Palmitoylation of Metazoan Carotenoid Oxygenases
Abstract
1. Introduction
2. Results
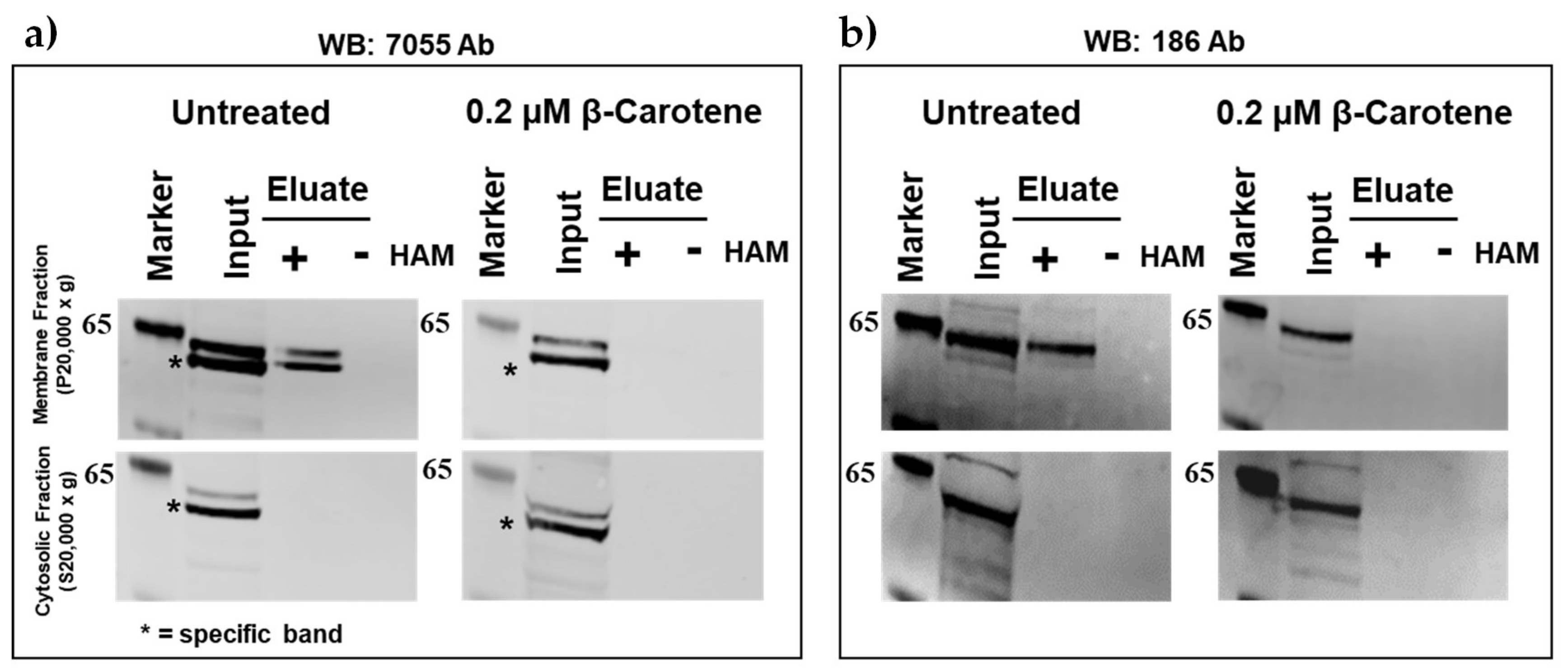
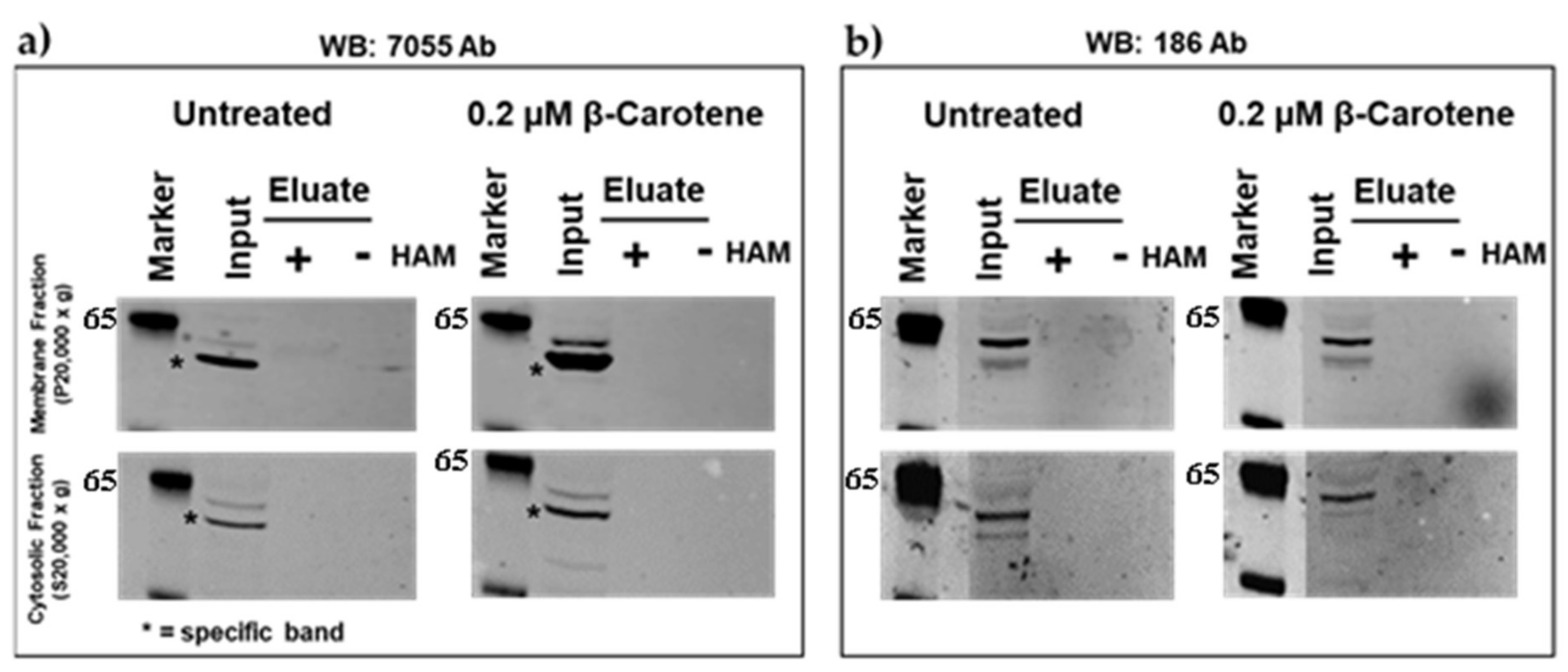
2.1. Palmitoylation of Mouse BCO2 (mBCO2) (without Tag and with Tag) in the Absence and Presence of Its Substrate
