Larvicidal Activities of 2-Aryl-2,3-Dihydroquinazolin -4-ones against Malaria Vector Anopheles arabiensis, In Silico ADMET Prediction and Molecular Target Investigation
Abstract
1. Introduction
2. Results and Discussion
2.1. Chemistry
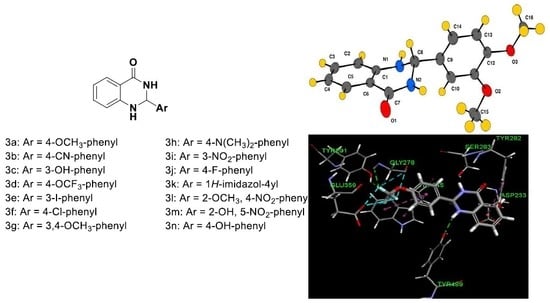
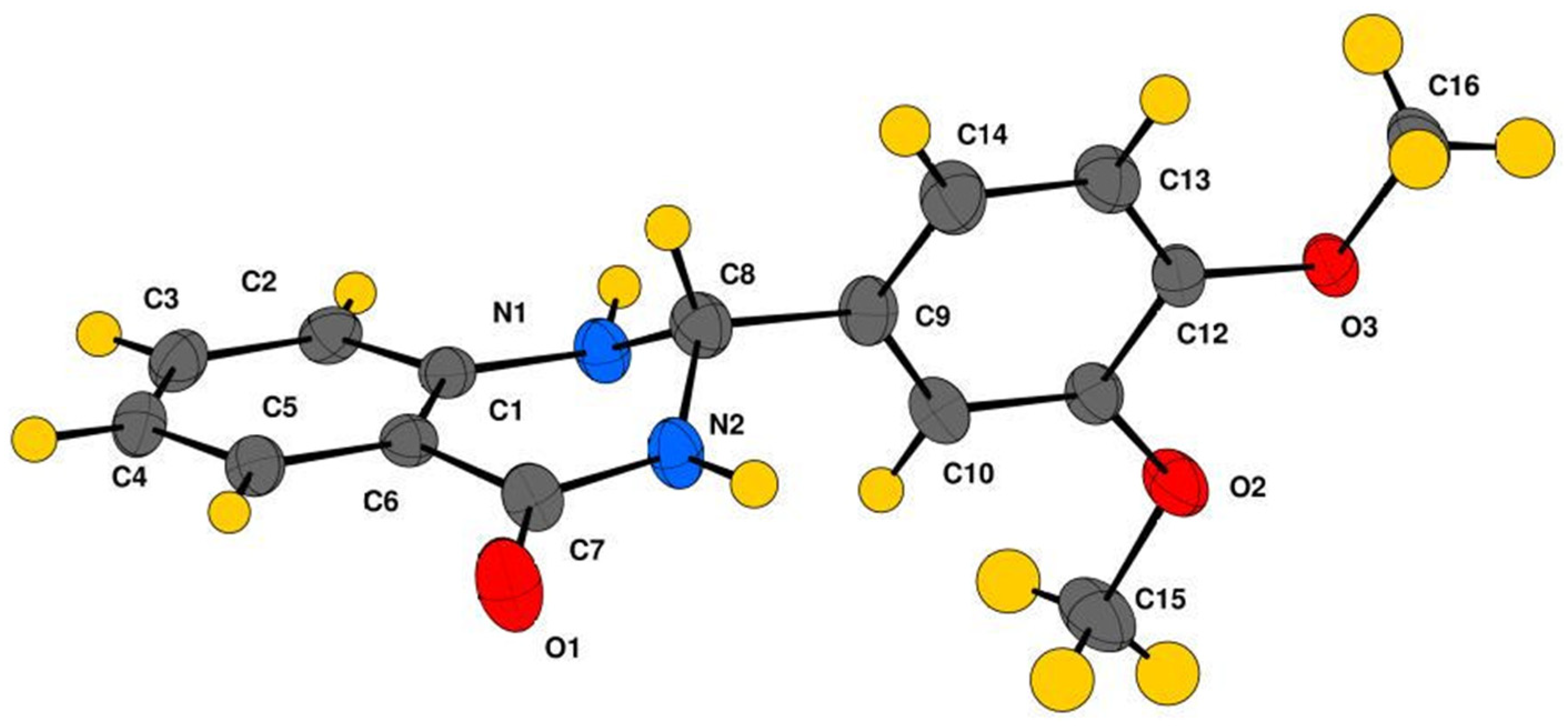
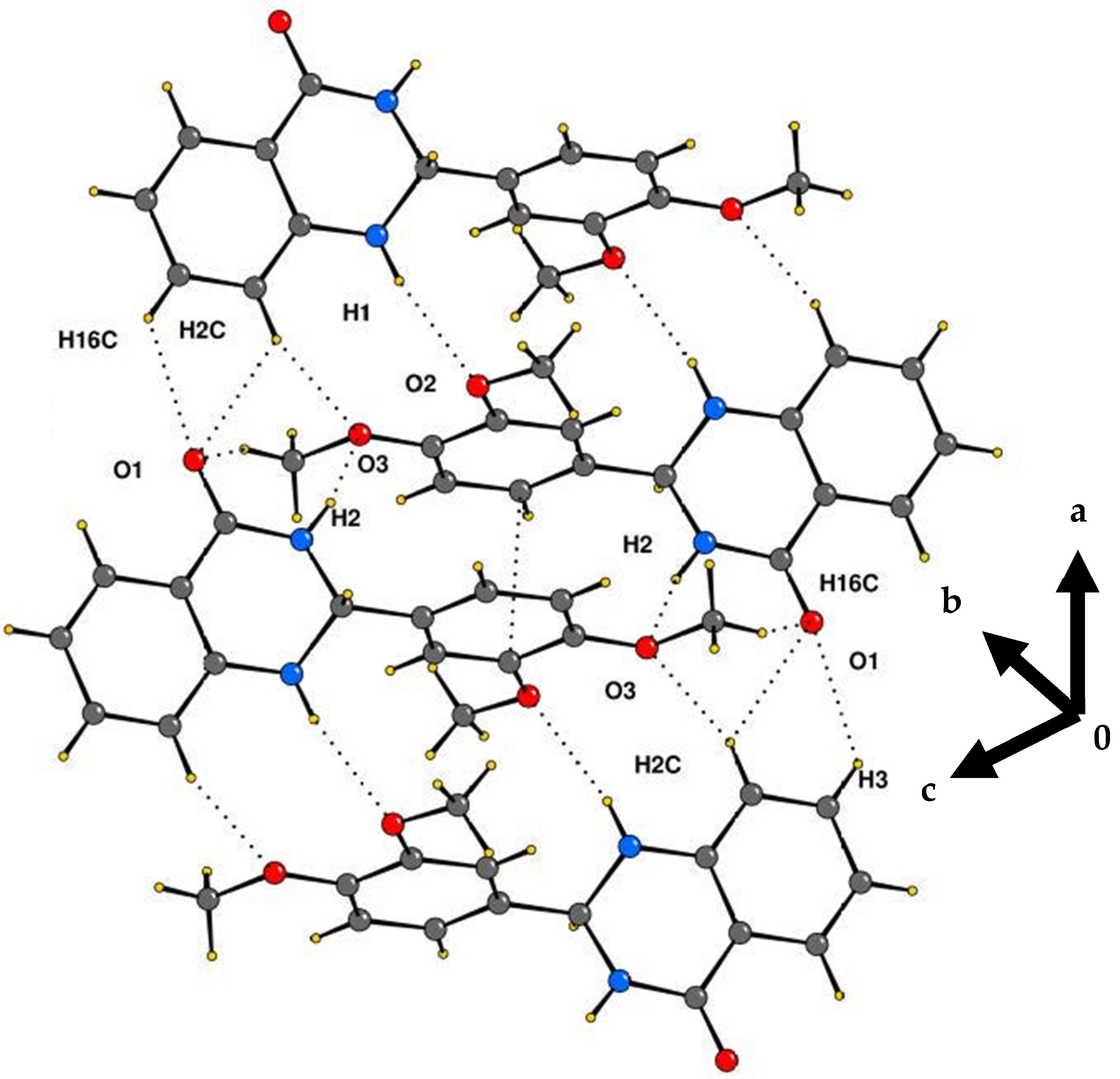
2.2. Crystallography
Crystal Structure Analysis of 2-(3,4-Dimethoxyphenyl)-2,3-dihydroquinazolin-4(1H)-one (3g)
2.3. Larvicidal Activity
2.4. In Silico Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) Analysis
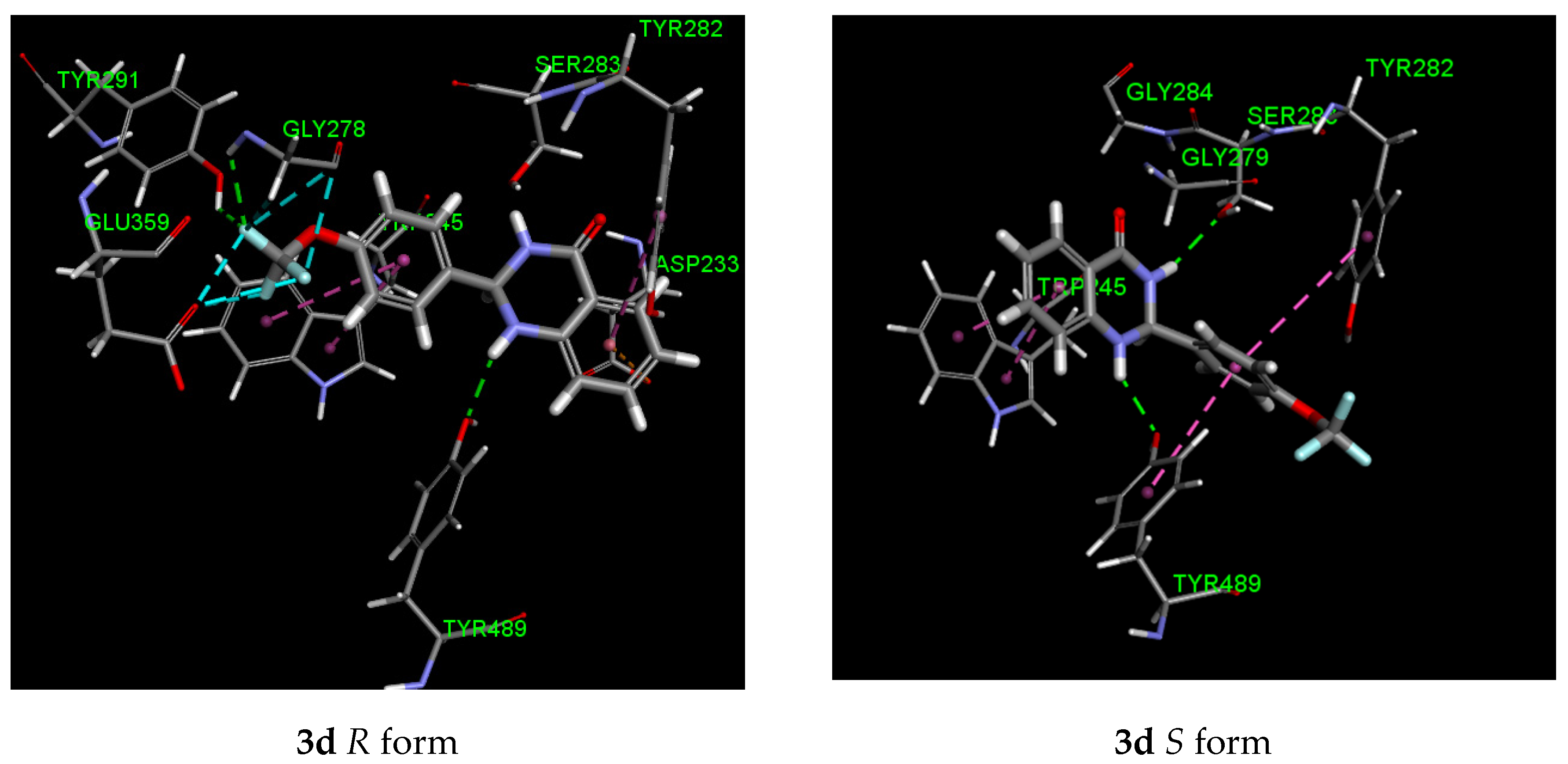
2.5. Molecular Modeling
3. Materials and Methods
3.1. General
3.2. General Procedure for the Synthesis of 2-(Substituted phenyl)-2,3-dihydroquinazolin-4(1H)-ones (3a–n)
3.2.1. 2-(4-Methoxyphenyl)-2, 3-dihydroquinazolin-4(1H)-one (3a)
3.2.2. 4-(4-Oxo-1,2,3,4-tetrahydroquinazolin-2-yl)benzonitrile (3b)
3.2.3. 2-(3-Hydroxyphenyl)-2,3-dihydroquinazolin-4(1H)-one (3c)
3.2.4. 2-(4-(Trifluoromethoxy)phenyl)-2,3-dihydroquinazolin-4(1H)-one (3d)
3.2.5. 2-(3-Iodophenyl)-2,3-dihydroquinazolin-4(1H)-one (3e)
3.2.6. 2-(4-Chlorophenyl)-2,3-dihydroquinazolin-4(1H)-one (3f)
3.2.7. 2-(3,4-Dimethoxyphenyl)-2,3-dihydroquinazolin-4(1H)-one (3g)
3.2.8. 2-(4-(Dimethylamino)phenyl)-2,3-dihydroquinazolin-4(1H)-one (3h)
3.2.9. 2-(3-Nitrophenyl)-2,3-dihydroquinazolin-4(1H)-one (3i)
3.2.10. 2-(4-Fluorophenyl)-2,3-dihydroquinazolin-4(1H)-one (3j)
3.2.11. 2-(1H-Imidazol-4-yl)-2,3-dihydroquinazolin-4(1H)-one (3k)
3.2.12. 2-(2-Methoxy-4-nitrophenyl)-2,3-dihydroquinazolin-4(1H)-one (3l)
3.2.13. 2-(2-Hydroxy-5-nitrophenyl)-2,3-dihydroquinazolin-4(1H)-one (3m)
3.2.14. 2-(4-Hydroxyphenyl)-2,3-dihydroquinazolin-4(1H)-one (3n)
3.3. Crystallographic Studies
3.4. Larvicidal Activity
3.5. Data Analysis
3.6. In silico ADMET Prediction
3.7. Molecular Modeling
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- WHO. World Malaria Report; WHO: Geneva, Switzerland, 2019; Available online: https://www.who.int/publications-detail/world-malaria-report-2019 (accessed on 11 December 2019).
- Venugopala, K.N.; Gleiser, R.M.; Chalannavar, R.K.; Odhav, B. Antimosquito properties of 2-substituted phenyl/benzylamino-6-(4-chlorophenyl)-5-methoxycarbonyl-4-methyl-3,6-dihydropyrimidin-1-ium chlorides against Anopheles arabiensis. Med. Chem. 2014, 10, 211–219. [Google Scholar]
- Mohankumar, T.K.; Shivanna, K.S.; Achuttan, V.V. Screening of Methanolic Plant extracts against larvae of Aedes aegypti and Anopheles stephensi in Mysore. J. Arthropod Borne Dis. 2016, 10, 303–314. [Google Scholar] [PubMed]
- Hemingway, J.; Ranson, H. Insecticide resistance in insect vectors of human disease. Annu. Rev. Entomol. 2000, 45, 371–391. [Google Scholar] [CrossRef] [PubMed]
