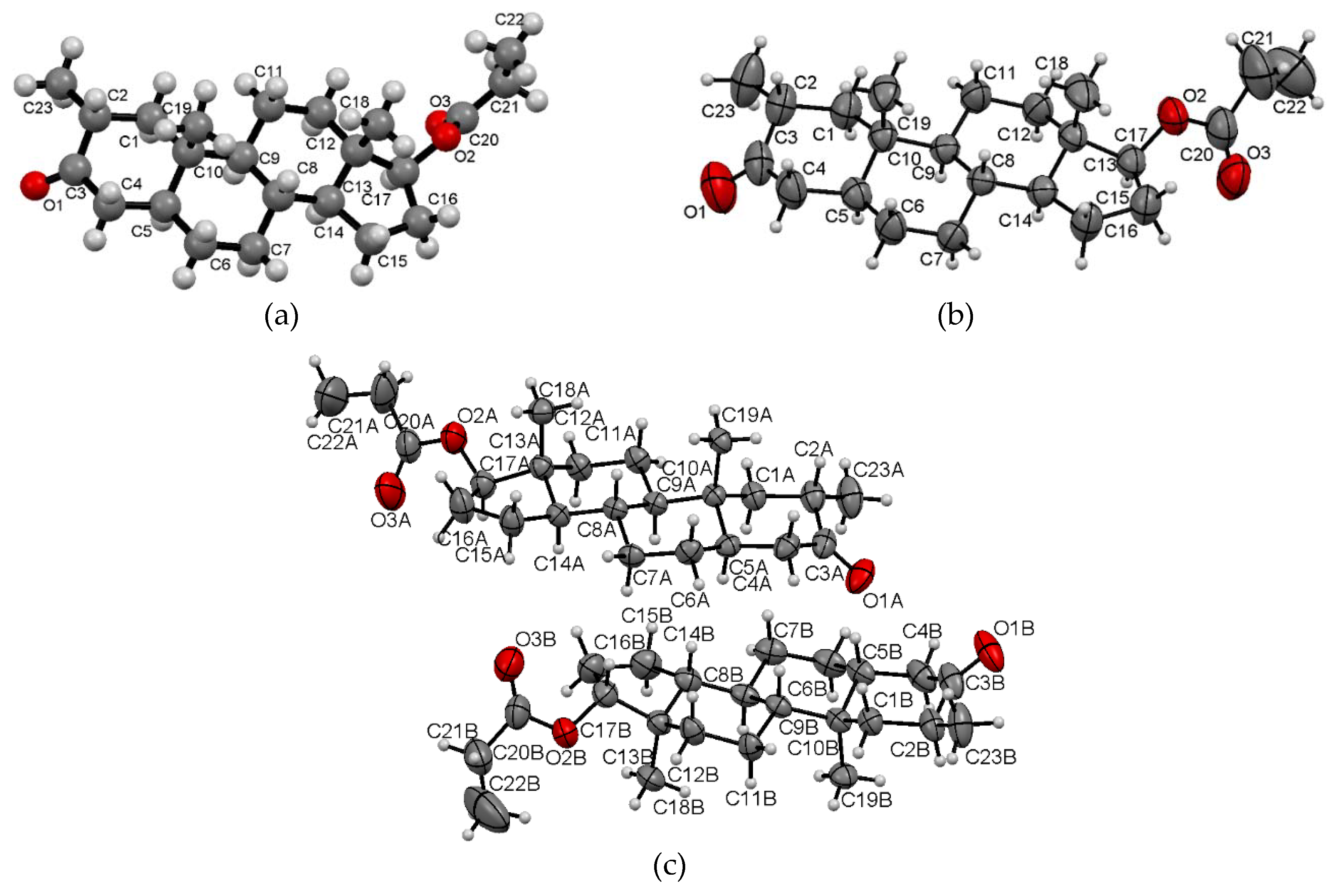
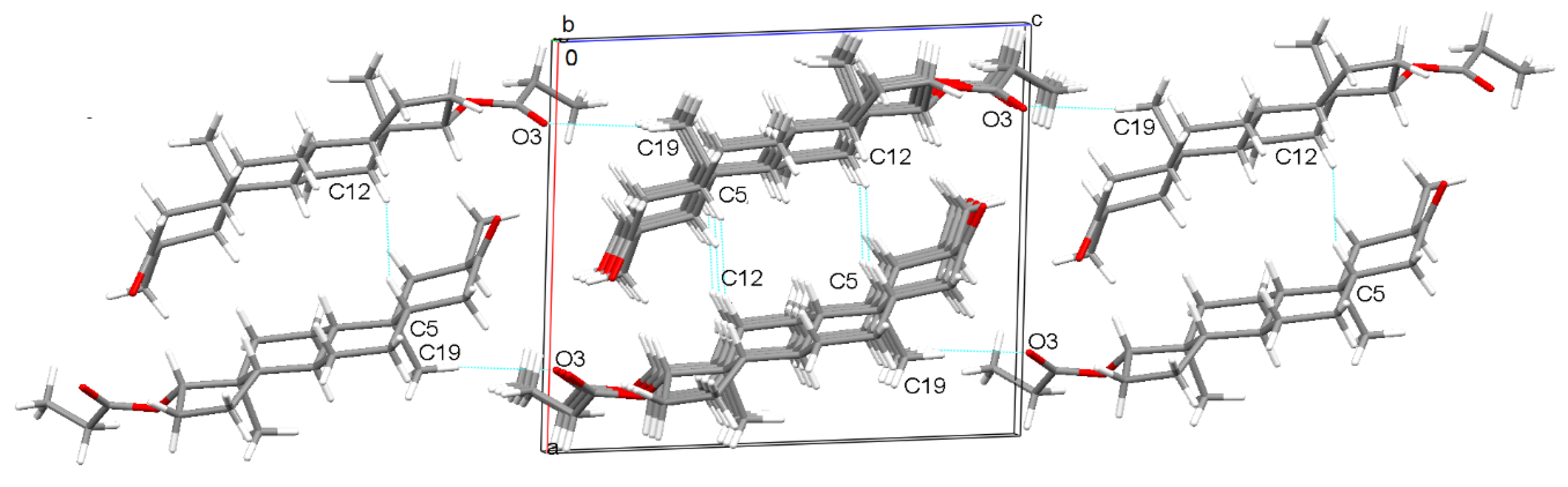
2.4.1. Lattice Energy Evaluation by the Coulomb-London-Pauli (CLP) Method
The CLP method (See details at
Section 3.6), which is based on atom-atom type potentials, has been shown that the formation of two drostanolone propionate polymorphs has led to structures that display similar lattice energies (−156.3 kJ/mol in Drost 1; −159.2 kJ/mol in Drost 2 and −151.6 kJ/mol in Drost 3, respectively).
The partitioned packing energies calculated for the polymorphs driven by slow evaporation in ethanol (Drost 2) and acetone (Drost3) shows similar values as the start compound (Drost 1). The total lattice energy breakdown in individual terms is given in
