In vitro Anti-Inflammatory and Anti-Oxidative Stress Activities of Kushenol C Isolated from the Roots of Sophora flavescens
Abstract
1. Introduction
2. Material and Methods
2.1. Materials
2.2. Cell Culture
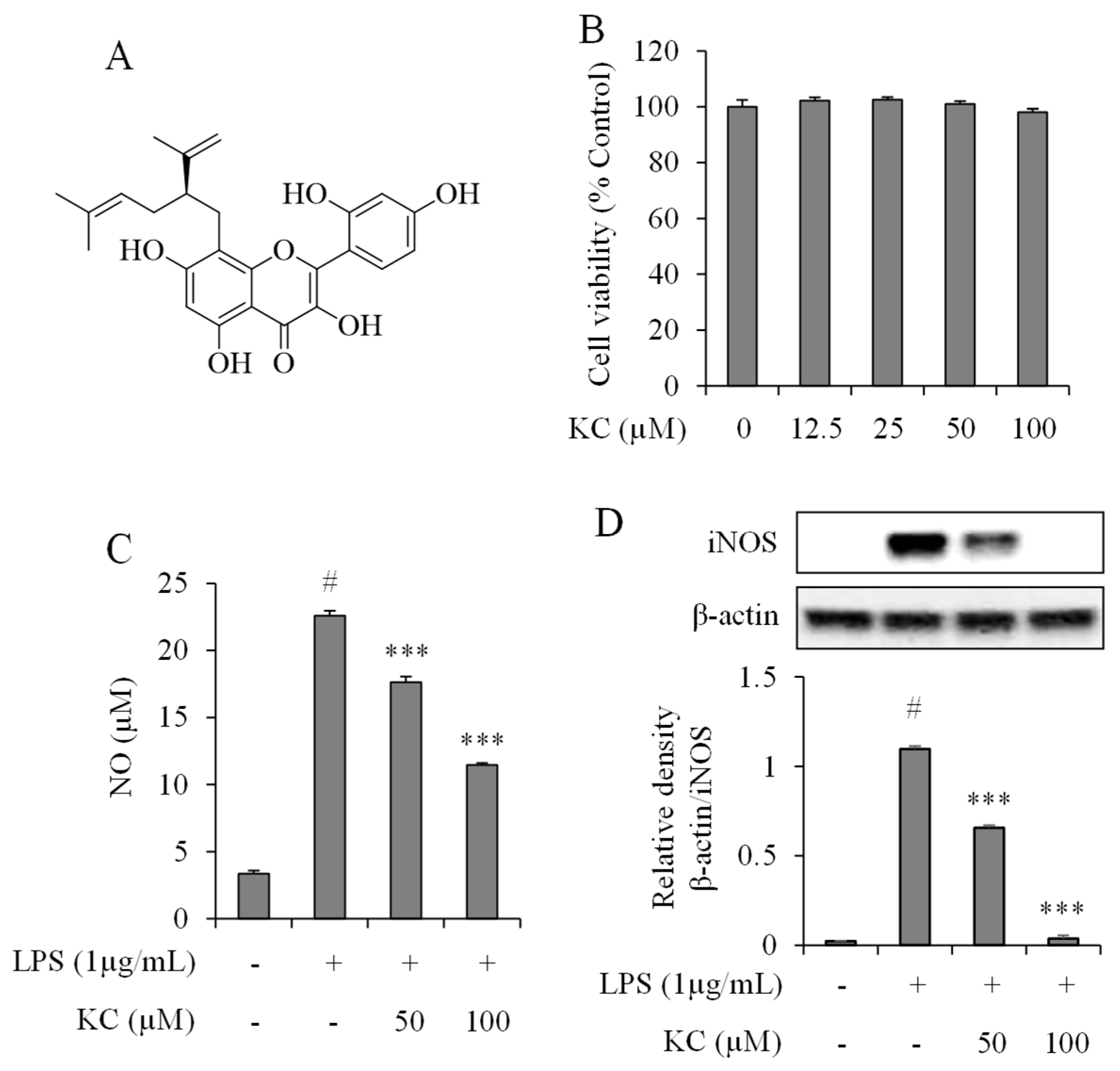
2.3. Cell Viability
2.4. Cell Proliferation Assay
2.5. Nitric Oxide Production Assay
2.6. ELISA
2.7. Total Protein Extraction
2.8. Cytosol and Nuclear Protein Extraction
2.9. Western Blotting
2.10. GSH, SOD, HO-1, and Catalase Activity Assay
2.11. NF-κB and Nrf2 Activity Assay
2.12. Intracellular ROS Measurement
2.13. Statistical Analysis
3. Results
3.1. Evaluation of NO Production and iNOS Expression Levels in LPS-Stimulated RAW264.7 Cells Treated with KC
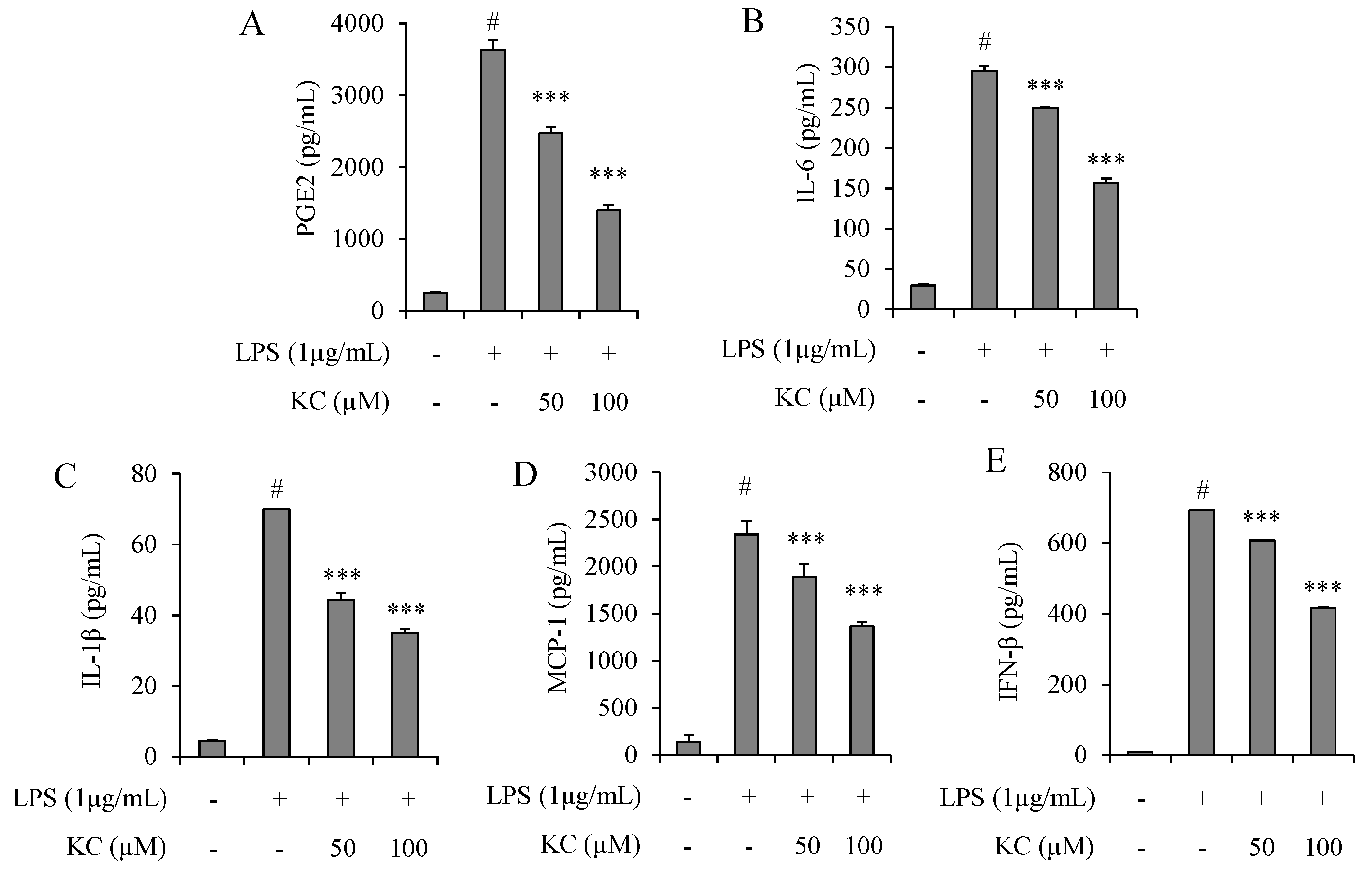
3.2. Evaluation of PGE2, IL-6, IL-1β, IFN-β, and MCP-1 Production in LPS-Stimulated RAW264.7 Cells Treated with KC
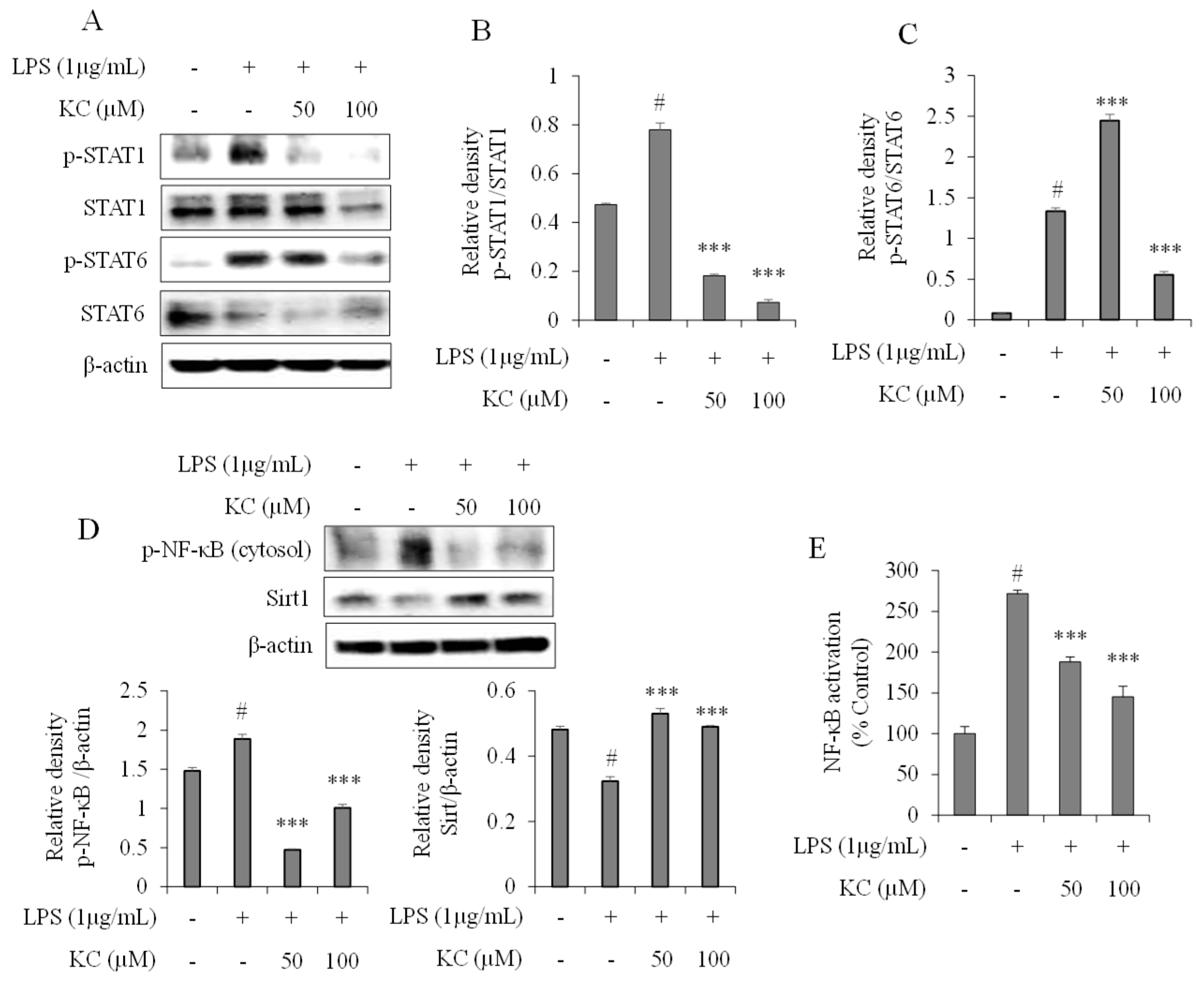
3.3. Evaluation of STAT and NF-κB Pathways in LPS-Stimulated RAW264.7 Cells Treated with KC
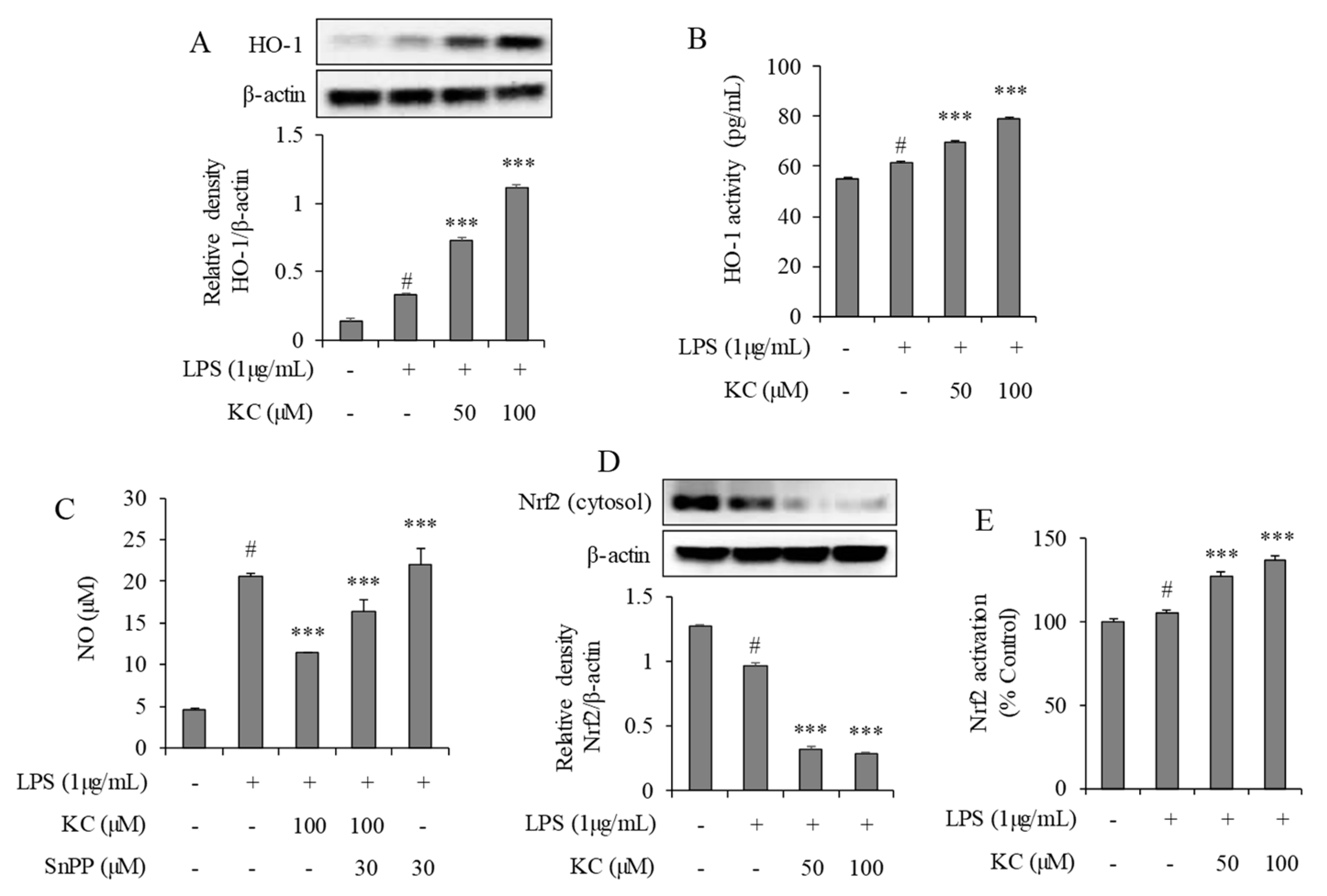
3.4. Evaluation of HO-1 Induction and Nrf2 Activation in LPS-Stimulated RAW264.7 Cells Treated with KC
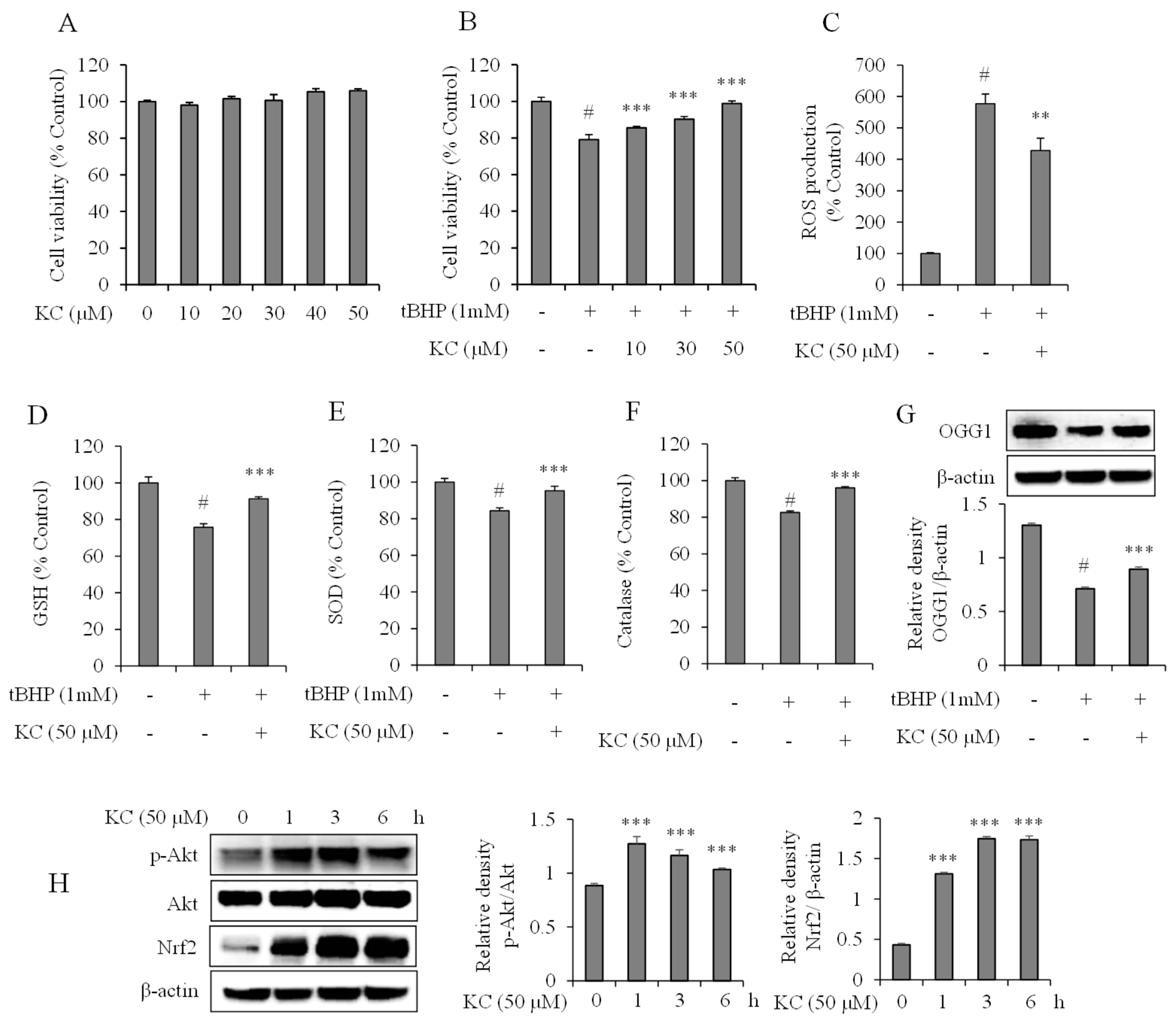
3.5. Evaluation of KC Treatment on Oxidative Stress in HaCaT Cells
3.6. Evaluation of KC Treatment on Nrf2 and Akt Expression in HaCaT Cells
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Jung, H.A.; Jeong, D.-M.; Chung, H.Y.; Lim, H.A.; Kim, J.Y.; Yoon, N.Y.; Choi, J.S. Re-evaluation of the antioxidant prenylated flavonoids from the roots of Sophora flavescens. Biol. Pharm. Bull. 2008, 31, 908–915. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, H.; Yang, J.; Hao, J.; Lv, Y.; Chen, L.; Lin, Q.; Yuan, J.; Yang, X. A Novel Flavonoid Kushenol Z from Sophora flavescens Mediates mTOR Pathway by Inhibiting Phosphodiesterase and Akt Activity to Induce Apoptosis in Non-Small-Cell Lung Cancer Cells. Molecules 2019, 24, 4425. [Google Scholar] [CrossRef] [PubMed]