2.2. Mutations in C111 Change Palmitoylation Status of the Protein
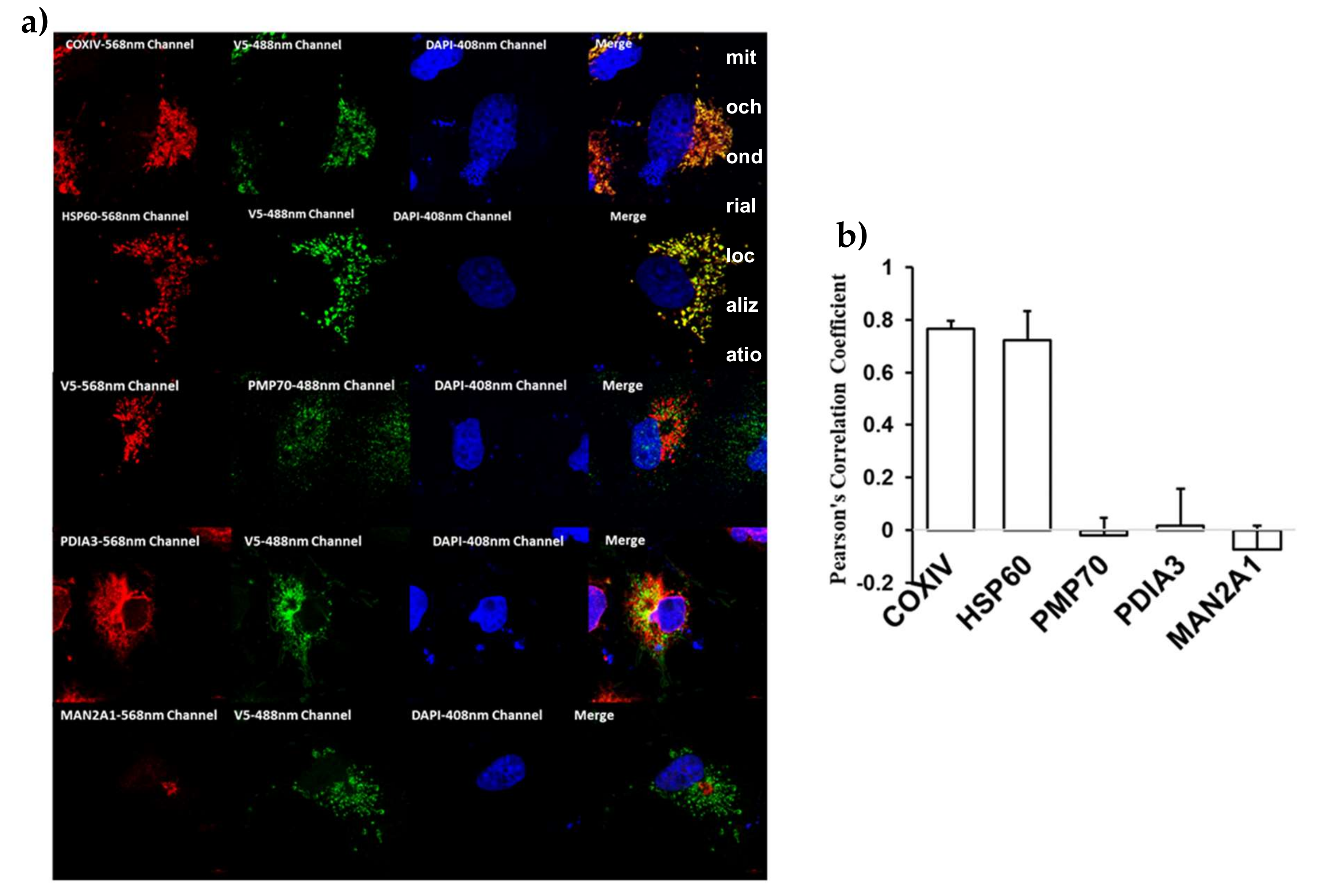
2.3. Cellular Localization of Mouse BCO2 with and without Substrate
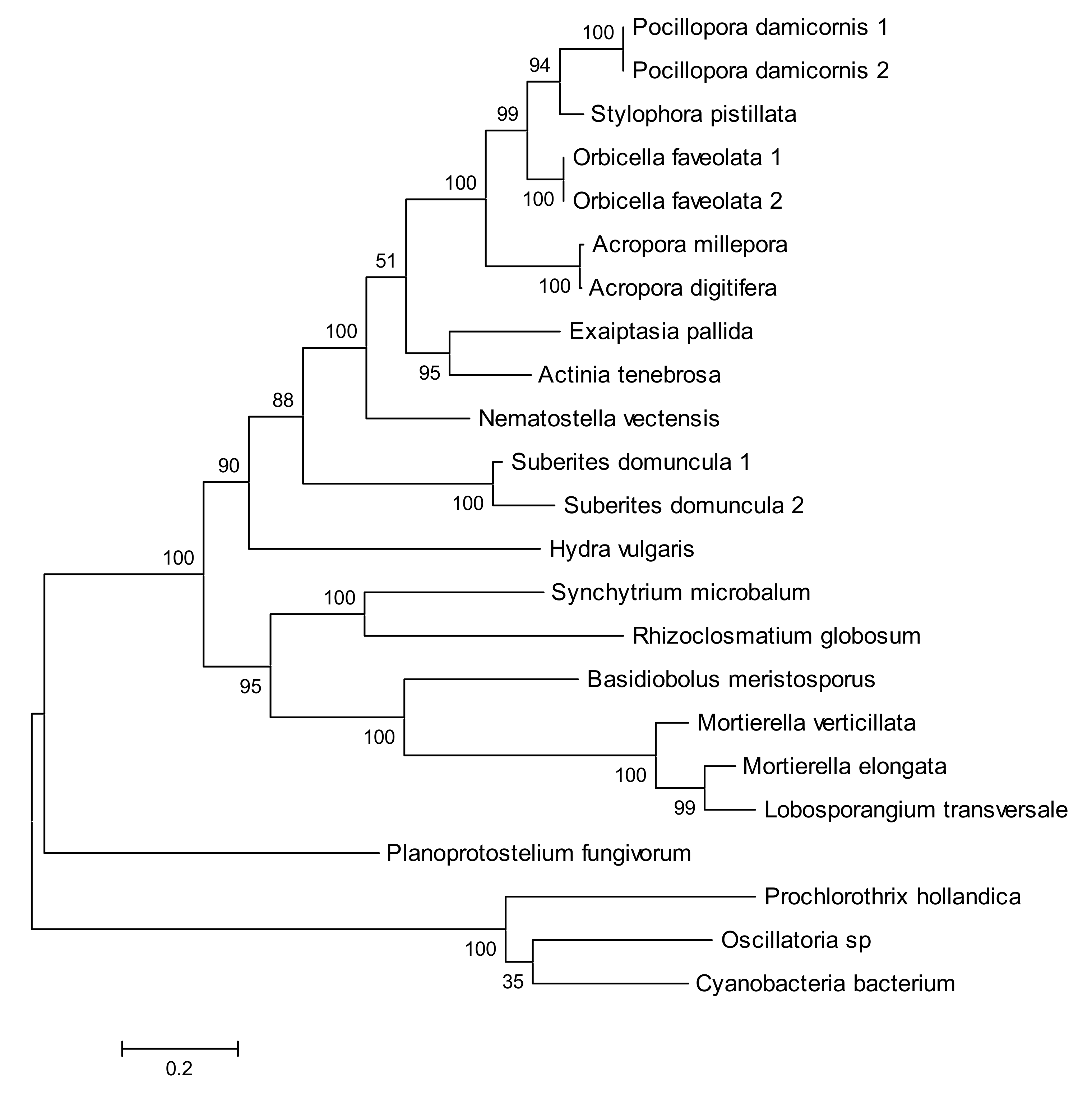
2.4. Phylogenetic Analysis of New Metazoan Carotenoid Oxygenase Subfamily without PDPCK Motif (ACOL)
