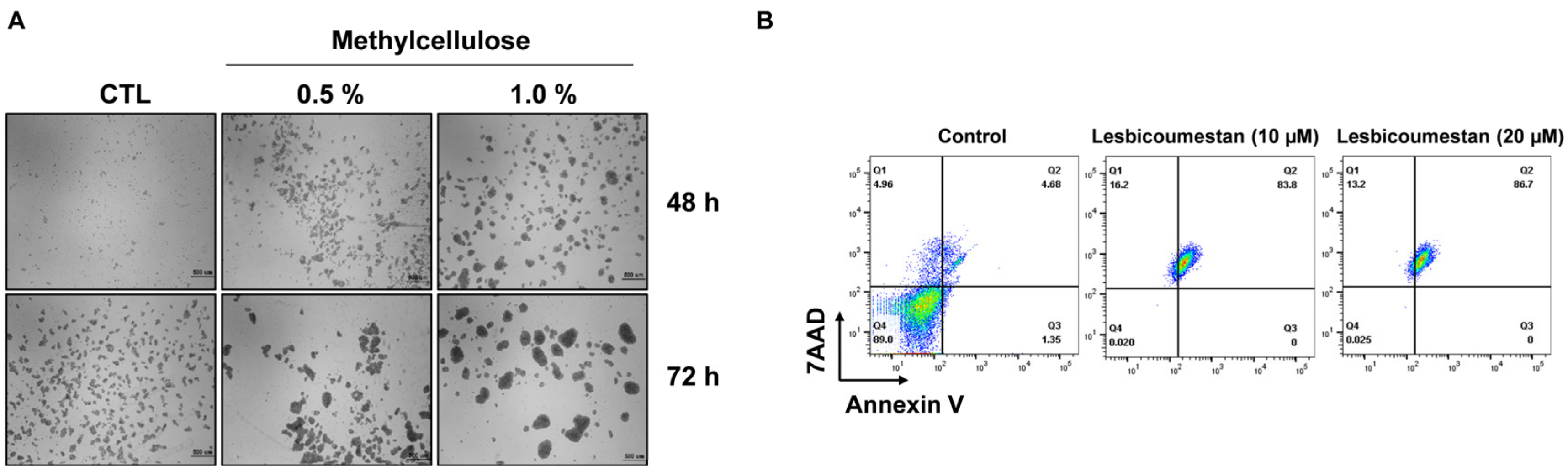
2.4. Sensitivity of Lesbicoumestan on 3D Jurkat Cell Models
3. Discussion
4. Materials and Methods
4.1. Plant Material, Extraction, and Isolation
4.2. Cell Viability Assay
4.3. Apoptosis Assay
4.4. Mitochondrial Membrane Potential (MMP) and ROS Assay
4.5. Western Blotting
4.6. RT-qPCR
4.7. Ubiquitination of MALT1
4.8. Confocal Imaging
4.9. Generation of Spheroids
4.10. Molecular Docking
4.11. Statistical Analysis
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Schlapbach, A.; Revesz, L.; Pissot Soldermann, C.; Zoller, T.; Régnier, C.H.; Bornancin, F.; Radimerski, T.; Blank, J.; Schuffenhauer, A.; Renatus, M.; et al. N-aryl-piperidine-4-carboxamides as a novel class of potent inhibitors of MALT1 proteolytic activity. Bioorg. Med. Chem. Lett. 2018, 28, 2153–2158. [Google Scholar] [CrossRef]
- Ruland, J.; Duncan, G.S.; Wakeham, A.; Mak, T.W. Differential requirement for Malt1 in T and B cell antigen receptor signaling. Immunity 2003, 19, 749–758. [Google Scholar] [CrossRef]
- Ishikawa, C.; Mori, N. MALT-1 as a novel therapeutic target for adult T-cell leukemia. Eur. J. Haematol. 2020, 105, 460–467. [Google Scholar] [CrossRef]
- Nagel, D.; Spranger, S.; Vincendeau, M.; Grau, M.; Raffegerst, S.; Kloo, B.; Hlahla, D.; Neuenschwander, M.; Peter von Kries, J.; Hadian, K.; et al. Pharmacologic inhibition of MALT1 protease by phenothiazines as a therapeutic approach for the treatment of aggressive ABC-DLBCL. Cancer Cell 2012, 22, 825–837. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.D.; Wilson BA, P.; Henrich, C.J.; Staudt, L.M.; Krumpe LR, H.; Smith, E.A.; King, J.; Wendt, K.L.; Stchigel, A.M.; Miller, A.N.; et al. Secondary Metabolites from the Fungus Dictyosporium sp. and Their MALT1 Inhibitory Activities. J. Nat. Prod. 2019, 82, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Do, M.H.; Lee, J.H.; Wahedi, H.M.; Pak, C.; Lee, C.H.; Yeo, E.J.; Lim, Y.; Ha, S.K.; Choi, I.; Kim, S.Y. Lespedeza bicolor ameliorates endothelial dysfunction induced by methylglyoxal glucotoxicity. Phytomedicine 2017, 36, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Hossaine, M.D.; Park, S.C. A potential anti-inflammation activity and depigmentation effect of Lespedeza bicolor extract and its fractions. Saudi J. Biol. Sci. 2016, 23, 9–14. [Google Scholar] [CrossRef]
- Thuy NT, T.; Lee, J.E.; Yoo, H.M.; Cho, N. Antiproliferative Pterocarpans and Coumestans from Lespedeza bicolor. J. Nat. Prod. 2019, 82, 3025–3032. [Google Scholar] [CrossRef]
- Jian, K.L.; Zhang, C.; Shang, Z.C.; Yang, L.; Kong, L.Y. Eucalrobusone C suppresses cell proliferation and induces ROS-dependent mitochondrial apoptosis via the p38 MAPK pathway in hepatocellular carcinoma cells. Phytomedicine 2017, 25, 71–82. [Google Scholar] [CrossRef]