- Yim, D.; Kim, M.J.; Shin, Y.; Lee, S.-J.; Shin, J.G.; Kim, D.H. Inhibition of Cytochrome P450 Activities by Sophora flavescens Extract and Its Prenylated Flavonoids in Human Liver Microsomes. Evid Based Complement Alternat Med. 2019, 2019, 2673769. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Cho, I.S.; So, Y.K.; Kim, H.-H.; Kim, Y.H. Kushenol A and 8-prenylkaempferol, tyrosinase inhibitors, derived from Sophora flavescens. J. Enzyme Inhib. Med. Chem. 2018, 33, 1048–1054. [Google Scholar] [CrossRef]
- Ma, H.; Huang, Q.; Qu, W.; Li, L.; Wang, M.; Li, S.; Chu, F. In vivo and in vitro anti-inflammatory effects of Sophora flavescens residues. J. Ethnopharmacol. 2018, 224, 497–503. [Google Scholar] [CrossRef]
- Jin, J.H.; Kim, J.S.; Kang, S.S.; Son, K.H.; Chang, H.W.; Kim, H.P. Anti-inflammatory and anti-arthritic activity of total flavonoids of the roots of Sophora flavescens. J. Ethnopharmacol. 2010, 127, 589–595. [Google Scholar] [CrossRef]
- Han, C.-C.; Wei, H.; Guo, J. Anti-inflammatory effects of fermented and non-fermented Sophora flavescens: A comparative study. BMC Complementary Alternative Med. 2011, 11, 100. [Google Scholar] [CrossRef]
- Han, C.-C.; Wang, Y. Anti-inflammation Effects of Sophora flavescens Nanoparticles. Inflammation 2012, 35, 1262–1268. [Google Scholar] [CrossRef]
- Chen, L.; Deng, H.; Cui, H.; Fang, J.; Zuo, Z.; Deng, J.; Li, Y.; Wang, X.; Zhao, L. Inflammatory responses and inflammation-associated diseases in organs. Oncotarget 2017, 9, 7204–7218. [Google Scholar] [CrossRef]
