PF74 and Its Novel Derivatives Stabilize Hexameric Lattice of HIV-1 Mature-Like Particles
Abstract
:1. Introduction
2. Results and Discussion
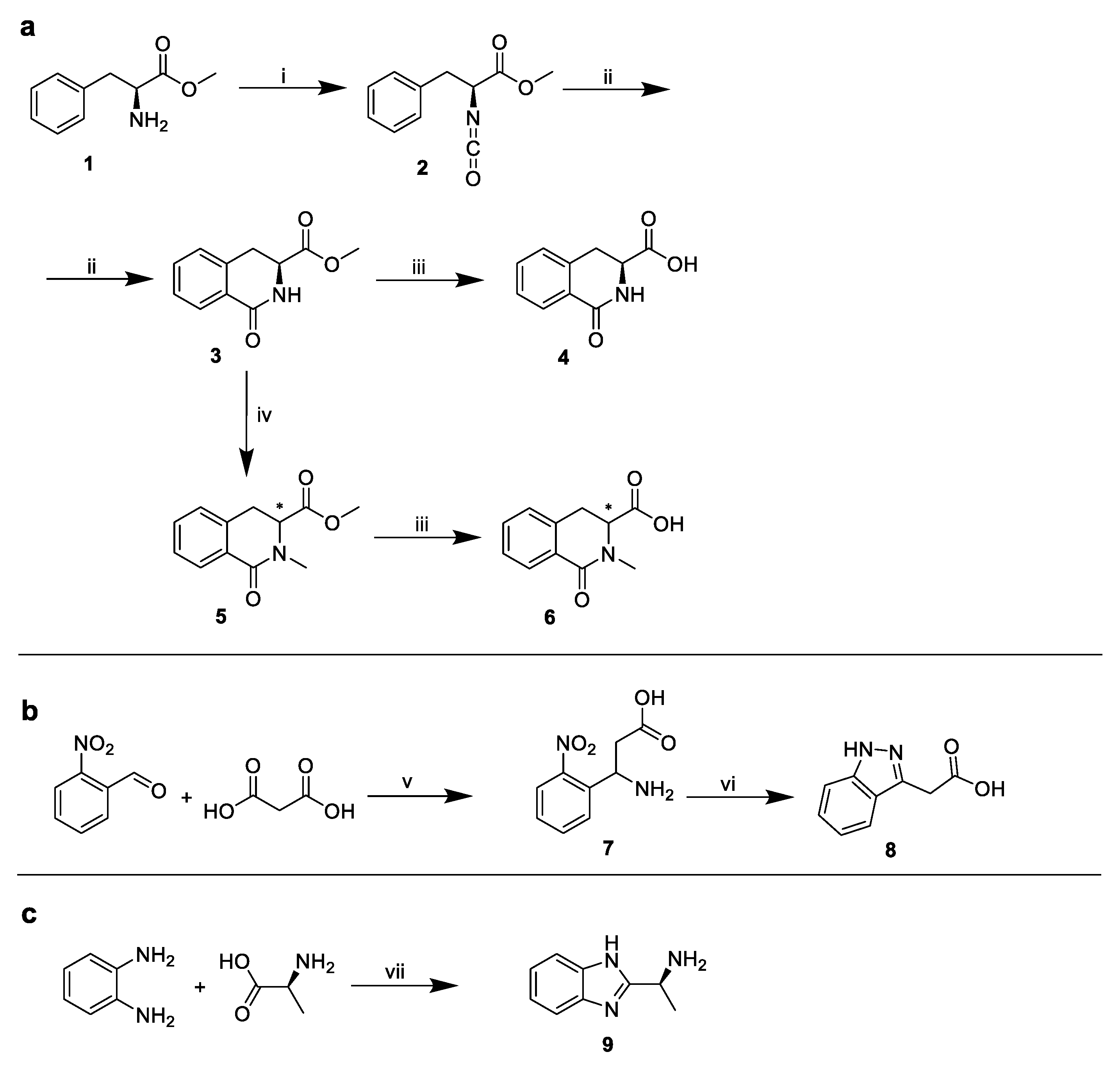
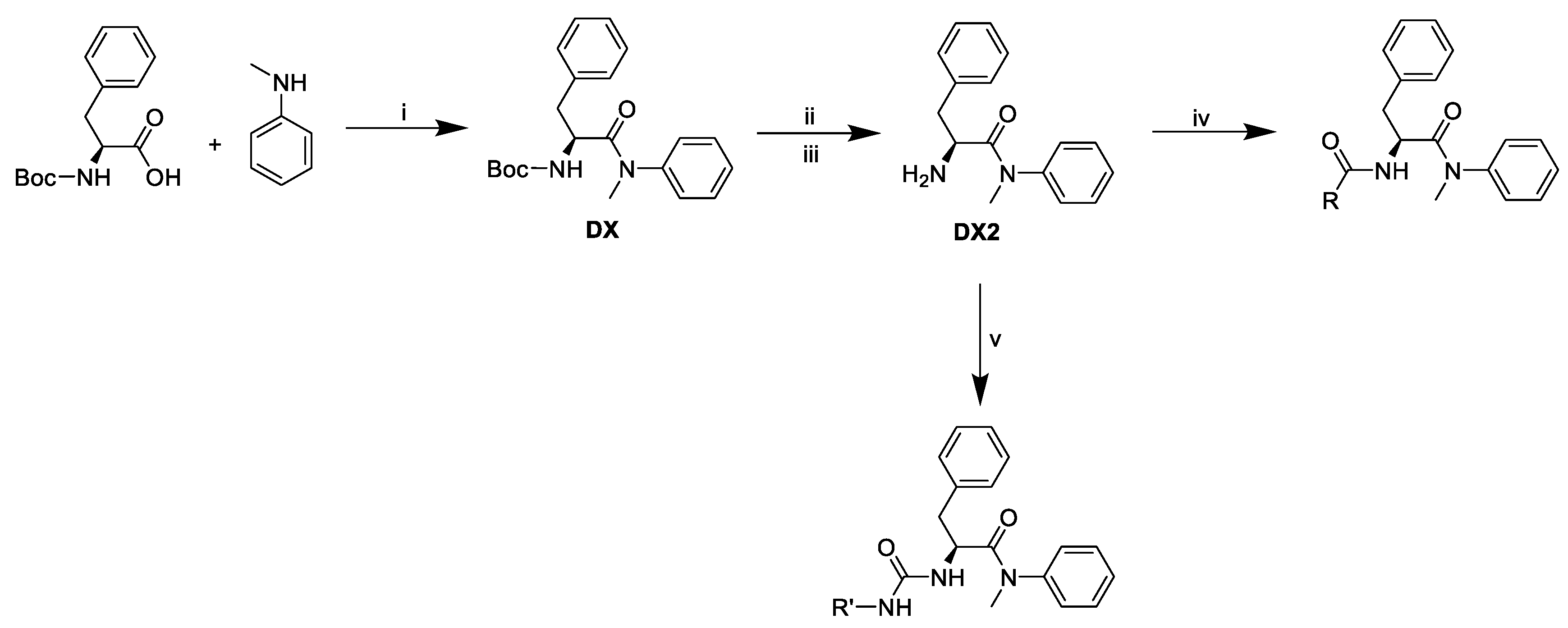
2.1. The Design and Synthesis of PF74 Derivatives
2.2. Analysis of PF74 Derivatives Activity Using in Vitro Protein-Based Methods
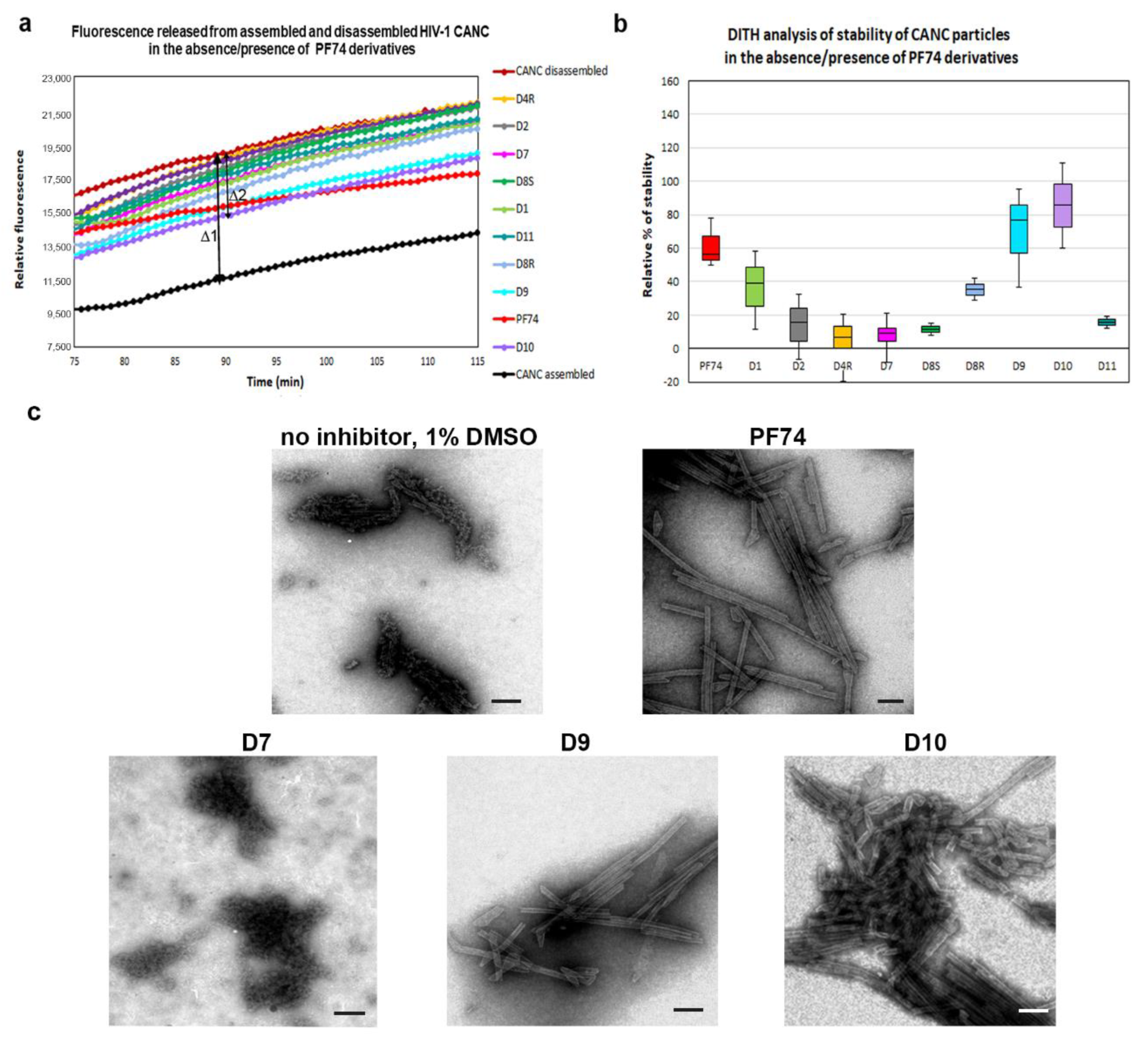
2.2.1. Fast Assembly Inhibitor Test for HIV (FAITH) Analysis
2.2.2. DITH Analysis
2.3. Analysis of PF74 Derivatives Using Cell-Based Methods
2.3.1. Effect of PF74 Derivatives on HIV-1 Infectivity