- Abdalla, H.; Wilding, C.S.; Nardini, L.; Pignatelli, P.; Koekemoer, L.L.; Ranson, H.; Coetzee, M. Insecticide resistance in Anopheles arabiensis in Sudan: Temporal trends and underlying mechanisms. Parasites Vectors 2014, 7, 213. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Sun, R.; Liu, Y.; Chen, P.; Li, Y.; Yang, S.; Wang, Q. Synthesis and insecticidal activity studies of novel phenylpyrazole derivatives containing arylimine or carbimidate moiety. Bioorg. Med. Chem. 2019, 27, 115092. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Liu, Y.; Liu, Y.; Song, H.; Wang, Q. C ring may be dispensable for β-carboline: Design, synthesis, and bioactivities evaluation of tryptophan analog derivatives based on the biosynthesis of β-carboline alkaloids. Bioorg. Med. Chem. 2016, 24, 462–473. [Google Scholar] [CrossRef] [PubMed]
- da Silva, J.B.; Navarro, D.M.; da Silva, A.G.; Santos, G.K.; Dutra, K.A.; Moreira, D.R.; Ramos, M.N.; Espindola, J.W.; de Oliveira, A.D.; Brondani, D.J.; et al. Thiosemicarbazones as Aedes aegypti larvicidal. Eur. J. Med. Chem. 2015, 100, 162–175. [Google Scholar] [CrossRef]
- Castelino, P.A.; Naik, P.; Dasappa, J.P.; Sujayraj, R.S.; Sharath Chandra, K.; Chaluvaiah, K.; Nair, R.; Sandya Kumari, M.V.; Kalthur, G.; Adiga, S.K. Synthesis of novel thiadiazolotriazin-4-ones and study of their mosquito-larvicidal and antibacterial properties. Eur. J. Med. Chem. 2014, 84, 194–199. [Google Scholar] [CrossRef]
- Oliveira, V.S.; Pimenteira, C.; da Silva-Alves, D.C.; Leal, L.L.; Neves-Filho, R.A.; Navarro, D.M.; Santos, G.K.; Dutra, K.A.; dos Anjos, J.V.; Soares, T.A. The enzyme 3-hydroxykynurenine transaminase as potential target for 1,2,4-oxadiazoles with larvicide activity against the dengue vector Aedes aegypti. Bioorg. Med. Chem. 2013, 21, 6996–7003. [Google Scholar] [CrossRef]
- Mao, M.; Li, Y.; Liu, Q.; Zhou, Y.; Zhang, X.; Xiong, L.; Li, Y.; Li, Z. Synthesis and insecticidal evaluation of novel N-pyridylpyrazolecarboxamides containing cyano substituent in the ortho-position. Bioorg. Med. Chem. Lett. 2013, 23, 42–46. [Google Scholar] [CrossRef]
- Abass, M.; Mostafa, B.B. Synthesis and evaluation of molluscicidal and larvicidal activities of some novel enaminones derived from 4-hydroxyquinolinones: Part IX. Bioorg. Med. Chem. 2005, 13, 6133–6144. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.L.; Cha, H.C.; Jahng, Y. Recent advances in the studies on luotonins. Molecules 2011, 16, 4861–4883. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kim, D.-C.; Baek, H.Y.; Lee, K.-D.; Kim, Y.-C.; Oh, H. Anti-neuroinflammatory effects of tryptanthrin from Polygonum tinctorium Lour. in lipopolysaccharide-stimulated BV2 microglial cells. Arch. Pharmacal Res. 2018, 41, 419–430. [Google Scholar] [CrossRef] [PubMed]
- Tian, K.-M.; Li, J.-J.; Xu, S.-W. Rutaecarpine: A promising cardiovascular protective alkaloid from Evodia rutaecarpa (Wu Zhu Yu). Pharmacol. Res. 2019, 141, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Estlin, E.J.; Pinkerton, C.R.; Lewis, I.J.; Lashford, L.; McDowell, H.; Morland, B.; Kohler, J.; Newell, D.R.; Boddy, A.V.; Taylor, G.A.; et al. A phase I study of nolatrexed dihydrochloride in children with advanced cancer. A United Kingdom Children’s Cancer Study Group Investigation. Br. J. Cancer 2001, 84, 11–18. [Google Scholar] [CrossRef][Green Version]