- Li, J.; Casanova, J.L.; Puel, A. Mucocutaneous IL-17 immunity in mice and humans: Host defense vs. excessive inflammation. Mucosal Immunol. 2018, 11, 581–589. [Google Scholar] [CrossRef]
- Libby, P. Inflammatory mechanisms: The molecular basis of inflammation and disease. Nutr. Rev. 2007, 65, S140–S146. [Google Scholar] [CrossRef] [PubMed]
- Laskin, D.L.; Pendino, K.J. Macrophages and inflammatory mediators in tissue injury. Annu. Rev. Pharmacol. Toxicol. 1995, 35, 655–677. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-M.; An, J. Cytokines, inflammation, and pain. Int. Anesthesiol. Clin. 2007, 45, 27–37. [Google Scholar] [CrossRef]
- Rinnerthaler, M.; Bischof, J.; Streubel, M.K.; Trost, A.; Richter, K. Oxidative stress in aging human skin. Biomolecules 2015, 5, 545–589. [Google Scholar] [CrossRef] [PubMed]
- Prasad, A.S.; Bao, B. Molecular Mechanisms of Zinc as a Pro-Antioxidant Mediator: Clinical Therapeutic Implications. Antioxidants 2019, 8, 164. [Google Scholar] [CrossRef] [PubMed]
- Sostres, C.; Gargallo, C.J.; Arroyo, M.T.; Lanas, A. Adverse effects of non-steroidal anti-inflammatory drugs (NSAIDs, aspirin and coxibs) on upper gastrointestinal tract. Best Pract. Res. Clin. Gastroenterol. 2010, 24, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Schäcke, H.; Döcke, W.D.; Asadullah, K. Mechanisms involved in the side effects of glucocorticoids. Pharmacol. Ther. 2002, 96, 23–43. [Google Scholar] [CrossRef]
- Lin, C.-Y.; Wang, W.-H.; Chen, S.-H.; Chang, Y.-W.; Hung, L.-C.; Chen, C.-Y.; Chen, Y.-H. Lipopolysaccharide-Induced Nitric Oxide, Prostaglandin E2, and Cytokine Production of Mouse and Human Macrophages Are Suppressed by Pheophytin-b. Int. J. Mol. Sci. 2017, 18, 2637. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.-C. NF-κB signaling in inflammation. Signal Transduction Targeted Ther. 2017, 23, 17023. [Google Scholar] [CrossRef]