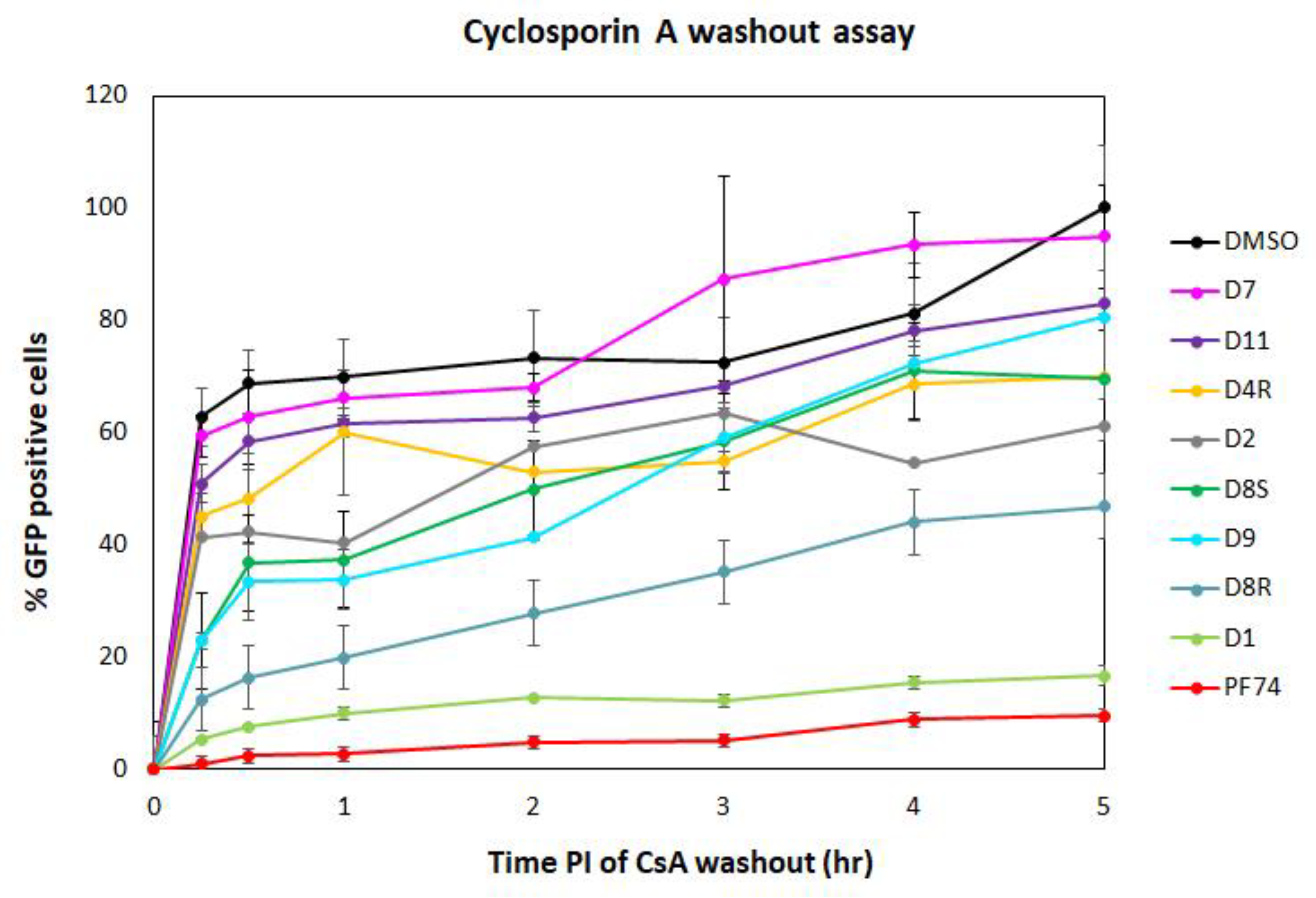
2.3.2. Cyclosporin A (CsA)-Washout Assay
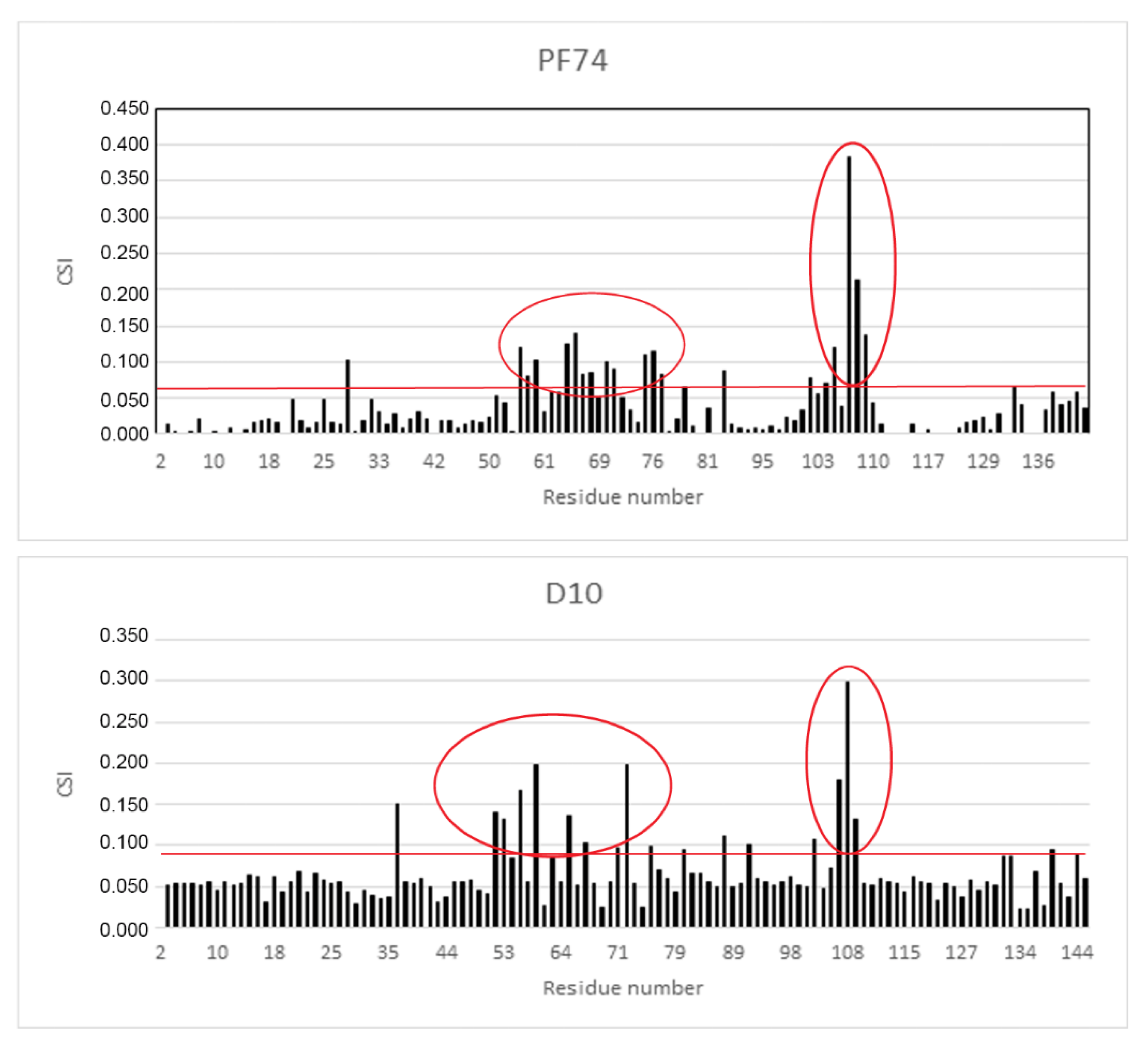
2.4. NMR Analysis of the Binding Mode of D10
3. Materials and Methods
3.1. Expression Vector Preparation
3.2. Expression and Purification of HIV-1 CA-Derived Proteins
3.3. Fast Assembly Inhibitor Test for HIV (FAITH)
3.4. Stabilization Fast Assembly Inhibitor Test for HIV (DITH)
3.5. Determination of Cytotoxicity Using Resazurin Assay
3.6. Production of VSV-G Pseudotyped HIV-1 Particles
3.7. Enzyme-Linked Immunosorbent Assay (ELISA)
3.8. Single-Round Infectivity Assay
3.9. Flow Cytometry
3.10. Synthesis of PF74 Derivatives
3.10.1. Methyl (S)-2-isocyanato-3-phenylpropanoate (1)
3.10.2. Methyl (S)-1-oxo-1,2,3,4-tetrahydroisoquinoline-3-carboxylate (2)
3.10.3. Triethylammonium Salt of (S)-1-oxo-1,2,3,4-tetrahydroisoquinoline-3-carboxylic Acid (3)
3.10.4. Methyl 2-methyl-1-oxo-1,2,3,4-tetrahydroisoquinoline-3-carboxylate (4)
3.10.5. Triethylammonium Salt of 2-methyl-1-oxo-1,2,3,4-tetrahydroisoquinoline-3-carboxylic Acid (5)
3.10.6. 3-Amino-3-(2-nitrophenyl)propanoic Acid (6)
3.10.7. 2-(1. H-indazol-3-yl)acetic Acid (7)
3.10.8. (S)-1-(1H-benzo[d]imidazol-2-yl)ethan-1-amine (8)