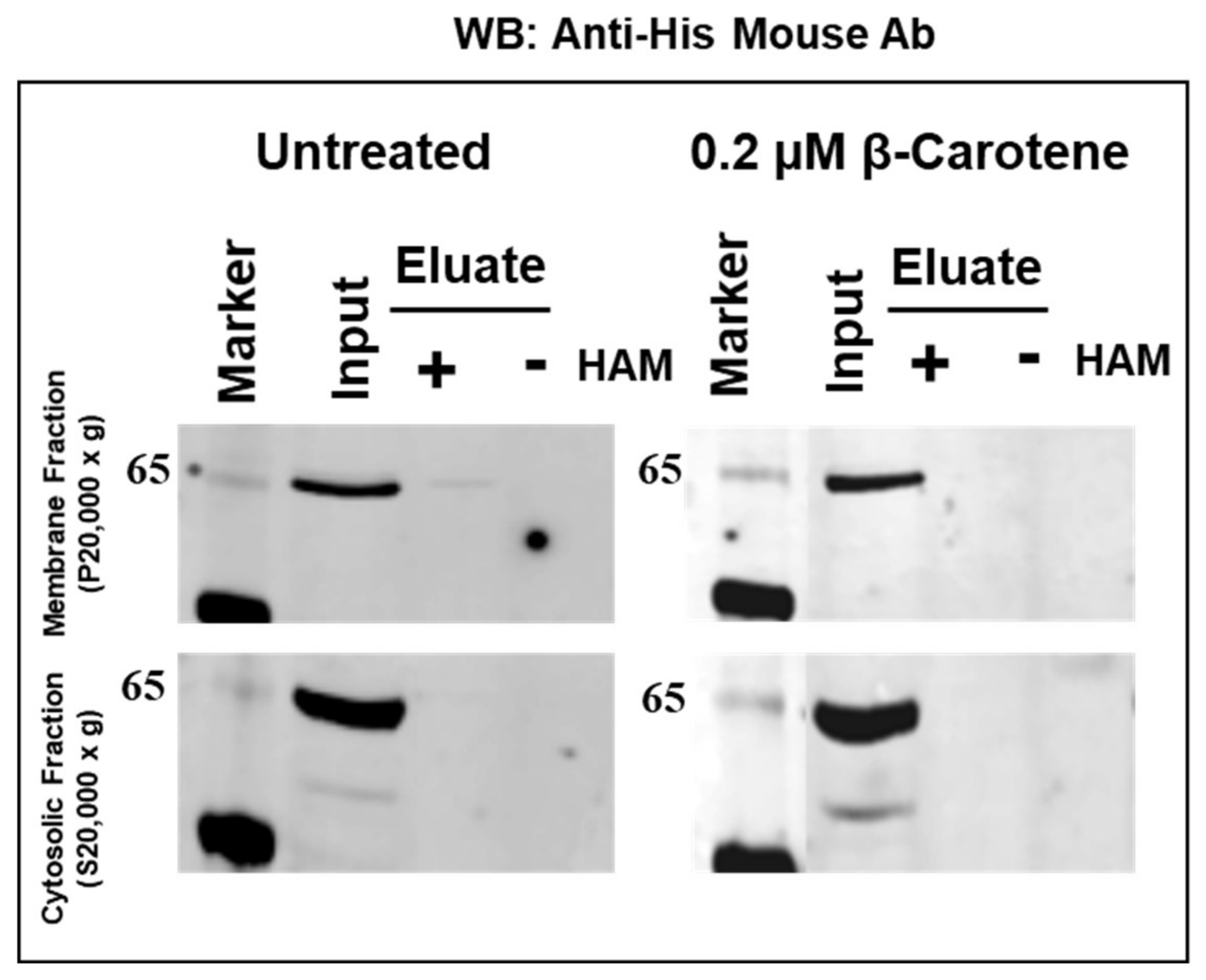
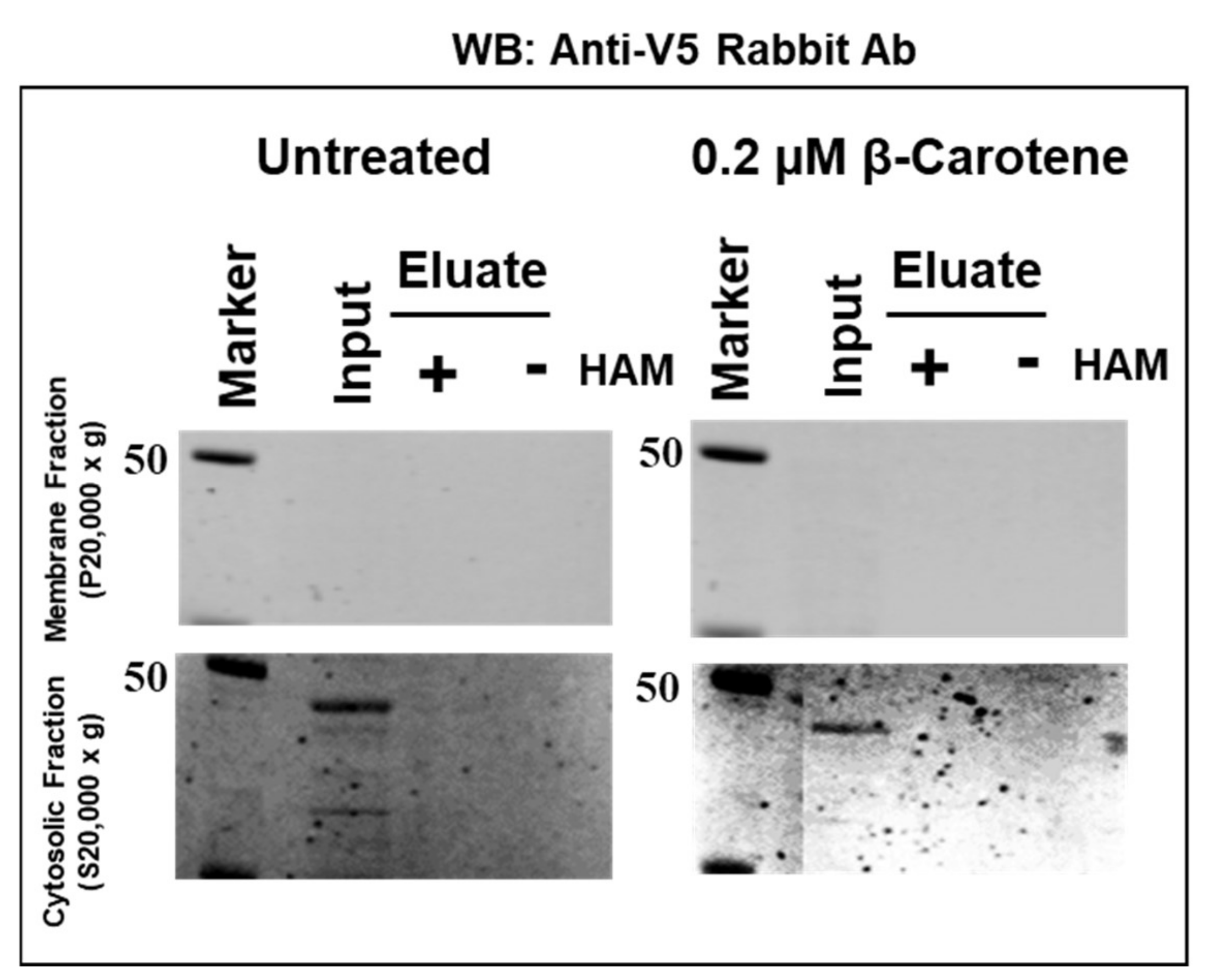
2.5. Absence of Palmitoylation in Carotenoid Oxygenase Metazoan Family Members without PDPCK Motif
3. Discussion
4. Materials and Methods
4.1. Generation of Expression Vectors and Site-Directed Mutagenesis