- Rhee, S.H.; Jones, B.W.; Toshchakov, V.; Vogel, S.N.; Fenton, M.J. Toll-like receptors 2 and 4 activate STAT1 serine phosphorylation by distinct mechanisms in macrophages. J. Biol. Chem. 2003, 278, 22506–22512. [Google Scholar] [CrossRef]
- Tang, X.; Marciano, D.L.; Leeman, S.E.; Amar, S. LPS induces the interaction of a transcription factor, LPS-induced TNF-alpha factor, and STAT6(B) with effects on multiple cytokines. Proc. Natl. Aca. Sci. USA 2005, 102, 5132–5137. [Google Scholar] [CrossRef] [PubMed]
- Xue, Q.; Yan, Y.; Zhang, R.; Xiong, H. Regulation of iNOS on Immune Cells and Its Role in Diseases. Int. J. Mol. Sci. 2018, 19, 3805. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Cheon, B.S.; Kim, Y.H.; Kim, S.Y.; Kim, H.P. Effects of naturally occurring flavonoids on nitric oxide production in the macrophage cell line RAW 264.7 and their structure-activity relationships. Biochem. Pharmacol. 1999, 58, 759–765. [Google Scholar] [CrossRef]
- Kim, A.R.; Cho, J.Y.; Zou, Y.; Choi, J.S.; Chung, H.Y. Flavonoids differentially modulate nitric oxide production pathways in lipopolysaccharide-activated RAW264.7 cells. Arch. Pharmacal. Res. 2005, 28, 297–304. [Google Scholar] [CrossRef]
- Hirano, T. Interleukin 6 in autoimmune and inflammatory diseases: A personal memoir. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2010, 86, 717–730. [Google Scholar] [CrossRef]
- Fattahi, M.J.; Mirshafiey, A. Prostaglandins and rheumatoid arthritis. Arthritis 2012, 2012, 239310. [Google Scholar] [CrossRef]