3.10.9. Tert-butyl (S)-(1-(methyl(phenyl)amino)-1-oxo-3-phenylpropan-2-yl)carbamate (DX).
3.10.10. (S)-2-amino-N-methyl-N,3-diphenylpropanamide (DX2)
3.10.11. (S)-2-(2-(1H-indol-3-yl)acetamido)-N-methyl-N,3-diphenylpropanamide (D1)
3.10.12. (S)-2-(2-(1-(4-chlorobenzoyl)-5-methoxy-2-methyl-1H-indol-3-yl)acetamido)-N-methyl--N,3-diphenylpropanamide (D2)
3.10.13. (S)-2-((R)-2-(4-isobutylphenyl)propanamido)-N-methyl-N,3-diphenylpropanamide (D4(R))
3.10.14. (S)-N-((S)-1-(methyl(phenyl)amino)-1-oxo-3-phenylpropan-2-yl)-1-oxo-1,2,3,4--tetrahydroisoquinoline-3-carboxamide (D7)
3.10.15. (R)-2-methyl-N-((S)-1-(methyl(phenyl)amino)-1-oxo-3-phenylpropan-2-yl)-1-oxo--1,2,3,4-tetrahydroisoquinoline-3-carboxamide (D8(R))
3.10.16. (S)-2-methyl-N-((S)-1-(methyl(phenyl)amino)-1-oxo-3-phenylpropan-2-yl)-1-oxo--1,2,3,4-tetrahydroisoquinoline-3-carboxamide (D8(S))
3.10.17. (S)-2-(2-(1H-indazol-3-yl)acetamido)-N-methyl-N,3-diphenylpropanamide (D9)
3.10.18. (S)-2-(3-((1H-benzo[d]imidazol-2-yl)methyl)ureido)-N-methyl-N,3-diphenylpropanamide (D10)(General procedure of urea-linked derivatives preparation)
3.10.19. (S)-2-(3-((S)-1-(1H-benzo[d]imidazol-2-yl)ethyl)ureido)-N-methyl-N,3--diphenylpropanamide (D11)
3.11. The Cyclosporin A (CsA) Washout Assay
3.12. NMR Titration
Author Contributions
Funding
Conflicts of Interest
References
- Qu, K.; Glass, B.; Dolezal, M.; Schur, F.K.M.; Murciano, B.; Rein, A.; Rumlova, M.; Ruml, T.; Krausslich, H.G.; Briggs, J.A.G. Structure and architecture of immature and mature murine leukemia virus capsids. Proc. Natl. Acad. Sci. USA 2018, 115, E11751–E11760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schur, F.K.; Hagen, W.J.; Rumlova, M.; Ruml, T.; Muller, B.; Krausslich, H.G.; Briggs, J.A. Structure of the immature HIV-1 capsid in intact virus particles at 8.8 A resolution. Nature 2015, 517, 505–508. [Google Scholar] [CrossRef] [PubMed]
- Schur, F.K.; Dick, R.A.; Hagen, W.J.; Vogt, V.M.; Briggs, J.A. The Structure of Immature Virus-Like Rous Sarcoma Virus Gag Particles Reveals a Structural Role for the p10 Domain in Assembly. J. Virol. 2015, 89, 10294–10302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bharat, T.A.M.; Davey, N.E.; Ulbrich, P.; Riches, J.D.; de Marco, A.; Rumlová, M.; Sachse, C.; Ruml, T.; Briggs, J.A.G. Structure of the immature retroviral capsid at 8 Å resolution by cryo-electron microscopy. Nature 2012, 487, 385–389. [Google Scholar] [CrossRef]
- Briggs, J.A.G.; Riches, J.D.; Glass, B.; Bartonova, V.; Zanetti, G.; Kräusslich, H.G. Structure and assembly of immature HIV. Proc. Natl. Acad. Sci. USA 2009, 106, 11090–11095. [Google Scholar] [CrossRef] [Green Version]
- Mattei, S.; Glass, B.; Hagen, W.J.; Krausslich, H.G.; Briggs, J.A. The structure and flexibility of conical HIV-1 capsids determined within intact virions. Science 2016, 354, 1434–1437. [Google Scholar] [CrossRef]
- Cao, S.; Maldonado, J.O.; Grigsby, I.F.; Mansky, L.M.; Zhang, W. Analysis of human T-cell leukemia virus type 1 particles by using cryo-electron tomography. J. Virol. 2015, 89, 2430–2435. [Google Scholar] [CrossRef] [Green Version]
- Zhao, G.; Perilla, J.R.; Yufenyuy, E.L.; Meng, X.; Chen, B.; Ning, J.; Ahn, J.; Gronenborn, A.M.; Schulten, K.; Aiken, C.; et al. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature 2013, 497, 643–646. [Google Scholar] [CrossRef] [PubMed]
- Butan, C.; Winkler, D.C.; Heymann, J.B.; Craven, R.C.; Steven, A.C. RSV Capsid Polymorphism Correlates with Polymerization Efficiency and Envelope Glycoprotein Content: Implications that Nucleation Controls Morphogenesis. J. Mol. Biol. 2008, 376, 1168–1181. [Google Scholar] [CrossRef] [Green Version]
- Ganser-Pornillos, B.K.; von Schwedler, U.K.; Stray, K.M.; Aiken, C.; Sundquist, W.I. Assembly properties of the human immunodeficiency virus type 1 CA protein. J. Virol. 2004, 78, 2545–2552. [Google Scholar] [CrossRef] [Green Version]