Table 5a. The electrostatic terms, which account for Coulombic and polarization term have a small contribution, while the dispersion energy plays the major role. Specific interatomic interactions that contribute mainly into the total intermolecular interaction energy are given in
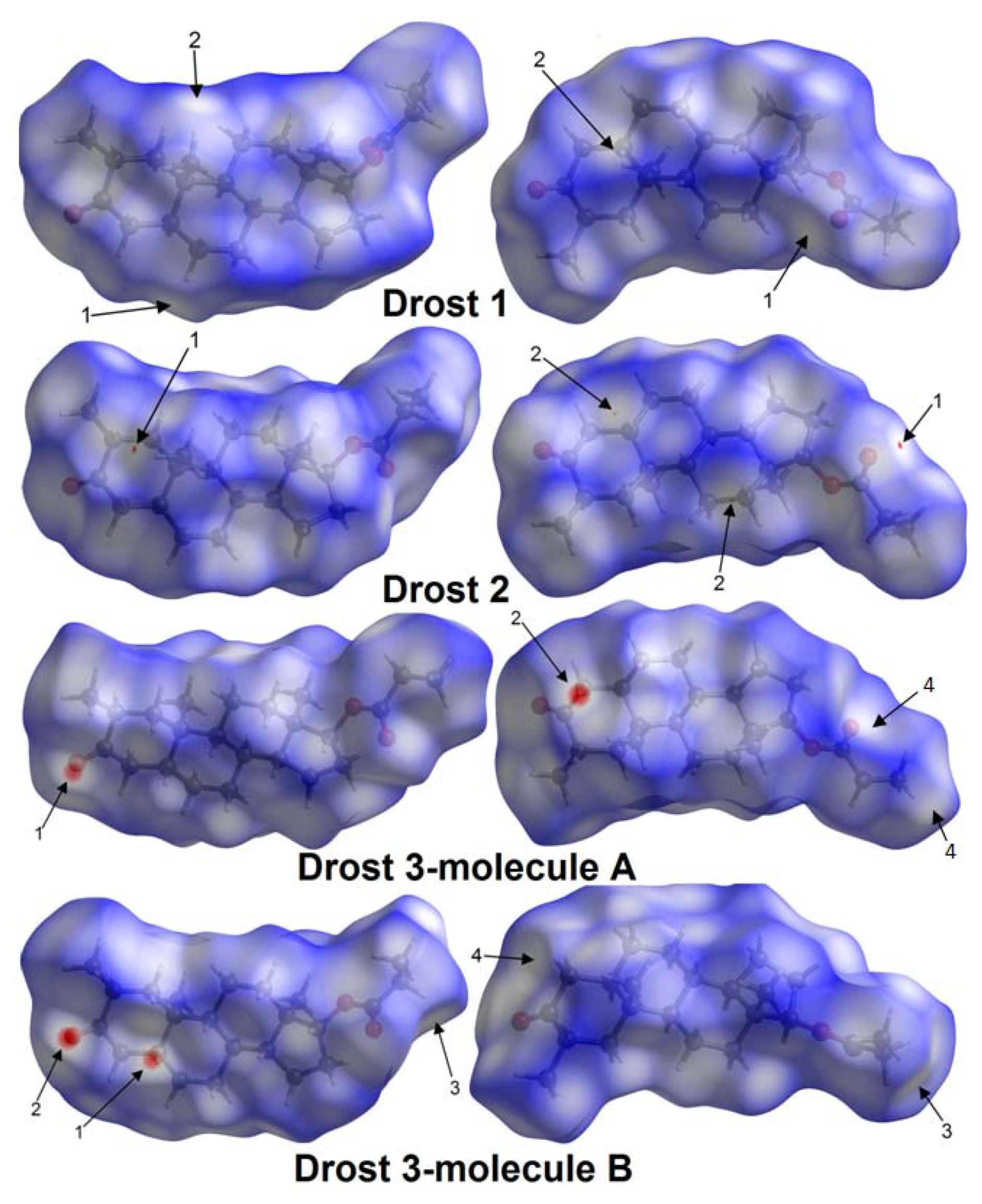
Table 3. Although the O...H contacts have a lower preponderance than the H...H contacts (
Table 4), they have a high energy and contribute in the greatest extent to lattice energy.
Similar behavior regarding lattice stability through weak H...H and O...H intermolecular interactions was found in compounds of corticosteroid class [
28]. Such compounds were investigated by adsorption and Raman scattering, which is assigned to intermolecular interaction and specific effects of crystal packing by comparing experimental spectra with quantum chemical calculations [
29].
The Kitaigorodskii packing index [
30] is a measure of packing efficiency and usually has a value of 65%. It was evaluated by PLATON software [
31] and the results obtained are as follows: 60.13% for Drost 3, 60.25% for Drost 1, and the highest value of 61.64% for Drost 2. Between the packing index and total CLP lattice energy, there is a correlation. The higher the packing index is, the greater the absolute value of the lattice energy is. From
Table 5a, it is observed that this correlation between packing index and lattice energy is noticed.