- Lu, R.; Munroe, M.E.; Guthridge, J.M.; Bean, K.M.; Fife, D.A.; Chen, H.; Slight-Webb, S.R.; Keith, M.P.; Harley, J.B.; James, J.A. Dysregulation of innate and adaptive serum mediators precedes systemic lupus erythematosus classification and improves prognostic accuracy of autoantibodies. J. Autoimmun. 2016, 74, 182–193. [Google Scholar] [CrossRef]
- Dinarello, C.A. Interleukin-1 in the pathogenesis and treatment of inflammatory diseases. Blood 2011, 117, 3720–3732. [Google Scholar] [CrossRef]
- Borges da Silva, H.; Fonseca, R.; Alvarez, J.M.; D’Império Lima, M.R. IFN-γ Priming Effects on the Maintenance of Effector Memory CD4(+) T Cells and on Phagocyte Function: Evidences from Infectious Diseases. J. Immunol. Res. 2015, 2015, 202816. [Google Scholar] [CrossRef]
- Deshmane, S.L.; Kremlev, S.; Amini, S.; Sawaya, B.E. Monocyte chemoattractant protein-1 (MCP-1): An overview. J. Interferon Cytokine. Res. 2009, 29, 313–326. [Google Scholar] [CrossRef]
- Zhou, H.; Lutterodt, H.; Cheng, Z.; Yu, L.L. Anti-Inflammatory and antiproliferative activities of trifolirhizin, a flavonoid from Sophora flavescens roots. J. Agric. Food Chem. 2009, 57, 4580–4585. [Google Scholar] [CrossRef] [PubMed]
- Hong, M.H.; Lee, J.Y.; Jung, H.; Jin, D.-H.; Go, H.Y.; Kim, J.H.; Jang, B.-H.; Shin, Y.-C.; Ko, S.-G. Sophora flavescens Aiton inhibits the production of pro-inflammatory cytokines through inhibition of the NF kappaB/IkappaB signal pathway in human mast cell line (HMC-1). Toxicol. Vitro 2009, 23, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Yeung, F.; Hoberg, J.E.; Ramsey, C.S.; Keller, M.D.; Jones, D.R.; Frye, R.A.; Mayo, M.W. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004, 23, 2369–2380. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.G.; Ahern, M.J.; Coleman, M.; Weedon, H.; Papangelis, V.; Beroukas, D.; Roberts-Thomson, P.J.; Smith, M.D. Expression of Jak3, STAT1, STAT4, and STAT6 in inflammatory arthritis: Unique Jak3 and STAT4 expression in dendritic cells in seropositive rheumatoid arthritis. Ann. Rheumatic Dis. 2006, 65, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Kuuliala, K.; Kuuliala, A.; Koivuniemi, R.; Kautiainen, H.; Repo, H.; Leirisalo-Repo, M. STAT6 and STAT1 Pathway Activation in Circulating Lymphocytes and Monocytes as Predictor of Treatment Response in Rheumatoid Arthritis. PLoS ONE 2016, 11, e0167975. [Google Scholar] [CrossRef]
- Seeliger, C.; Schyschka, L.; Kronbach, Z.; Wottge, A.; van Griensven, M.; Wildemann, B.; Vester, H. Signaling pathway STAT1 is strongly activated by IFN-β in the pathogenesis of osteoporosis. Eur. J. Med. Res. 2015, 20, 1. [Google Scholar] [CrossRef]
- Ryter, S.W.; Alam, J.; Choi, A.M.K. Heme oxygenase-1/carbon monoxide: From basic science to therapeutic applications. Physiol. Rev. 2006, 86, 583–650. [Google Scholar] [CrossRef]
- Kim, K.-S.; Cui, X.; Lee, D.-S.; Ko, W.; Sohn, J.H.; Yim, J.H.; An, R.-B.; Kim, Y.-C.; Oh, H. Inhibitory effects of benzaldehyde derivatives from the marine fungus Eurotium sp. SF-5989 on inflammatory mediators via the induction of heme oxygenase-1 in lipopolysaccharide-stimulated RAW264.7 macrophages. Int. J. Mol. Sci. 2014, 15, 23749–23765. [Google Scholar] [CrossRef]
- Shinkai, Y.; Yamanaka, I.; Duong, H.H.T.; Quynh, N.T.; Kanaho, Y.; Kumagai, Y. Garcinia vilersiana bark extract activates the Nrf2/HO-1 signaling pathway in RAW264.7 cells. J. Toxicol. Sci. 2013, 38, 875–878. [Google Scholar] [CrossRef][Green Version]
- Hussain, T.; Tan, B.; Yin, Y.; Blachier, F.; Tossou, M.C.B.; Rahu, N. Oxidative Stress and Inflammation: What Polyphenols Can Do for Us ? Oxidative Med. Cell. Longevity 2016, 2016, 7432797. [Google Scholar] [CrossRef]