- Ganser-Pornillos, B.K.; Yeager, M.; Sundquist, W.I. The structural biology of HIV assembly. Curr.Opin.Struct. Biol. 2008, 18, 203–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Cao, S.; Martin, J.L.; Mueller, J.D.; Mansky, L.M. Morphology and ultrastructure of retrovirus particles. AIMS Biophys. 2015, 2, 343–369. [Google Scholar] [CrossRef] [PubMed]
- Campbell, E.M.; Hope, T.J. HIV-1 capsid: The multifaceted key player in HIV-1 infection. Nat. Rev. Microbiol. 2015, 13, 471–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rasaiyaah, J.; Tan, C.P.; Fletcher, A.J.; Price, A.J.; Blondeau, C.; Hilditch, L.; Jacques, D.A.; Selwood, D.L.; James, L.C.; Noursadeghi, M.; et al. HIV-1 evades innate immune recognition through specific cofactor recruitment. Nature 2013, 503, 402. [Google Scholar] [CrossRef] [PubMed]
- Francis, A.C.; Melikyan, G.B. Single HIV-1 Imaging Reveals Progression of Infection through CA-Dependent Steps of Docking at the Nuclear Pore, Uncoating, and Nuclear Transport. Cell Host Microbe 2018, 23, 536–548.e6. [Google Scholar] [CrossRef] [Green Version]
- Burdick, R.C.; Delviks-Frankenberry, K.A.; Chen, J.; Janaka, S.K.; Sastri, J.; Hu, W.S.; Pathak, V.K. Dynamics and regulation of nuclear import and nuclear movements of HIV-1 complexes. PLoS Pathog. 2017, 13, e1006570. [Google Scholar] [CrossRef]
- Matreyek, K.A.; Engelman, A. The requirement for nucleoporin NUP153 during human immunodeficiency virus type 1 infection is determined by the viral capsid. J. Virol. 2011, 85, 7818–7827. [Google Scholar] [CrossRef] [Green Version]
- Hilditch, L.; Towers, G.J. A model for cofactor use during HIV-1 reverse transcription and nuclear entry. Curr. Opin. Virol. 2014, 4, 32–36. [Google Scholar] [CrossRef] [Green Version]
- Hulme, A.E.; Perez, O.; Hope, T.J. Complementary assays reveal a relationship between HIV-1 uncoating and reverse transcription. Proc. Natl. Acad. Sci. USA 2011, 108, 9975–9980. [Google Scholar] [CrossRef] [Green Version]
- Mamede, J.I.; Cianci, G.C.; Anderson, M.R.; Hope, T.J. Early cytoplasmic uncoating is associated with infectivity of HIV-1. Proc. Natl. Acad. Sci. USA 2017, 114, E7169–E7178. [Google Scholar] [CrossRef] [Green Version]
- Lukic, Z.; Dharan, A.; Fricke, T.; Diaz-Griffero, F.; Campbell, E.M. HIV-1 uncoating is facilitated by dynein and kinesin 1. J. Virol. 2014, 88, 13613–13625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Price, A.J.; Jacques, D.A.; McEwan, W.A.; Fletcher, A.J.; Essig, S.; Chin, J.W.; Halambage, U.D.; Aiken, C.; James, L.C. Host cofactors and pharmacologic ligands share an essential interface in HIV-1 capsid that is lost upon disassembly. PLoS Pathog. 2014, 10, e1004459. [Google Scholar] [CrossRef] [PubMed]
- Stremlau, M.; Owens, C.M.; Perron, M.J.; Kiessling, M.; Autissier, P.; Sodroski, J. The cytoplasmic body component TRIM5 alpha restricts HIV-1 infection in Old World monkeys. Nature 2004, 427, 848–853. [Google Scholar] [CrossRef] [PubMed]
- Matreyek, K.A.; Engelman, A. Viral and cellular requirements for the nuclear entry of retroviral preintegration nucleoprotein complexes. Viruses 2013, 5, 2483–2511. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Franks, T.; Gibson, G.; Huber, K.; Rahm, N.; Strambio De Castillia, C.; Luban, J.; Aiken, C.; Watkins, S.; Sluis-Cremer, N.; et al. Evidence for biphasic uncoating during HIV-1 infection from a novel imaging assay. Retrovirology 2013, 10, 70. [Google Scholar] [CrossRef] [Green Version]
- Blanco-Rodriguez, G.; Gazi, A.; Monel, B.; Frabetti, S.; Scoca, V.; Mueller, F.; Schwartz, O.; Krijnse-Locker, J.; Charneau, P.; Di Nunzio, F. Remodeling of the core leads HIV-1 pre-integration complex in the nucleus of human lymphocytes. J. Virol. 2020. [Google Scholar] [CrossRef]
- Yang, Y.; Fricke, T.; Diaz-Griffero, F. Inhibition of reverse transcriptase activity increases stability of the HIV-1 core. J. Virol. 2013, 87, 683–687. [Google Scholar] [CrossRef] [Green Version]
- Cosnefroy, O.; Murray, P.J.; Bishop, K.N. HIV-1 capsid uncoating initiates after the first strand transfer of reverse transcription. Retrovirology 2016, 13, 58. [Google Scholar] [CrossRef] [Green Version]
- Franke, E.K.; Yuan, H.E.; Luban, J. Specific incorporation of cyclophilin A into HIV-1 virions. Nature 1994, 372, 359–362. [Google Scholar] [CrossRef]