- Gudimella, K.K.; Bonige, K.B.; Gundla, R.; Katari, N.K.; Yamajala, B.; Battula, V.R. 2,4-Diphenyl-1,2-dihydroquinazoline derivatives: Synthesis, anticancer activity and docking studies. ChemistrySelect 2019, 4, 12528–12533. [Google Scholar] [CrossRef]
- Iyer, K.A.; Alix, K.; Eltit, J.M.; Solis, E., Jr.; Pan, X.; Argade, M.D.; Khatri, S.; De Felice, L.J.; Sweet, D.H.; Schulte, M.K.; et al. Multi-modal antidepressant-like action of 6- and 7-chloro-2-aminodihydroquinazolines in the mouse tail suspension test. Psychopharmacology (Berl) 2019, 236, 2093–2104. [Google Scholar] [CrossRef]
- Barmak, A.; Niknam, K.; Mohebbi, G. Synthesis, structural studies, and α-Glucosidase inhibitory, antidiabetic, and antioxidant activities of 2,3-Dihydroquinazolin-4(1H)-ones derived from Pyrazol-4-carbaldehyde and anilines. ACS Omega 2019, 4, 18087–18099. [Google Scholar] [CrossRef]
- Guillon, R.; Pagniez, F.; Picot, C.; Hédou, D.; Tonnerre, A.; Chosson, E.; Duflos, M.; Besson, T.; Logé, C.; Le Pape, P. Discovery of a novel broad-spectrum antifungal agent derived from albaconazole. ACS Med. Chem. Lett. 2013, 4, 288–292. [Google Scholar] [CrossRef]
- Mujeeb Ur, R.; Rathore, A.; Siddiqui, A.A.; Parveen, G.; Yar, M.S. Synthesis and characterization of quinazoline derivatives: Search for hybrid molecule as diuretic and antihypertensive agents. J. Enzym. Inhib Med. Chem. 2014, 29, 733–743. [Google Scholar] [CrossRef]
- Obase, H.; Takai, H.; Teranishi, M.; Nakamizo, N. Synthesis of (1-substituted piperidin-4-yl)-1H-benzimidazoles and (1-substituted piperidin-4-yl)-3,4-dihydroquinazolines as possible antihypertensive agents. J. Heterocycl. Chem. 1983, 20, 565–573. [Google Scholar] [CrossRef]
- El-Sabbagh, O.I.; Ibrahim, S.M.; Baraka, M.M.; Kothayer, H. Synthesis of new 2,3-dihydroquinazolin-4(1H)-one derivatives for analgesic and anti-inflammatory evaluation. Arch. Der Pharm. 2010, 343, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Clissold, S.P.; Beresford, R. Proquazone. A review of its pharmacodynamic and pharmacokinetic properties, and therapeutic efficacy in rheumatic diseases and pain states. Drugs 1987, 33, 478–502. [Google Scholar] [CrossRef] [PubMed]
- Kamal, A.; Babu, K.S.; Poornachandra, Y.; Nagaraju, B.; Ali Hussaini, S.M.; Shaik, S.P.; Ganesh Kumar, C.; Alarifi, A. Efficient and green sulfamic acid catalyzed synthesis of new 1,2-dihydroquinazoline derivatives with antibacterial potential. Arab. J. Chem. 2015. [Google Scholar] [CrossRef]
- Salehi, P.; Ayyari, M.; Bararjanian, M.; Ebrahimi, S.N.; Aliahmadi, A. Synthesis, antibacterial and antioxidant activity of novel 2,3-dihydroquinazolin-4(1H)-one derivatives of dehydroabietylamine diterpene. J. Iran. Chem. Soc. 2014, 11, 607–613. [Google Scholar] [CrossRef]
- Chung, C.K.; Liu, Z.; Lexa, K.W.; Andreani, T.; Xu, Y.; Ji, Y.; DiRocco, D.A.; Humphrey, G.R.; Ruck, R.T. Asymmetric hydrogen bonding catalysis for the synthesis of dihydroquinazoline-containing antiviral, letermovir. J. Am. Chem. Soc. 2017, 139, 10637–10640. [Google Scholar] [CrossRef]