4.2. Cell Culture
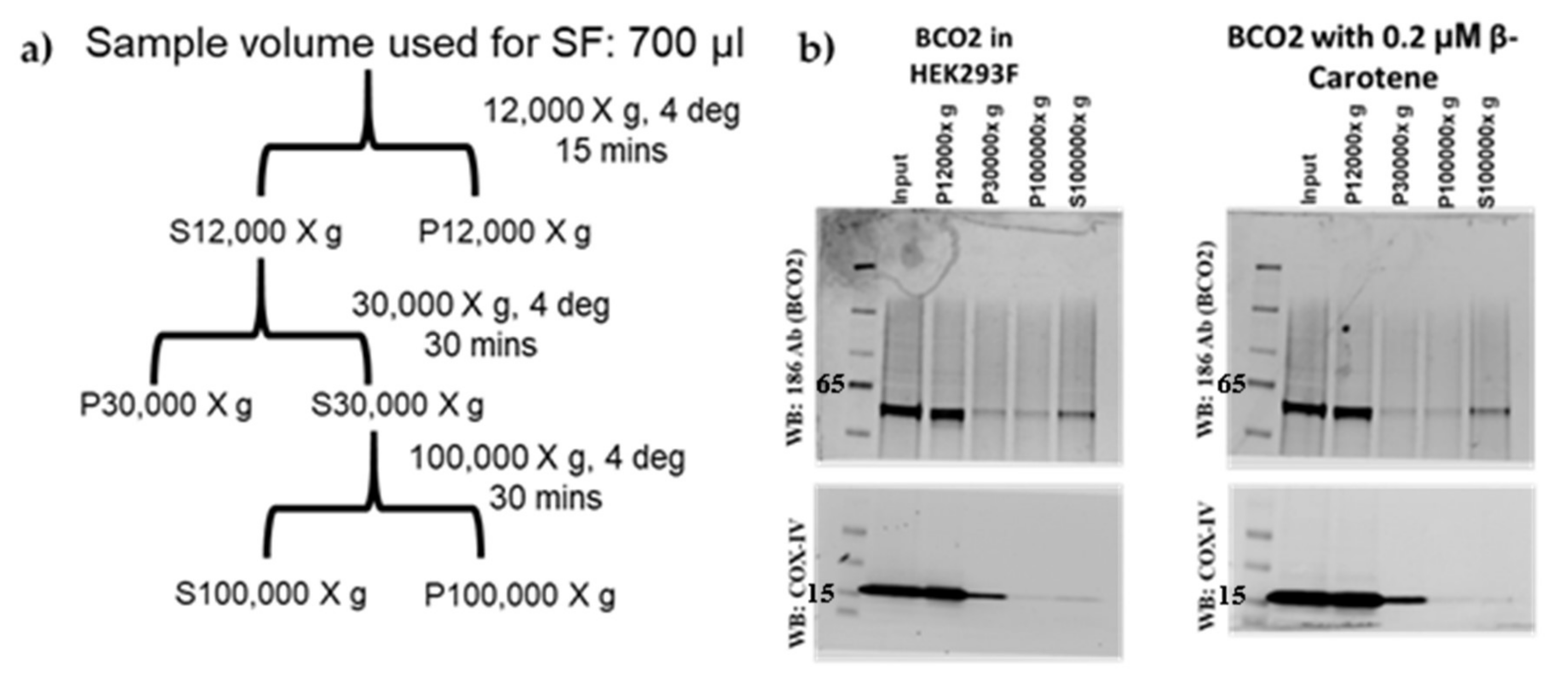
4.3. Sucrose Sub-Cellular Fractionation HEK293F-Overexpressed Mouse BCO2
4.4. Antibodies
4.5. Determination of BCO2 Palmitoylation by Acyl-RAC Method
4.6. Immunofluorescence, Confocal Microscopy and Analysis of BCO2 Colocalization in Different Subcellular Organelles (ER, Golgi, Mitochondria, and Peroxisomes)
4.7. Phylogenetic Analysis of Functionality in Carotenoid Oxygenase Family
- 1)
- We reconstructed an ML tree for protein sequences (Figure 6 and Supplementary Figure S4) and reconstructed ancestral sequences using the ML method.