- Brass, A.L.; Dykxhoorn, D.M.; Benita, Y.; Yan, N.; Engelman, A.; Xavier, R.J.; Lieberman, J.; Elledge, S.J. Identification of host proteins required for HIV infection through a functional genomic screen. Science 2008, 319, 921–926. [Google Scholar] [CrossRef]
- Krishnan, L.; Matreyek, K.A.; Oztop, I.; Lee, K.; Tipper, C.H.; Li, X.; Dar, M.J.; Kewalramani, V.N.; Engelman, A. The requirement for cellular transportin 3 (TNPO3 or TRN-SR2) during infection maps to human immunodeficiency virus type 1 capsid and not integrase. J. Virol. 2010, 84, 397–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.; Ambrose, Z.; Martin, T.D.; Oztop, I.; Mulky, A.; Julias, J.G.; Vandegraaff, N.; Baumann, J.G.; Wang, R.; Yuen, W.; et al. Flexible Use of Nuclear Import Pathways by HIV-1. Cell Host Microbe 2010, 7, 221–233. [Google Scholar] [CrossRef] [Green Version]
- Price, A.J.; Fletcher, A.J.; Schaller, T.; Elliott, T.; Lee, K.; KewalRamani, V.N.; Chin, J.W.; Towers, G.J.; James, L.C. CPSF6 defines a conserved capsid interface that modulates HIV-1 replication. PLoS Pathog. 2012, 8, e1002896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bushman, F.D.; Malani, N.; Fernandes, J.; D’Orso, I.; Cagney, G.; Diamond, T.L.; Zhou, H.; Hazuda, D.J.; Espeseth, A.S.; König, R.; et al. Host Cell Factors in HIV Replication: Meta-Analysis of Genome-Wide Studies. PLoS Pathog. 2009, 5, e1000437. [Google Scholar] [CrossRef] [PubMed]
- Konig, R.; Zhou, Y.; Elleder, D.; Diamond, T.L.; Bonamy, G.M.; Irelan, J.T.; Chiang, C.Y.; Tu, B.P.; De Jesus, P.D.; Lilley, C.E.; et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 2008, 135, 49–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dick, R.A.; Mallery, D.L.; Vogt, V.M.; James, L.C. IP6 Regulation of HIV Capsid Assembly, Stability, and Uncoating. Viruses 2018, 10, 640. [Google Scholar] [CrossRef] [Green Version]
- Dick, R.A.; Zadrozny, K.K.; Xu, C.; Schur, F.K.M.; Lyddon, T.D.; Ricana, C.L.; Wagner, J.M.; Perilla, J.R.; Ganser-Pornillos, B.K.; Johnson, M.C.; et al. Inositol phosphates are assembly co-factors for HIV-1. Nature 2018, 560, 509–512. [Google Scholar] [CrossRef]
- Mallery, D.L.; Marquez, C.L.; McEwan, W.A.; Dickson, C.F.; Jacques, D.A.; Anandapadamanaban, M.; Bichel, K.; Towers, G.J.; Saiardi, A.; Bocking, T.; et al. IP6 is an HIV pocket factor that prevents capsid collapse and promotes DNA synthesis. Elife 2018, 7, e35335. [Google Scholar] [CrossRef]
- Rihn, S.J.; Wilson, S.J.; Loman, N.J.; Alim, M.; Bakker, S.E.; Bhella, D.; Gifford, R.J.; Rixon, F.J.; Bieniasz, P.D. Extreme genetic fragility of the HIV-1 capsid. PLoS Pathog. 2013, 9, e1003461. [Google Scholar] [CrossRef] [Green Version]
- Forshey, B.M.; von Schwedler, U.; Sundquist, W.I.; Aiken, C. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J. Virol. 2002, 76, 5667–5677. [Google Scholar] [CrossRef] [Green Version]
- Novikova, M.; Zhang, Y.; Freed, E.O.; Peng, K. Multiple Roles of HIV-1 Capsid during the Virus Replication Cycle. Virol. Sin. 2019, 34, 119–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thenin-Houssier, S.; Valente, S.T. HIV-1 Capsid Inhibitors as Antiretroviral Agents. Curr. HIV Res. 2016, 14, 270–282. [Google Scholar] [CrossRef] [Green Version]
- Rumlova, M.; Ruml, T. In vitro methods for testing antiviral drugs. Biotechnol. Adv. 2017, 36, 557–576. [Google Scholar] [CrossRef] [PubMed]
- Carnes, S.K.; Sheehan, J.H.; Aiken, C. Inhibitors of the HIV-1 capsid, a target of opportunity. Curr. Opin. HIV AIDS 2018, 13, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Yant, S.R.; Mulato, A.; Hansen, D.; Tse, W.C.; Niedziela-Majka, A.; Zhang, J.R.; Stepan, G.J.; Jin, D.; Wong, M.H.; Perreira, J.M.; et al. A highly potent long-acting small-molecule HIV-1 capsid inhibitor with efficacy in a humanized mouse model. Nat. Med. 2019, 25, 1377–1384. [Google Scholar] [CrossRef]
- McArthur, C.; Gallazzi, F.; Quinn, T.P.; Singh, K. HIV Capsid Inhibitors Beyond PF74. Diseases 2019, 7, 56. [Google Scholar] [CrossRef] [Green Version]