- Mehta, D.R.; Naravane, J.S.; Desai, R.M. Vasicinone. A Bronchodilator Principle from Adhatoda vasica Nees (N. O. Acanthaceae). J. Org. Chem. 1963, 28, 445–448. [Google Scholar] [CrossRef]
- Yamamura, M.; Ochiai, T.; Ishida, R. [Effects of afloqualone, a new centrally acting muscle relaxant, on DRL response and CER in rats (author’s transl)]. Nihon Yakurigaku Zasshi 1981, 78, 381–392. [Google Scholar] [CrossRef]
- Ferrando, C.; Foy, J.M.; Pratt, C.N.F.W.; Purvis, J.R. On the pharmacological actions of a diuretic, fenquizone, with particular reference to its site of action. J. Pharm. Pharmacol. 1981, 33, 219–222. [Google Scholar] [CrossRef]
- Van Zyl, E.F. A survey of reported synthesis of methaqualone and some positional and structural isomers. Forensic Sci. Int. 2001, 122, 142–149. [Google Scholar] [CrossRef]
- Jafari, E.; Khajouei, M.R.; Hassanzadeh, F.; Hakimelahi, G.H.; Khodarahmi, G.A. Quinazolinone and quinazoline derivatives: Recent structures with potent antimicrobial and cytotoxic activities. Res. Pharm. Sci. 2016, 11, 1–14. [Google Scholar] [PubMed]
- Li, W.J.; Li, Q.; Liu, D.L.; Ding, M.W. Synthesis, fungicidal activity, and sterol 14alpha-demethylase binding interaction of 2-azolyl-3,4-dihydroquinazolines on Penicillium digitatum. J. Agric. Food Chem. 2013, 61, 1419–1426. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Feng, Q.; Di, F.; Liu, Q.; Wang, D.; Chen, Y.; Xiong, L.; Song, H.; Li, Y.; Li, Z. Synthesis and insecticidal activities of 2,3-dihydroquinazolin-4(1H)-one derivatives targeting calcium channel. Bioorg. Med. Chem. 2013, 21, 4968–4975. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekharappa, S.; Venugopala, K.N.; Nayak, S.K.; Gleiser, R.M.; García, D.A.; Kumalo, H.M.; Kulkarni, R.S.; Mahomoodally, F.M.; Venugopala, R.; Mohan, M.K.; et al. One-pot microwave assisted synthesis and structural elucidation of novel ethyl 3-substituted-7-methylindolizine-1-carboxylates with larvicidal activity against Anopheles arabiensis. J. Mol. Struct. 2018, 1156, 377–384. [Google Scholar] [CrossRef]
- Bairagi, K.M.; Venugopala, K.N.; Mondal, P.K.; Gleiser, R.M.; Chopra, D.; García, D.; Odhav, B.; Nayak, S.K. Larvicidal study of tetrahydropyrimidine scaffolds against Anopheles arabiensis and structural insight by single crystal X-ray studies. Chem. Biol. Drug Des. 2018, 92, 1924–1932. [Google Scholar] [CrossRef] [PubMed]
- Dharma Rao, B.D.; Bhandary, S.; Chopra, D.; Venugopala, K.N.; Gleiser, R.M.; Kasumbwe, K.; Odhav, B. Synthesis and characterization of a novel series of 1,4-dihydropyridine analogues for larvicidal activity against Anopheles arabiensis. Chem. Biol. Drug Des. 2017, 90, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Borzone, M.E.; Mariani, M.E.; Miguel, V.; Gleiser, R.M.; Odhav, B.; Venugopala, K.N.; Garcia, D.A. Membrane effects of dihydropyrimidine analogues with larvicidal activity. Colloids Surf. B Biointerfaces 2017, 150, 106–113. [Google Scholar] [CrossRef]