- 2)
- 3)
- We counted the number of amino acid replacements (R1), we used sites where ancestral states were predicted with the probability > 0.5 to infer high-confidence ancestral states. We also estimated the difference for each amino acid replacement (using BDM values) and summed BDM values for all R1 (BDM1).
- 4)
- We estimated BDM and R variables for three or four neighboring branches (depending on the local tree topology) and sum results for those branches producing R2 and BDM2. We avoided terminal branches because all tested branches (leading to RPE65, BCOL, and ACOL protein families) were internal ones.
- 5)
- We compared BDM1, R1, BDM2 and R2 using the one-tail Fisher exact test that produced a probability value P. We did not use any bulk analysis of all branches thus we did not apply the Bonferroni correction for multiple tests. However, obtained P values should be treated with caution anyway. We performed two control experiments using RPE65 and BCOL protein families (Figure 6)
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- McNulty, H.P.; Byun, J.; Lockwood, S.F.; Jacob, R.F.; Mason, R.P. Differential effects of carotenoids on lipid peroxidation due to membrane interactions: X-ray diffraction analysis. Biochim. Biophys. Acta 2007, 1768, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Miller, N.J.; Sampson, J.; Candeias, L.P.; Bramley, P.M.; Rice-Evans, C.A. Antioxidant activities of carotenes and xanthophylls. FEBS Lett. 1996, 384, 240–242. [Google Scholar] [CrossRef]
- Stahl, W.; Sies, H. Antioxidant activity of carotenoids. Mol. Aspects Med. 2003, 24, 345–351. [Google Scholar] [CrossRef]
- Hammond, B.R.; Fletcher, L.M.; Roos, F.; Wittwer, J.; Schalch, W. A double-blind, placebo-controlled study on the effects of lutein and zeaxanthin on photostress recovery, glare disability, and chromatic contrast. Invest. Ophthalmol. Vis. Sci. 2014, 55, 8583–8589. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.C.; Schwartz, S.H.; Zeevaart, J.A.; McCarty, D.R. Genetic control of abscisic acid biosynthesis in maize. Proc. Natl. Acad. Sci. USA 1997, 94, 12235–12240. [Google Scholar] [CrossRef] [PubMed]
- Zeevaart, J.A.; Heath, T.G.; Gage, D.A. Evidence for a universal pathway of abscisic Acid biosynthesis in higher plants from o incorporation patterns. Plant. Physiol. 1989, 91, 1594–1601. [Google Scholar] [CrossRef]
- Alder, A.; Jamil, M.; Marzorati, M.; Bruno, M.; Vermathen, M.; Bigler, P.; Ghisla, S.; Bouwmeester, H.; Beyer, P.; Al-Babili, S. The path from beta-carotene to carlactone, a strigolactone-like plant hormone. Science 2012, 335, 1348–1351. [Google Scholar] [CrossRef]
- Wyss, A. Carotene oxygenases: A new family of double bond cleavage enzymes. J. Nutr. 2004, 134, 246S–250S. [Google Scholar] [CrossRef]
- Gu, G.; Yang, J.; Mitchell, K.A.; O’Tousa, J.E. Drosophila ninaB and ninaD act outside of retina to produce rhodopsin chromophore. J. Biol. Chem. 2004, 279, 18608–18613. [Google Scholar] [CrossRef] [PubMed]
- Oberhauser, V.; Voolstra, O.; Bangert, A.; von Lintig, J.; Vogt, K. NinaB combines carotenoid oxygenase and retinoid isomerase activity in a single polypeptide. Proc. Natl. Acad. Sci. USA 2008, 105, 19000–19005. [Google Scholar] [CrossRef]
- Eroglu, A.; Hruszkewycz, D.P.; Curley, R.W., Jr.; Harrison, E.H. The eccentric cleavage product of beta-carotene, beta-apo-13-carotenone, functions as an antagonist of RXRalpha. Arch. Biochem. Biophys. 2010, 504, 11–16. [Google Scholar] [CrossRef]
- Poliakov, E.; Soucy, J.; Gentleman, S.; Rogozin, I.B.; Redmond, T.M. Phylogenetic analysis of the metazoan carotenoid oxygenase superfamily: A new ancestral gene assemblage of BCO-like (BCOL) proteins. Sci. Rep. 2017, 7, 13192. [Google Scholar] [CrossRef] [PubMed]
- Kiser, P.D.; Golczak, M.; Lodowski, D.T.; Chance, M.R.; Palczewski, K. Crystal structure of native RPE65, the retinoid isomerase of the visual cycle. Proc. Natl. Acad. Sci. USA 2009, 106, 17325–17330. [Google Scholar] [CrossRef] [PubMed]