- Blair, W.S.; Pickford, C.; Irving, S.L.; Brown, D.G.; Anderson, M.; Bazin, R.; Cao, J.; Ciaramella, G.; Isaacson, J.; Jackson, L.; et al. HIV capsid is a tractable target for small molecule therapeutic intervention. PLoS Pathog. 2010, 6, e1001220. [Google Scholar] [CrossRef] [Green Version]
- Saito, A.; Ferhadian, D.; Sowd, G.A.; Serrao, E.; Shi, J.; Halambage, U.D.; Teng, S.; Soto, J.; Siddiqui, M.A.; Engelman, A.N.; et al. Roles of Capsid-Interacting Host Factors in Multimodal Inhibition of HIV-1 by PF74. J. Virol. 2016, 90, 5808–5823. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; Zhou, J.; Shah, V.B.; Aiken, C.; Whitby, K. Small-molecule inhibition of human immunodeficiency virus type 1 infection by virus capsid destabilization. J. Virol. 2011, 85, 542–549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Price, A.J.; Halambage, U.D.; James, L.C.; Aiken, C. HIV-1 Resistance to the Capsid-Targeting Inhibitor PF74 Results in Altered Dependence on Host Factors Required for Virus Nuclear Entry. J. Virol. 2015, 89, 9068–9079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhattacharya, A.; Alam, S.L.; Fricke, T.; Zadrozny, K.; Sedzicki, J.; Taylor, A.B.; Demeler, B.; Pornillos, O.; Ganser-Pornillos, B.K.; Diaz-Griffero, F.; et al. Structural basis of HIV-1 capsid recognition by PF74 and CPSF6. Proc. Natl. Acad. Sci. USA 2014, 111, 18625–18630. [Google Scholar] [CrossRef] [Green Version]
- Rankovic, S.; Ramalho, R.; Aiken, C.; Rousso, I. PF74 Reinforces the HIV-1 Capsid To Impair Reverse Transcription-Induced Uncoating. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [Green Version]
- Fricke, T.; Buffone, C.; Opp, S.; Valle-Casuso, J.; Diaz-Griffero, F. BI-2 destabilizes HIV-1 cores during infection and Prevents Binding of CPSF6 to the HIV-1 Capsid. Retrovirology 2014, 11, 120. [Google Scholar] [CrossRef] [PubMed]
- Buffone, C.; Martinez-Lopez, A.; Fricke, T.; Opp, S.; Severgnini, M.; Cifola, I.; Petiti, L.; Frabetti, S.; Skorupka, K.; Zadrozny, K.K.; et al. Nup153 Unlocks the Nuclear Pore Complex for HIV-1 Nuclear Translocation in Nondividing Cells. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.P.; Francis, A.C.; Meuser, M.E.; Mankowski, M.; Ptak, R.G.; Rashad, A.A.; Melikyan, G.B.; Cocklin, S. Exploring Modifications of an HIV-1 Capsid Inhibitor: Design, Synthesis, and Mechanism of Action. J. Drug Des. Res. 2018, 5, 1070. [Google Scholar] [PubMed]
- Wu, G.; Zalloum, W.A.; Meuser, M.E.; Jing, L.; Kang, D.; Chen, C.H.; Tian, Y.; Zhang, F.; Cocklin, S.; Lee, K.H.; et al. Discovery of phenylalanine derivatives as potent HIV-1 capsid inhibitors from click chemistry-based compound library. Eur. J. Med. Chem. 2018, 158, 478–492. [Google Scholar] [CrossRef] [PubMed]
- Dostalkova, A.; Hadravova, R.; Kaufman, F.; Krizova, I.; Skach, K.; Flegel, M.; Hrabal, R.; Ruml, T.; Rumlova, M. A simple, high-throughput stabilization assay to test HIV-1 uncoating inhibitors. Sci. Rep. 2019, 9, 17076. [Google Scholar] [CrossRef] [Green Version]
- Hadravova, R.; Rumlova, M.; Ruml, T. FAITH—Fast Assembly Inhibitor Test for HIV. Virology 2015, 486, 78–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sticht, J.; Humbert, M.; Findlow, S.; Bodem, J.; Muller, B.; Dietrich, U.; Werner, J.; Krausslich, H.G. A peptide inhibitor of HIV-1 assembly in vitro. Nat. Struct. Mol. Biol. 2005, 12, 671–677. [Google Scholar] [CrossRef] [PubMed]
- Hulme, A.E.; Hope, T.J. The cyclosporin A washout assay to detect HIV-1 uncoating in infected cells. Methods Mol. Biol. 2014, 1087, 37–46. [Google Scholar]
- Ulbrich, P.; Haubova, S.; Nermut, M.V.; Hunter, E.; Rumlova, M.; Ruml, T. Distinct roles for nucleic acid in in vitro assembly of purified Mason-Pfizer monkey virus CANC proteins. J. Virol. 2006, 80, 7089–7099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rumlova, M.; Krizova, I.; Zelenka, J.; Weber, J.; Ruml, T. Does BCA3 Play a Role in the HIV-1 Replication Cycle? Viruses 2018, 10, 212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campbell, S.; Vogt, V.M. Self-Assembly In-Vitro of Purified Ca-Nc Proteins from Rous-Sarcoma Virus and Human-Immunodeficiency-Virus Type-1. J. Virol. 1995, 69, 6487–6497. [Google Scholar] [CrossRef] [Green Version]