- Saikia, S.; Bordoloi, M. Molecular Docking: Challenges, Advances and its Use in Drug Discovery Perspective. Curr. Drug Targets 2019, 20, 501–521. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Zhao, C.; Yao, R.; Sui, A.; Sun, L.; Liu, X.; Wu, S.; Su, Z.; Li, T.; Liu, S.; et al. 3D culture enhances chemoresistance of ALL Jurkat cell line by increasing DDR1 expression. Exp. Ther. Med. 2019, 17, 1593–1600. [Google Scholar] [CrossRef] [PubMed]
- Quancard, J.; Klein, T.; Fung, S.Y.; Renatus, M.; Hughes, N.; Israel, L.; Priatel, J.J.; Kang, S.; Blank, M.A.; Viner, R.I.; et al. An allosteric MALT1 inhibitor is a molecular corrector rescuing function in an immunodeficient patient. Nat. Chem. Biol. 2019, 15, 304–313. [Google Scholar] [CrossRef]
- Ruland, J.; Hartjes, L. CARD-BCL-10-MALT1 signalling in protective and pathological immunity. Nat. Rev. Immunol. 2019, 19, 118–134. [Google Scholar] [CrossRef]
- Emmerich, C.H.; Cohen, P. Optimising methods for the preservation, capture and identification of ubiquitin chains and ubiquitylated proteins by immunoblotting. Biochem. Biophys. Res. Commun. 2015, 466, 1–14. [Google Scholar] [CrossRef]
- Yu, J.W.; Jeffrey, P.D.; Ha, J.Y.; Yang, X.; Shi, Y. Crystal structure of the mucosa-associated lymphoid tissue lymphoma translocation 1 (MALT1) paracaspase region. Proc. Natl. Acad. Sci. USA 2011, 108, 21004–21009. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Burley, S.K.; Berman, H.M.; Bhikadiya, C.; Bi, C.; Chen, L.; Di Costanzo, L.; Christie, C.; Dalenberg, K.; Duarte, J.M.; Dutta, S.; et al. RCSB Protein Data Bank: Biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2018, 47, D464–D474. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Hughes, N.; Erbel, P.; Bornancin, F.; Wiesmann, C.; Schiering, N.; Villard, F.; Decock, A.; Rubi, B.; Melkko, S.; Spanka, C.; et al. Stabilizing Inactive Conformations of MALT1 as an Effective Approach to Inhibit Its Protease Activity. Adv. Ther. 2020, 3, 8. [Google Scholar] [CrossRef]
- Lim, S.M.; Jeong, Y.; Lee, S.; Im, H.; Tae, H.S.; Kim, B.G.; Park, H.D.; Park, J.; Hong, S. Identification of β-Lapachone Analogs as Novel MALT1 Inhibitors to Treat an Aggressive Subtype of Diffuse Large B-Cell Lymphoma. J. Med. Chem. 2015, 58, 8491–8502. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Lee, J.E.; Kim, S.; Kang, D.; Yoo, H.M. Evaluating Cell Death Using Cell-Free Supernatant of Probiotics in Three-Dimensional Spheroid Cultures of Colorectal Cancer Cells. J. Vis. Exp. 2020, 160, e61285. [Google Scholar]