- Kloer, D.P.; Ruch, S.; Al-Babili, S.; Beyer, P.; Schulz, G.E. The structure of a retinal-forming carotenoid oxygenase. Science 2005, 308, 267–269. [Google Scholar] [CrossRef] [PubMed]
- McAndrew, R.P.; Sathitsuksanoh, N.; Mbughuni, M.M.; Heins, R.A.; Pereira, J.H.; George, A.; Sale, K.L.; Fox, B.G.; Simmons, B.A.; Adams, P.D. Structure and mechanism of NOV1, a resveratrol-cleaving dioxygenase. Proc. Natl. Acad. Sci. USA 2016, 113, 14324–14329. [Google Scholar] [CrossRef] [PubMed]
- Messing, S.A.; Gabelli, S.B.; Echeverria, I.; Vogel, J.T.; Guan, J.C.; Tan, B.C.; Klee, H.J.; McCarty, D.R.; Amzel, L.M. Structural insights into maize viviparous14, a key enzyme in the biosynthesis of the phytohormone abscisic acid. Plant. Cell 2010, 22, 2970–2980. [Google Scholar] [CrossRef]
- Poliakov, E.; Gentleman, S.; Cunningham, F.X., Jr.; Miller-Ihli, N.J.; Redmond, T.M. Key role of conserved histidines in recombinant mouse beta-carotene 15,15’-monooxygenase-1 activity. J. Biol. Chem. 2005, 280, 29217–29223. [Google Scholar] [CrossRef]
- Redmond, T.M.; Poliakov, E.; Yu, S.; Tsai, J.Y.; Lu, Z.; Gentleman, S. Mutation of key residues of RPE65 abolishes its enzymatic role as isomerohydrolase in the visual cycle. Proc. Natl. Acad. Sci. USA 2005, 102, 13658–13663. [Google Scholar] [CrossRef]
- Poliakov, E.; Uppal, S.; Rogozin, I.B.; Gentleman, S.; Redmond, T.M. Evolutionary aspects and enzymology of metazoan carotenoid cleavage oxygenases. Biochim. Biophys. Acta Mol. Cell. Biol. Lipids 2020, 158665. [Google Scholar] [CrossRef]
- Jin, M.; Li, S.; Moghrabi, W.N.; Sun, H.; Travis, G.H. Rpe65 is the retinoid isomerase in bovine retinal pigment epithelium. Cell 2005, 122, 449–459. [Google Scholar] [CrossRef]
- Moiseyev, G.; Chen, Y.; Takahashi, Y.; Wu, B.X.; Ma, J.X. RPE65 is the isomerohydrolase in the retinoid visual cycle. Proc. Natl. Acad Sci. USA 2005, 102, 12413–12418. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Moiseyev, G.; Ablonczy, Z.; Chen, Y.; Crouch, R.K.; Ma, J.X. Identification of a novel palmitylation site essential for membrane association and isomerohydrolase activity of RPE65. J. Biol. Chem. 2009, 284, 3211–3218. [Google Scholar] [CrossRef]
- Uppal, S.; Liu, T.; Poliakov, E.; Gentleman, S.; Redmond, T.M. The dual roles of RPE65 S-palmitoylation in membrane association and visual cycle function. Sci. Rep. 2019, 9, 5218. [Google Scholar] [CrossRef]
- Palczewski, G.; Amengual, J.; Hoppel, C.L.; von Lintig, J. Evidence for compartmentalization of mammalian carotenoid metabolism. FASEB J. 2014, 28, 4457–4469. [Google Scholar] [CrossRef] [PubMed]
- Raghuvanshi, S.; Reed, V.; Blaner, W.S.; Harrison, E.H. Cellular localization of beta-carotene 15,15’ oxygenase-1 (BCO1) and beta-carotene 9’,10’ oxygenase-2 (BCO2) in rat liver and intestine. Arch. Biochem. Biophys. 2015, 572, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Amengual, J.; Lobo, G.P.; Golczak, M.; Li, H.N.; Klimova, T.; Hoppel, C.L.; Wyss, A.; Palczewski, K.; von Lintig, J. A mitochondrial enzyme degrades carotenoids and protects against oxidative stress. FASEB J. 2011, 25, 948–959. [Google Scholar] [CrossRef] [PubMed]
- Poliakov, E.; Gubin, A.N.; Stearn, O.; Li, Y.; Campos, M.M.; Gentleman, S.; Rogozin, I.B.; Redmond, T.M. Origin and evolution of retinoid isomerization machinery in vertebrate visual cycle: Hint from jawless vertebrates. PLoS One 2012, 7, e49975. [Google Scholar] [CrossRef]
- Muller, W.E.; Wang, X.; Binder, M.; von Lintig, J.; Wiens, M.; Schroder, H.C. Differential expression of the demosponge (Suberites domuncula) carotenoid oxygenases in response to light: Protection mechanism against the self-produced toxic protein (Suberitine). Mar. Drugs 2012, 10, 177–199. [Google Scholar] [CrossRef]