- Rumlova, M.; Ruml, T.; Pohl, J.; Pichova, I. Specific in vitro cleavage of Mason-Pfizer monkey virus capsid protein: Evidence for a potential role of retroviral protease in early stages of infection. Virology 2003, 310, 310–318. [Google Scholar] [CrossRef] [Green Version]
- Dostalkova, A.; Kaufman, F.; Krizova, I.; Kultova, A.; Strohalmova, K.; Hadravova, R.; Ruml, T.; Rumlova, M. Mutations in the basic region of the Mason-Pfizer monkey virus nucleocapsid protein affect reverse transcription, gRNA packaging and the site of viral assembly. J. Virol. 2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krizova, I.; Hadravova, R.; Stokrova, J.; Gunterova, J.; Dolezal, M.; Ruml, T.; Rumlova, M.; Pichova, I. The G-patch domain of Mason-Pfizer monkey virus is a part of reverse transcriptase. J. Virol. 2012, 86, 1988–1998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strohalmova-Bohmova, K.; Spiwok, V.; Lepsik, M.; Hadravova, R.; Krizova, I.; Ulbrich, P.; Pichova, I.; Bednarova, L.; Ruml, T.; Rumlova, M. Role of Mason-Pfizer monkey virus CA-NC spacer peptide-like domain in assembly of immature particles. J. Virol. 2014, 88, 14148–14160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keprova, A.; Korinkova, L.; Krizova, I.; Hadravova, R.; Kaufman, F.; Pichova, I.; Ruml, T.; Rumlova, M. Various AKIP1 expression levels affect its subcellular localization but have no effect on NF-kappaB activation. Physiol. Res. 2019, 68, 431–443. [Google Scholar] [CrossRef]
- Kirakci, K.; Demel, J.; Hynek, J.; Zelenka, J.; Rumlova, M.; Ruml, T.; Lang, K. Phosphinate Apical Ligands: A Route to a Water-Stable Octahedral Molybdenum Cluster Complex. Inorg. Chem. 2019, 58, 16546–16552. [Google Scholar] [CrossRef]
- Kirakci, K.; Zelenka, J.; Rumlova, M.; Cvacka, J.; Ruml, T.; Lang, K. Cationic octahedral molybdenum cluster complexes functionalized with mitochondria-targeting ligands: Photodynamic anticancer and antibacterial activities. Biomater. Sci. 2019, 7, 1386–1392. [Google Scholar] [CrossRef]
- Rumlova, M.; Krizova, I.; Keprova, A.; Hadravova, R.; Dolezal, M.; Strohalmova, K.; Pichova, I.; Hajek, M.; Ruml, T. HIV-1 protease-induced apoptosis. Retrovirology 2014, 11, 37. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of all compounds are available from the authors. |
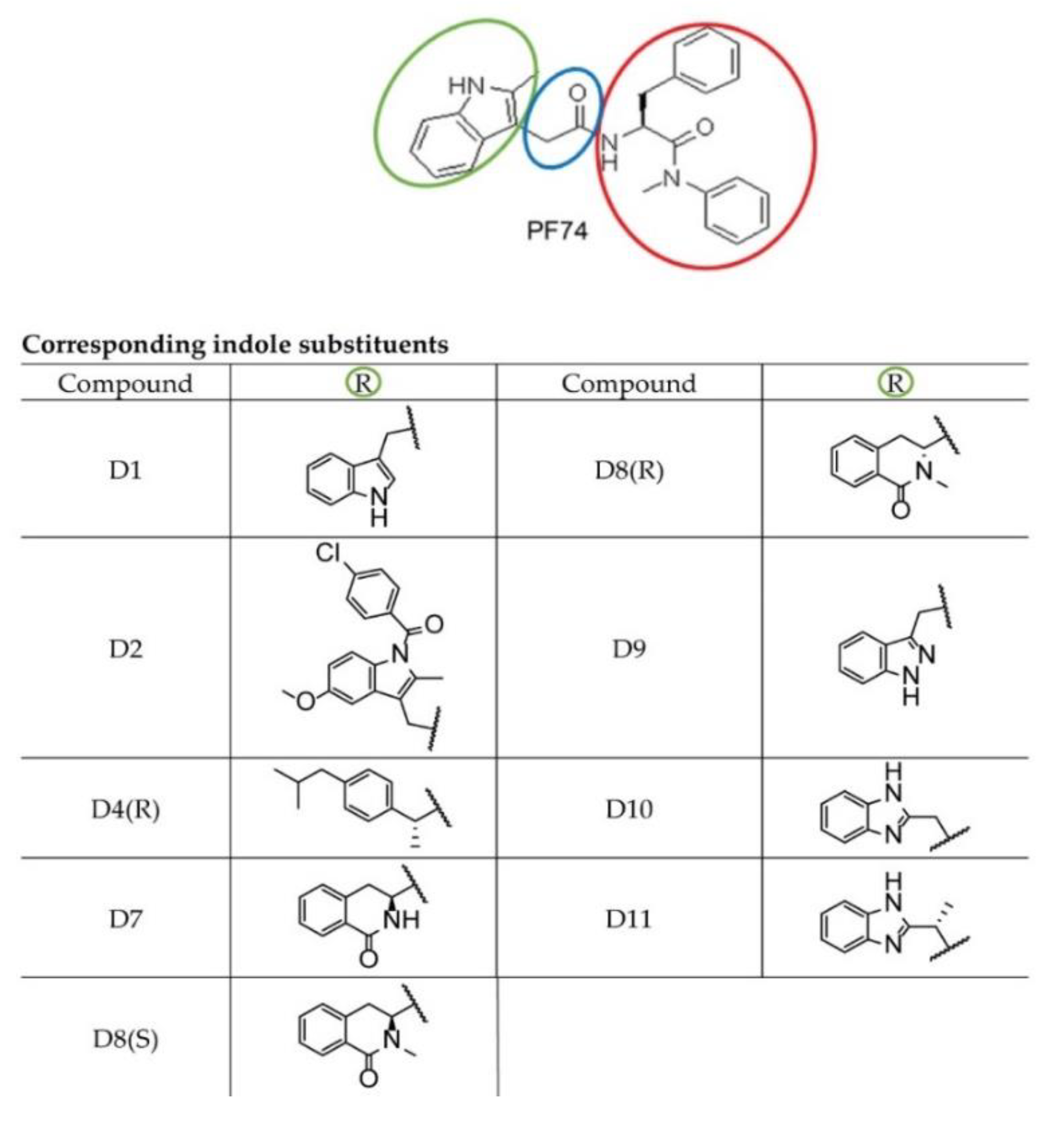

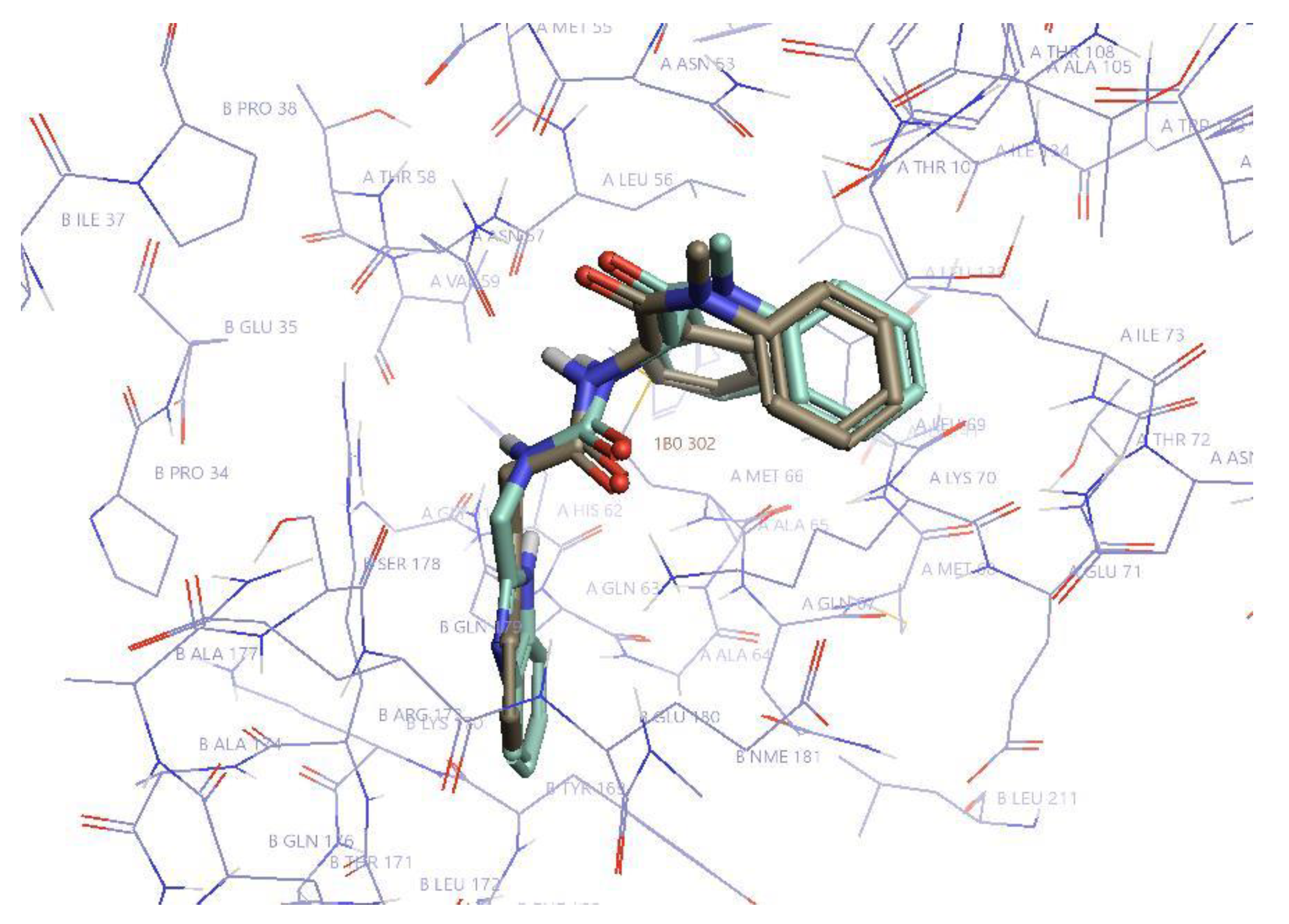
| Compound | PF74 | D1 | D2 | D4R | D7 | D8S | D8R | D9 | D10 | D11 |
|---|---|---|---|---|---|---|---|---|---|---|
| CC50 (µM) | >40 | >40 | >40 | 27 ± 3.5 | >40 | >40 | >40 | >40 | 10 ± 1.6 | >40 |
| IC50 (µM) | 1.75 ± 0.5 | 3.5 ± 0.6 | 8.0 ± 1.2 | >20 | >20 | >20 | 4.5 ± 1.5 | 3.4 ± 0.9 | 0.5 ± 0.3 | 15 ± 3 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dostálková, A.; Škach, K.; Kaufman, F.; Křížová, I.; Hadravová, R.; Flegel, M.; Ruml, T.; Hrabal, R.; Rumlová, M. PF74 and Its Novel Derivatives Stabilize Hexameric Lattice of HIV-1 Mature-Like Particles. Molecules 2020, 25, 1895. https://doi.org/10.3390/molecules25081895
Dostálková A, Škach K, Kaufman F, Křížová I, Hadravová R, Flegel M, Ruml T, Hrabal R, Rumlová M. PF74 and Its Novel Derivatives Stabilize Hexameric Lattice of HIV-1 Mature-Like Particles. Molecules. 2020; 25(8):1895. https://doi.org/10.3390/molecules25081895
Chicago/Turabian StyleDostálková, Alžběta, Kryštof Škach, Filip Kaufman, Ivana Křížová, Romana Hadravová, Martin Flegel, Tomáš Ruml, Richard Hrabal, and Michaela Rumlová. 2020. "PF74 and Its Novel Derivatives Stabilize Hexameric Lattice of HIV-1 Mature-Like Particles" Molecules 25, no. 8: 1895. https://doi.org/10.3390/molecules25081895
APA StyleDostálková, A., Škach, K., Kaufman, F., Křížová, I., Hadravová, R., Flegel, M., Ruml, T., Hrabal, R., & Rumlová, M. (2020). PF74 and Its Novel Derivatives Stabilize Hexameric Lattice of HIV-1 Mature-Like Particles. Molecules, 25(8), 1895. https://doi.org/10.3390/molecules25081895