- Sandeep, C.; Venugopala, K.N.; Gleiser, R.M.; Chetram, A.; Padmashali, B.; Kulkarni, R.S.; Venugopala, R.; Odhav, B. Greener synthesis of indolizine analogues using water as a base and solvent: Study for larvicidal activity against Anopheles arabiensis. Chem. Biol. Drug Des. 2016, 88, 899–904. [Google Scholar] [CrossRef]
- Venugopala, K.N.; Krishnappa, M.; Nayak, S.K.; Subrahmanya, B.K.; Vaderapura, J.P.; Chalannavar, R.K.; Gleiser, R.M.; Odhav, B. Synthesis and antimosquito properties of 2,6-substituted benzo[d]thiazole and 2,4-substituted benzo[d]thiazole analogues against Anopheles arabiensis. Eur. J. Med. Chem. 2013, 65, 295–303. [Google Scholar] [CrossRef]
- Venugopala, K.N. Synthesis and structural elucidation of novel 2, 4-disubstituted 1, 3-oxazole analogues for pharmacological properties. Asian J. Chem. 2018, 30, 684–688. [Google Scholar] [CrossRef]
- Venugopala, K.N. Design, microwave assisted synthesis and characterization of substituted 1, 2, 4-oxadiazole analogues as promising pharmacological agents. Asian J. Chem. 2017, 29, 1767–1770. [Google Scholar] [CrossRef]
- Venugopala, K.N.; Jayashree, B.S. Microwave-induced synthesis of Schiff bases of aminothiazolyl bromocoumarins as antibacterials. Indian J. Pharm. Sci. 2008, 70, 88–91. [Google Scholar] [CrossRef] [PubMed]
- Kausar, N.; Roy, I.; Chattopadhyay, D.; Das, A.R. Synthesis of 2,3-dihydroquinazolinones and quinazolin-4(3H)-ones catalyzed by graphene oxide nanosheets in an aqueous medium: “on-water” synthesis accompanied by carbocatalysis and selective C–C bond cleavage. RSC Adv. 2016, 6, 22320–22330. [Google Scholar] [CrossRef]
- Bahekar, S.P.; Dahake, N.D.; Sarode, P.B.; Chandak, H.S. Efficient access to 2, 3-dihydroquinazolin-4 (1H)-ones by environmentally benign l-proline nitrate as recyclable catalyst. Synlett 2015, 26, 2575–2577. [Google Scholar] [CrossRef]
- Shiri, L.; Heidari, L.; Kazemi, M. Magnetic Fe3O4 nanoparticles supported imine/Thiophene-nickel (II) complex: A new and highly active heterogeneous catalyst for the synthesis of polyhydroquinolines and 2, 3-dihydroquinazoline-4(1H)-ones. Appl. Organomet. Chem. 2018, 32, e3943. [Google Scholar] [CrossRef]
- Bunce, R.A.; Nammalwar, B. New conditions for synthesis of (±)-2-monosubstituted and (±)-2,2-disubstituted 2,3-dihydro-4(1H)-quinazolinones from 2-nitro- and 2-aminobenzamide. J. Heterocycl. Chem. 2011, 48, 991–997. [Google Scholar] [CrossRef]
- Shiri, L.; Ghorbani-Choghamarani, A.; Kazemi, M. Synthesis and characterization of bromine source supported on magnetic Fe3O4 nanoparticles: A new, versatile and efficient magnetically separable catalyst for organic synthesis. Appl. Organomet. Chem. 2017, 31, e3634. [Google Scholar] [CrossRef]
- Ramesh, R.; Sankar, G.; Małecki, J.; Lalitha, A. Carbon–SO3H derived from glycerol: A green recyclable catalyst for synthesis of 2,3-dihydroquinazolin-4(1H)-ones. J. Iran. Chem. Soc. 2017, 15. [Google Scholar] [CrossRef]
- Dhanunjaya Rao, A.V.; Vykunteswararao, B.P.; Bhaskarkumar, T.; Jogdand, N.R.; Kalita, D.; Lilakar, J.K.D.; Siddaiah, V.; Douglas Sanasi, P.; Raghunadh, A. Sulfonic acid functionalized Wang resin (Wang-OSO3H) as polymeric acidic catalyst for the eco-friendly synthesis of 2,3-dihydroquinazolin-4(1H)-ones. Tetrahedron Lett. 2015, 56, 4714–4717. [Google Scholar] [CrossRef]