- Chang, C.-H.; Yu, F.-Y.; Wu, T.-S.; Wang, L.-T.; Liu, B.-H. Mycotoxin citrinin induced cell cycle G2/M arrest and numerical chromosomal aberration associated with disruption of microtubule formation in human cells. Toxicol. Sci. 2011, 119, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Schuller Levis, G.B.; Lee, E.B.; Levis, W.R.; Lee, D.W.; Kim, B.S.; Park, S.Y.; Park, E. Platycodin D and D3 isolated from the root of Platycodon grandiflorum modulate the production of nitric oxide and secretion of TNF-α in activated RAW 264.7 cells. Inter. Immunopharmacol. 2004, 4, 1039–1049. [Google Scholar] [CrossRef] [PubMed]
- Crane, D.; Häussinger, D.; Graf, P.; Sies, H. Decreased flux through pyruvate dehydrogenase by thiol oxidation during t-butyl hydroperoxide metabolism in perfused rat liver. Hoppe. Seylers Z Physiol. Chem. 1983, 364, 977–987. [Google Scholar] [CrossRef] [PubMed]
- Davies, M.J. Detection of peroxyl and alkoxyl radicals produced by reaction of hydroperoxides with rat liver microsomal fractions. Biochem. J. 1989, 257, 603–606. [Google Scholar] [CrossRef]
- Giudice, A.; Montella, M. Activation of the Nrf2-ARE signaling pathway: A promising strategy in cancer prevention. Bioessays 2006, 28, 169–181. [Google Scholar] [CrossRef]
- Hwang, Y.P.; Choi, J.H.; Choi, J.M.; Chung, Y.C.; Jeong, H.G. Protective mechanisms of anthocyanins from purple sweet potato against tert-butyl hydroperoxide-induced hepatotoxicity. Food Chem. Toxicol. 2011, 49, 2081–2089. [Google Scholar] [CrossRef]