- Lee, J.E.; Lee, J.; Kim, J.H.; Cho, N.; Lee, S.H.; Park, S.B.; Koh, B.; Kang, D.; Kim, S.; Yoo, H.M. Characterization of the Anti-Cancer Activity of the Probiotic Bacterium Lactobacillus fermentum Using 2D vs. 3D Culture in Colorectal Cancer Cells. Biomolecules 2019, 9, 557. [Google Scholar] [CrossRef] [PubMed]
- Horvath, P.; Aulner, N.; Bickle, M.; Davies, A.M.; Nery, E.D.; Ebner, D.; Montoya, M.C.; Östling, P.; Pietiäinen, V.; Price, L.S.; et al. Screening out irrelevant cell-based models of disease. Nat. Rev. Drug Discov. 2016, 15, 751–769. [Google Scholar] [CrossRef]
- Nunes, A.S.; Barros, A.S.; Costa, E.C.; Moreira, A.F.; Correia, I.J. 3D tumor spheroids as in vitro models to mimic in vivo human solid tumors resistance to therapeutic drugs. Biotechnol. Bioeng. 2019, 116, 206–226. [Google Scholar] [CrossRef]
- Shamir, E.R.; Ewald, A.J. Three-dimensional organotypic culture: Experimental models of mammalian biology and disease. Nat. Rev. Mol. Cell Biol. 2014, 15, 647–664. [Google Scholar] [CrossRef]
- Hirose, M. Biology and modulation of multidrug resistance (MDR) in hematological malignancies. Int. J. Hematol. 2002, 76, 206–211. [Google Scholar] [CrossRef]
- Zhan, F.; Zangari, M.; Qiu, L. Drug resistance in hematologic malignancies: Induction mechanisms, genetics, and therapeutics. Biomed. Res. Int. 2015, 2015, 384575. [Google Scholar] [CrossRef]
- Lee, J.E.; Thuy NT, T.; Lee, J.; Cho, N.; Yoo, H.M. Platyphylloside Isolated from Betula platyphylla is Antiproliferative and Induces Apoptosis in Colon Cancer and Leukemic Cells. Molecules 2019, 24, 2960. [Google Scholar] [CrossRef]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995, 8, 127–134. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.-E.; Bo, F.; Thuy, N.T.T.; Hong, J.; Lee, J.S.; Cho, N.; Yoo, H.M. Anticancer Activity of Lesbicoumestan in Jurkat Cells via Inhibition of Oxidative Stress-Mediated Apoptosis and MALT1 Protease. Molecules 2021, 26, 185. https://doi.org/10.3390/molecules26010185
Lee J-E, Bo F, Thuy NTT, Hong J, Lee JS, Cho N, Yoo HM. Anticancer Activity of Lesbicoumestan in Jurkat Cells via Inhibition of Oxidative Stress-Mediated Apoptosis and MALT1 Protease. Molecules. 2021; 26(1):185. https://doi.org/10.3390/molecules26010185
Chicago/Turabian StyleLee, Joo-Eun, Fang Bo, Nguyen Thi Thanh Thuy, Jaewoo Hong, Ji Shin Lee, Namki Cho, and Hee Min Yoo. 2021. "Anticancer Activity of Lesbicoumestan in Jurkat Cells via Inhibition of Oxidative Stress-Mediated Apoptosis and MALT1 Protease" Molecules 26, no. 1: 185. https://doi.org/10.3390/molecules26010185
APA StyleLee, J.-E., Bo, F., Thuy, N. T. T., Hong, J., Lee, J. S., Cho, N., & Yoo, H. M. (2021). Anticancer Activity of Lesbicoumestan in Jurkat Cells via Inhibition of Oxidative Stress-Mediated Apoptosis and MALT1 Protease. Molecules, 26(1), 185. https://doi.org/10.3390/molecules26010185