- Chernikova, D.; Motamedi, S.; Csuros, M.; Koonin, E.V.; Rogozin, I.B. A late origin of the extant eukaryotic diversity: Divergence time estimates using rare genomic changes. Biol. Direct 2011, 6, 26. [Google Scholar] [CrossRef]
- Torres-Cortes, G.; Ghignone, S.; Bonfante, P.; Schussler, A. Mosaic genome of endobacteria in arbuscular mycorrhizal fungi: Transkingdom gene transfer in an ancient mycoplasma-fungus association. Proc. Natl. Acad. Sci. USA 2015, 112, 7785–7790. [Google Scholar] [CrossRef]
- Jensen, L.; Grant, J.R.; Laughinghouse, H.D.T.; Katz, L.A. Assessing the effects of a sequestered germline on interdomain lateral gene transfer in Metazoa. Evolution 2016, 70, 1322–1333. [Google Scholar] [CrossRef] [PubMed]
- Steele, R.E.; David, C.N.; Technau, U. A genomic view of 500 million years of cnidarian evolution. Trends Genet. 2011, 27, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Aicart-Ramos, C.; Valero, R.A.; Rodriguez-Crespo, I. Protein palmitoylation and subcellular trafficking. Biochim. Biophys. Acta 2011, 1808, 2981–2994. [Google Scholar] [CrossRef] [PubMed]
- Chamberlain, L.H.; Shipston, M.J. The physiology of protein S-acylation. Physiol. Rev. 2015, 95, 341–376. [Google Scholar] [CrossRef]
- Prakash, P.; Jackson, C.L.; Gerber, L.E. Subcellular accumulation of beta-carotene and retinoids in growth-inhibited NCI-H69 small cell lung cancer cells. Nutr. Cancer 1999, 34, 76–82. [Google Scholar] [CrossRef]
- Forrester, M.T.; Hess, D.T.; Thompson, J.W.; Hultman, R.; Moseley, M.A.; Stamler, J.S.; Casey, P.J. Site-specific analysis of protein S-acylation by resin-assisted capture. J. Lipid Res. 2011, 52, 393–398. [Google Scholar] [CrossRef]
- Manders, E.M.M.; Verbeek, F.J.; Aten, J.A. Measurement of co-localization of objects in dual-colour confocal images. J. Microsc. 1993, 169, 375–382. [Google Scholar] [CrossRef]
- Kumar, S.; Nei, M.; Dudley, J.; Tamura, K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 2008, 9, 299–306. [Google Scholar] [CrossRef]
- Huzurbazar, S.; Kolesov, G.; Massey, S.E.; Harris, K.C.; Churbanov, A.; Liberles, D.A. Lineage-specific differences in the amino acid substitution process. J. Mol. Biol. 2010, 396, 1410–1421. [Google Scholar] [CrossRef][Green Version]
- Wernegreen, J.J. Reduced selective constraint in endosymbionts: Elevation in radical amino acid replacements occurs genome-wide. PLoS ONE 2011, 6, e28905. [Google Scholar] [CrossRef]
- Zhang, J. Rates of conservative and radical nonsynonymous nucleotide substitutions in mammalian nuclear genes. J. Mol. Evol. 2000, 50, 56–68. [Google Scholar] [CrossRef] [PubMed]
- Dagan, T.; Talmor, Y.; Graur, D. Ratios of radical to conservative amino acid replacement are affected by mutational and compositional factors and may not be indicative of positive Darwinian selection. Mol. Biol. Evol. 2002, 19, 1022–1025. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Smith, N.G. Are radical and conservative substitution rates useful statistics in molecular evolution? J. Mol. Evol. 2003, 57, 467–478. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Genetic constructs are available from the authors according to NIH policy. |


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uppal, S.; Rogozin, I.B.; Redmond, T.M.; Poliakov, E. Palmitoylation of Metazoan Carotenoid Oxygenases. Molecules 2020, 25, 1942. https://doi.org/10.3390/molecules25081942
Uppal S, Rogozin IB, Redmond TM, Poliakov E. Palmitoylation of Metazoan Carotenoid Oxygenases. Molecules. 2020; 25(8):1942. https://doi.org/10.3390/molecules25081942
Chicago/Turabian StyleUppal, Sheetal, Igor B. Rogozin, T.Michael Redmond, and Eugenia Poliakov. 2020. "Palmitoylation of Metazoan Carotenoid Oxygenases" Molecules 25, no. 8: 1942. https://doi.org/10.3390/molecules25081942
APA StyleUppal, S., Rogozin, I. B., Redmond, T. M., & Poliakov, E. (2020). Palmitoylation of Metazoan Carotenoid Oxygenases. Molecules, 25(8), 1942. https://doi.org/10.3390/molecules25081942