- Varì, R.; D’Archivio, M.; Filesi, C.; Carotenuto, S.; Scazzocchio, B.; Santangelo, C.; Giovannini, C.; Masella, R. Protocatechuic acid induces antioxidant/detoxifying enzyme expression through JNK-mediated Nrf2 activation in murine macrophages. J. Nutritional Biochem. 2011, 22, 409–417. [Google Scholar] [CrossRef]
- Xing, H.-Y.; Liu, Y.; Chen, J.-H.; Sun, F.-J.; Shi, H.-Q.; Xia, P.-Y. Hyperoside attenuates hydrogen peroxide-induced L02 cell damage via MAPK-dependent Keap₁-Nrf₂-ARE signaling pathway. Biochem. Biophys. Res. Commun. 2011, 410, 759–765. [Google Scholar] [CrossRef]
Sample Availability: Samples of the Kushenol C are available from the authors. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cho, B.O.; Che, D.N.; Kim, J.-S.; Kim, J.H.; Shin, J.Y.; Kang, H.J.; Jang, S.I. In vitro Anti-Inflammatory and Anti-Oxidative Stress Activities of Kushenol C Isolated from the Roots of Sophora flavescens. Molecules 2020, 25, 1768. https://doi.org/10.3390/molecules25081768
Cho BO, Che DN, Kim J-S, Kim JH, Shin JY, Kang HJ, Jang SI. In vitro Anti-Inflammatory and Anti-Oxidative Stress Activities of Kushenol C Isolated from the Roots of Sophora flavescens. Molecules. 2020; 25(8):1768. https://doi.org/10.3390/molecules25081768
Chicago/Turabian StyleCho, Byoung Ok, Denis Nchang Che, Ji-Su Kim, Jang Hoon Kim, Jae Young Shin, Hyun Ju Kang, and Seon Il Jang. 2020. "In vitro Anti-Inflammatory and Anti-Oxidative Stress Activities of Kushenol C Isolated from the Roots of Sophora flavescens" Molecules 25, no. 8: 1768. https://doi.org/10.3390/molecules25081768
APA StyleCho, B. O., Che, D. N., Kim, J.-S., Kim, J. H., Shin, J. Y., Kang, H. J., & Jang, S. I. (2020). In vitro Anti-Inflammatory and Anti-Oxidative Stress Activities of Kushenol C Isolated from the Roots of Sophora flavescens. Molecules, 25(8), 1768. https://doi.org/10.3390/molecules25081768