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef]
- Loza-Mejia, M.A.; Salazar, J.R.; Sanchez-Tejeda, J.F. In Silico studies on compounds derived from Calceolaria: Phenylethanoid glycosides as potential multitarget inhibitors for the development of pesticides. Biomolecules 2018, 8, 121. [Google Scholar] [CrossRef] [PubMed]
- El Yazal, J.; Rao, S.N.; Mehl, A.; Slikker, W., Jr. Prediction of organophosphorus acetylcholinesterase inhibition using three-dimensional quantitative structure-activity relationship (3D-QSAR) methods. Toxicol. Sci. 2001, 63, 223–232. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cheung, J.; Mahmood, A.; Kalathur, R.; Liu, L.; Carlier, P.R. Structure of the G119S mutant acetylcholinesterase of the malaria vector Anopheles gambiae reveals basis of insecticide resistance. Structure 2018, 26, 130–136.e132. [Google Scholar] [CrossRef] [PubMed]
- Knutsson, S.; Engdahl, C.; Kumari, R.; Forsgren, N.; Lindgren, C.; Kindahl, T.; Kitur, S.; Wachira, L.; Kamau, L.; Ekström, F.; et al. Noncovalent inhibitors of mosquito acetylcholinesterase 1 with resistance-breaking potency. J. Med. Chem. 2018, 61, 10545–10557. [Google Scholar] [CrossRef] [PubMed]
- Dyer, D.H.; Lovell, S.; Thoden, J.B.; Holden, H.M.; Rayment, I.; Lan, Q. The structural determination of an insect sterol carrier protein-2 with a ligand-bound C16 fatty acid at 1.35-A resolution. J. Biol. Chem. 2003, 278, 39085–39091. [Google Scholar] [CrossRef]
- Kim, I.H.; Pham, V.; Jablonka, W.; Goodman, W.G.; Ribeiro, J.M.C.; Andersen, J.F. A mosquito hemolymph odorant-binding protein family member specifically binds juvenile hormone. J. Biol. Chem. 2017, 292, 15329–15339. [Google Scholar] [CrossRef]
- Mans, B.J.; Calvo, E.; Ribeiro, J.M.; Andersen, J.F. The crystal structure of D7r4, a salivary biogenic amine-binding protein from the malaria mosquito Anopheles gambiae. J. Biol. Chem. 2007, 282, 36626–36633. [Google Scholar] [CrossRef]
- Vidadala, R.S.; Ojo, K.K.; Johnson, S.M.; Zhang, Z.; Leonard, S.E.; Mitra, A.; Choi, R.; Reid, M.C.; Keyloun, K.R.; Fox, A.M.; et al. Development of potent and selective Plasmodium falciparum calcium-dependent protein kinase 4 (PfCDPK4) inhibitors that block the transmission of malaria to mosquitoes. Eur. J. Med. Chem. 2014, 74, 562–573. [Google Scholar] [CrossRef][Green Version]
- Taylor, E.A.; Rinaldo-Matthis, A.; Li, L.; Ghanem, M.; Hazleton, K.Z.; Cassera, M.B.; Almo, S.C.; Schramm, V.L. Anopheles gambiae purine nucleoside phosphorylase: Catalysis, structure, and inhibition. Biochemistry 2007, 46, 12405–12415. [Google Scholar] [CrossRef][Green Version]
- Han, Q.; Wong, D.M.; Robinson, H.; Ding, H.; Lam, P.C.H.; Totrov, M.M.; Carlier, P.R.; Li, J. Crystal structure of acetylcholinesterase catalytic subunits of the malaria vector Anopheles gambiae. Insect Sci. 2018, 25, 721–724. [Google Scholar] [CrossRef]
- SAINT Version 7.60a; Bruker AXS Inc.: Madison, WI, USA, 2012.