2.4.2. Lattice Energy Evaluation by a Density-Functional Tight-Binding Model
In order to validate our method based on the atom-atom potential-type CLP model, a higher level theoretical method was also considered. Accordingly, the density-functional tight-binding (DFTB) model in its self-consistent charge corrected the variant (SCC-DFTB) [
32] implemented in the DFTB+ code [
33] and applied in order to compute lattice energies for the three crystal configurations [
34]. Several supercells with different cell sizes (1 × 1 × 1, 1 × 1 × 2, 1 × 2 × 2, 2 × 2 × 2, 2 × 2 × 3, 2 × 3 × 3, 3 × 3 × 3) were generated [
35] and their electronic energies were computed, including the dispersion effects via the Slater-Kirkwood (SK) dispersion correction scheme [
36]. Based on our past experience [
37], compared with results computed using the second order Møller-Plesset perturbation theory, the SK-dispersion model can well reproduce the dispersion effects in the case of large molecular clusters. The largest supercell structure (3 × 3 × 3) for Drost 2 crystal conformation includes 87 monomers (5394 atoms) while those for Drost 1 and Drost 3 (2 × 3 × 3) contain 134 (8308 atoms) and 164 monomers (10168 atoms), respectively. Then, the total electronic energy values of the supercells with different cell sizes defined by the number of monomers in the supercell were extrapolated into the infinite large monomer numbers and the lattice energy, as the size independent parameter in the interpolation scheme (parameter
a), was obtained [
34]. The best performant fitting function (R
2 ≈ 0.94) was found as:
where
N is the number of the unit cells,
a is the fitting parameter, and
Elatt means the lattice energy for an infinite number of unit cells.
The dispersion component of the lattice energy considering the same interpolation scheme (See Equation (1)) was also computed. Accordingly, the lattice total energies and their dispersion parts computed for the three unit cell configurations with the above described calculation scheme are presented in
Table 5b. Comparing the results obtained based on the CLP model [
38,
39] and by SCC-DFTB theory, relatively good agreement between the two theories can be found for the lattice energy values. This agreement looks effective for the case of Drost 2 crystal configuration (the energy difference is 7.5 kJ/mol), while, for the Drost 1 and Drost 3, the energy deviations are a bit larger: 13.7 and 13.9 kJ/mol, respectively. Furthermore, both theoretical methods give the Drost 2 conformation as the most bounded crystal structure followed by Drost 1, while the Drost 3 crystal has the smallest cohesive energy value. Furthermore, both methods suggest that the main attractive forces, which keep the crystalline structures together are the dispersion effects.
In order to better understand the nature of the intermolecular forces, which govern the formation of the molecular crystals, higher level electronic structure theory was also considered. Accordingly, the molecular dimer energies for different dimer geometry conformations taken from the Drost 2 crystal supercell were computed using the second order Møller-Plesset perturbation theory based on localized molecular orbitals and density-fitting technique (DF-LMP2) [
40]. The local correlation treatment also offers the possibility to decompose the intermolecular interaction energy into intramolecular, dispersive, and ionic components of the correlation contribution. Five different dimer geometries, relevant for the unit cell configuration, were selected (see
Figure 9). The selection criterium is based on the Drost 2 unit cell configuration, where all close contacts (defined as the sum of the vdW radii + 0.2 Å) of the Drost 2 molecule from the unit cell were generated and, from the resulted oligomer cluster, all the possible dimer configurations were kept.