- Sheldrick, G. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Farrugia, L. WinGX and ORTEP for Windows: An update. J. Appl. Crystallogr. 2012, 45, 849–854. [Google Scholar] [CrossRef]
- Spek, A. PLATON SQUEEZE: A tool for the calculation of the disordered solvent contribution to the calculated structure factors. Acta Crystallogr. Sect. C 2015, 71, 9–18. [Google Scholar] [CrossRef]
- Service, M.W. Management of vectors. In Pest and Vectors Management in Tropics; Youdeowei, A., Service, M.W., Eds.; Longman: London, UK, 1983; pp. 265–280. [Google Scholar]
- DiRienzo, J.A.; Casanoves, F.; Balzarini, M.G.; Gonzalez, L.; Tabalada, M.; Robledo, C.W. InfoStat Version 2014; Group InfoStat, FCA, Universidad Nacional de Cordoba: Argentina. Available online: http://www.infostat.com.ar/ (accessed on 7 December 2019).
- Venugopala, K.N.; Tratrat, C.; Chandrashekharappa, S.; Attimarad, M.; Sreeharsha, N.; Nair, A.B.; Pottathil, S.; Venugopala, R.; Al-Attraqchi, O.H.A.; Morsy, M.A.; et al. Anti-tubercular potency and computationally-assessed drug-likeness and toxicology of diversely substituted indolizines. Indian J. Pharma. Educ. Res. 2019, 53, 545–552. [Google Scholar] [CrossRef]
- Pran Kishore, D.; Anuradha, K.; Poonam, P.; Raghuram Rao, A. Molecular docking and receptor specific 3D-QSAR studies of acetylcholinesterase Inhibitors. Mol. Diver. 2012, 16, 803–823. [Google Scholar]
- Chandrashekharappa, S.; Venugopala, K.N.; Tratrat, C.; Mahomoodally, F.M.; Aldhubiab, B.E.; Haroun, M.; Venugopala, R.; Mohan, M.K.; Kulkarni, R.S.; Attimarad, M.V. Efficient synthesis and characterization of novel indolizines: Exploration of in vitro COX-2 inhibitory activity and molecular modelling studies. New J. Chem. 2018, 42, 4893–4901. [Google Scholar] [CrossRef]
- Available online: https://www.rcsb.org/structure/6ary (accessed on 4 March 2020).
Sample Availability: Samples of the compounds 3a–n are available from the authors. |
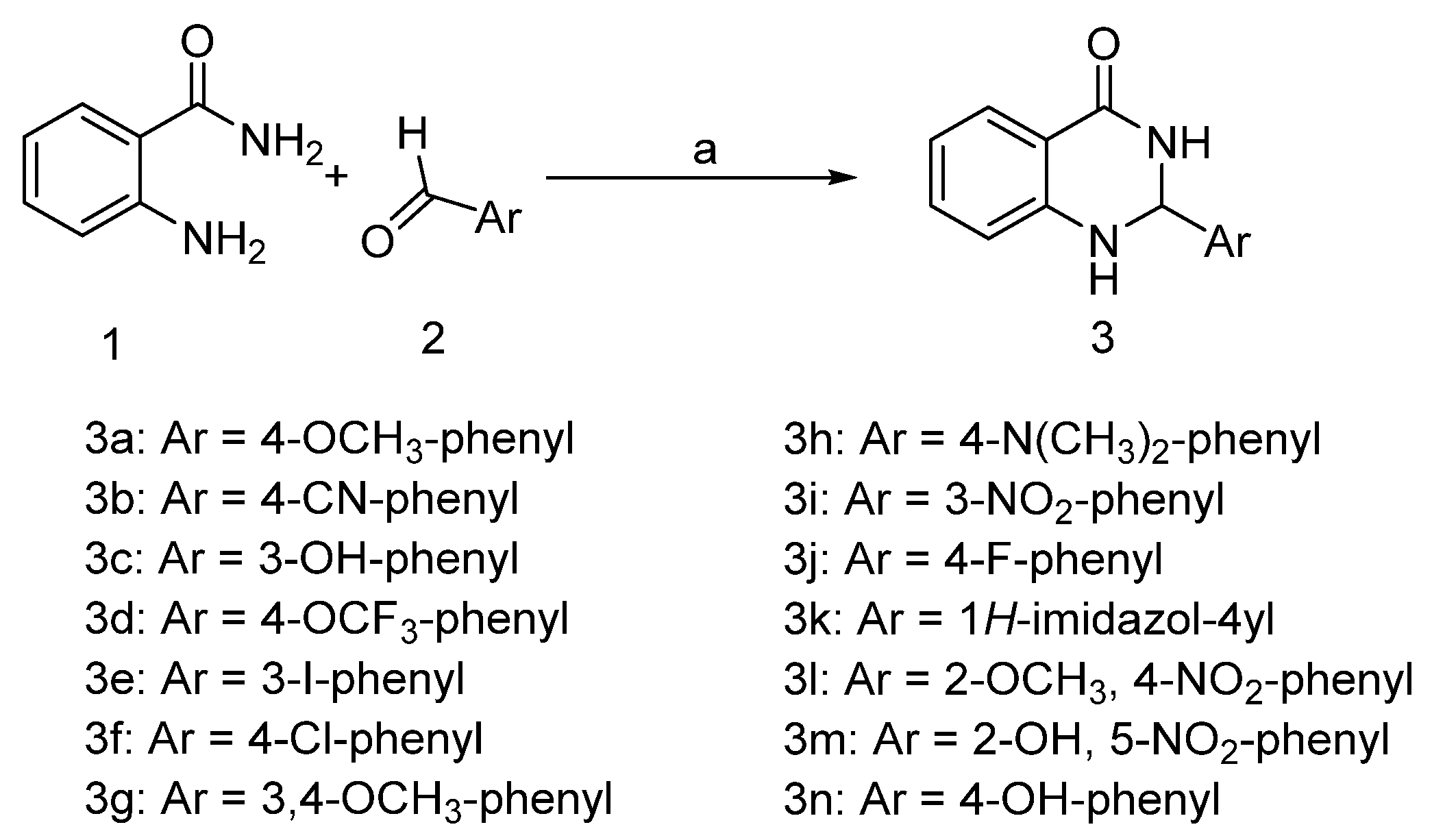
| Compound Code | Ar | Molecular Formula (Molecular Mass) | Yield (%) | m.p (°C) Reported | mp (°C) Found | cLogP | References | CAS Number |
|---|---|---|---|---|---|---|---|---|
| 3a | 4-OCH3-Ph | C15H14N2O2 (254) | 95 | 182–183 | 181–182 | 2.2388 | [45] | 61195-16-2 |
| 3b | 4-CN-Ph | C15H11N3O (249) | 92 | 178–180 | 178–180 | 1.7528 | [44] | 219619-59-7 |
| 3c | 3-OH-Ph | C14H12N2O2 (240) | 91 | 184–186 | 185–187 | 1.6528 | [46] | 107922-06-5 |
| 3d | 4-OCF3-Ph | C15H11F3N2O2 (308) | 94 | 194–196 | 195–196 | 3.3478 | [47] | 685505-75-3 |