The intermolecular interaction energies between the drostanolone propionate monomers were computed considering the DF-LMP2 method and using the def2-tzvp [
41] basis set, as implemented in the Molpro program package [
42]. Binding energies were estimated in the supramolecular approximation (i.e., as a difference between the energy of a given dimer complex and the sum of energies of the isolated molecules constituting it). The Drost 2 crystal structure is built by parallel layers grown in the aob crystal plane, where layers are kept together by the side-chain interactions of the drostanolone molecules along the oc crystal axis. Three dimer configurations (see
Figure 9a)–c)) were identified as relevant pair conformations for the layer structure stability (parallel stacking, T-shape, and antiparallel stacking). Their intermolecular interaction energies obtained at the DF-LMP2/def2-tzvp level of theory are: ΔE
a) = −21.67 kJ/mol, ΔE
b) = −15.37 kJ/mol and ΔE
c) = −18.08 kJ/mol, respectively. On the other hand, the interaction between the layers is mainly built by two characteristic pair configurations (See
Figure 9d,e). Their intermolecular interaction energies computed at DF-LMP2/def2-tzvp level of theory are: ΔE
d) = −8.25 kJ/mol and ΔE
e) = −14.13 kJ/mol, respectively. To estimate the contribution of higher (other than pair correlation) electron correlation effects to the intermolecular interaction energy, the coupled cluster level of theory based on localized molecular orbitals and density-fitting technique (DF-LCCSD(T)) for the a) dimer configuration was considered. The result shows that higher level electron correlation effects do not change the magnitude of the intermolecular interaction energy significantly, which gives only a small contribution (+1.18 kJ/mol) to the final energy value (−21.67 kJ/mol for DF-LMP2 versus −20.49 kJ/mol for DF-LCCSD(T)). Since advanced ab initio theories including high-level electron correlation effects are not suitable to directly compute the lattice energies due to the large amount of computation, they can be used indirectly to estimate the lattice energy as a sum of the energies of intermolecular (noncovalent) pairwise interactions between the considered molecule and its neighbors [
43]. Accordingly, in the case of the Drost 2 molecule, the total pairwise interaction energy between a monomer from the crystal and its neighbors (close contacts) can be computed as the sum of the binding energies of the five dimers but consider each of them twice, since in the close contact configuration, each of them occurs two times. In this way, the sum of the pairwise interaction energies is −155.00 kcal/mol, which is consistent with previous lattice energy calculations for the Drost 2 case based on the CLP and SCC-DFTB methods (−159.2 kcal/mol and −151.7 kcal/mol, respectively). On the other hand, the Espinoza’s empirical formula [
44] defined in the framework of QTAIM (or quantum theory of atoms in molecules) theory [
45] can be used as another possible solution for estimating the lattice energy. This solution has been successfully applied in several cases [
46,
47,
48] based on Density Functional Theory calculations, but applying it in the case of the DF-LMP2 theory is not a simple task.
Using the CLP and the SCC-DFTB simple models, only the global contribution of the dispersion effects was taken into account. However, it would be interesting to analyze how the dispersion effects manifest along the different crystal axes, since it is shown in
Figure 4 that different relative pair conformations can be observed along the oa and ob or oc axes. More precisely, the binding energies of dimers a)–c) build layers along the oab crystal plane, while interactions found in case of dimers d) and e) enhance the crystal cohesion along the oc direction. Accordingly, for the five dimer cases of the Drost 2 crystal configuration, the symmetry adapted perturbation theory (SAPT) method [
49] was applied to decompose their intermolecular interaction energy into physically meaningful energy components (electrostatic, exchange, induction, and dispersion) defined in the framework of the SAPT theory. Using the SAPT energy decomposition method, our goal was to get rather realistic values than to directly compare them with results obtained by other theoretical methods, like DF-LMP2. Due to the large numbers of atoms in the dimer geometry, applying the so-called “gold” and “silver” standards for the SAPT method are not feasible. Only the “bronze” standard is feasible. This standard is defined as the zero-order SAPT expansion combined with the exchange-scaling (sSAPT0) approximation [
50] and used together with the jun-cc-pVDZ basis set [
51]. The intermolecular energy values and their different energy components computed at sSAPT0 as well as the intermolecular energy values computed at DF-LMP2 levels of theory are presented in
Table 6.
In the case of dimer configurations having parallel stacking or a), T-shape or b) and antiparallel stacking or c) conformations, the most dominant energy component is the dispersion part, which is almost equal or even lower than the total sSAPT0 energy, but it is almost half balanced by the exchange repulsion effects. The electrostatic and induction contributions individually have a relatively small contribution to the sSAPT0 energy, but their common effect is no longer a negligible contribution. In the case of layer-layer interaction, which includes the d) and e) dimer configurations, in addition to dispersion and exchange interactions, electrostatic contributions have also become important. All these results show us that the layer structure defined by the a)–c) interaction configurations (oa and ob crystal axes directions) are much stronger bound than the interaction between the layers (along the oc crystal axis).