| 3e | 3-I-Ph | C14H11IN2O (349) | 92 | -- | 153–155 | 3.4428 | Novel Compound | |
| 3f | 4-Cl-Ph | C14H11ClN2O (258) | 87 | 197–198 | 198–199 | 3.0328 | [44] | 13165-11-2 |
| 3g | 3,4-diOCH3-Ph | C16H16N2O3 (284) | 88 | 214–216 | 213–215 | 1.9778 | [46] | 126492-17-9 |
| 3h | 4-N,N dimethyl-Ph | C16H17N3O (267) | 93 | 209–211 | 210–211 | 2.4848 | [48] | 66181-66-6 |
| 3i | 3-NO2-Ph | C14H11N3O3 (269) | 91 | 192–194 | 192–1193 | 2.0628 | [44] | 26029-30-1 |
| 3j | 4-F-Ph | C14H11FN2O (242) | 91 | 196–198 | 197–198 | 2.4628 | [48] | 359605-44-0 |
| 3k | Imidazole | C11H10N4O (214) | 87 | -- | 156–158 | 0.1488 | Novel Compound | |
| 3l | 2-OCH3-4-NO2-Ph | C15H13N3O4 (299) | 93 | -- | 149–151 | 2.2818 | Novel Compound | |
| 3m | 2-OH, 5-NO2-Ph | C14H11N3O4 (285) | 89 | 244–246 | 245–246 | 1.9818 | [49] | 1794721-69-9 |
| 3n | 4-OH-Ph | C14H12N2O2 (240) | 95 | 209–211 | 210–211 | 1.6528 | [50] | 107920-18-3 |
| CCDC Number | 1983734 |
|---|---|
| Molecular Formula | C16H16N2O3 |
| Molecular weight | 284.31 |
| Temperature | 173(2) |
| Crystal Size (mm) | 0.2, 0.110, 0.040 |
| Absorption coefficient (mm−1) | 0.096 |
| Tmin, Tmax | 0.981, 0.996 |
| Crystal system | Monoclinic |
| Lattice parameters: a (Å), b (Å), c (Å) | 7.8881(6), 20.2194 (13), 8.9638 (6) |
| α, β, γ (°) | 90, 104.887(3), 90 |
| Space Group, Density, Z, Z′ | P21/n, 1.367, 4, 1 |
| hmin, max; kmin, max; lmin, max; | −8, 10; −26, 26; −11, 11 |
| Number of total/unique/observed reflections | 13,538, 3403, 2321 |
| No. of parameters | 200 |
| Independent reflections, Rint | 3403, 0.0482 |
| Rall, Robs | 0.0879, 0.0544 |
| wR2all, wR2obs | 0.1425, 0.1221 |
| Δρmax, min (eÅ−3) | 0.541, −0.269 |
| goodness-of-fit (G.o.F) | 1.029 |
| Interaction | H···A (Å) | D···A (Å) | ∠D–H···A (°) | Symmetry Code |
|---|---|---|---|---|
| N1–H1···O2 | 2.30 | 3.176 | 170 | −x + 1, −y, −z + 2 |
| N2–H2···O3 | 2.30 | 3.070 | 148 | −x, −y, −z + 2 |
| C16–H16C···O1 | 2.66 | 3.441 | 137 | −x, −y, −z + 2 |
| C3–H3···O1 | 2.54 | 3.167 | 124 | x + 1,+y,+z |
| C2–H2C···O1 | 2.57 | 3.180 | 122 | x + 1,+y,+z |
| C2–H2C···O3 | 2.64 | 3.421 | 140 | −x + 1, −y, −z + 2 |
| Compound | 24-h Mortality | 48-h Mortality |
|---|---|---|
| 3a (4-OCH3) | 0.77 ± 0.04 d,e,f | 0.83 ± 0.04 c,d,e |
| 3b (4-CN) | 0.86 ± 0.04 b,c,d,e | 0.88 ± 0.03 b,c,d |
| 3c (3-OH) | 0.51 ± 0.05 h,i,j,k,l,m | 0.57 ± 0.05 h,i,j,k,l |
| 3d (4-OCF3) | 0.91 ± 0.03 b,c | 0.93 ± 0.03 a,b |
| 3e (3-I) | 0.82 ± 0.04 c,d,e | 0.87 ± 0.04 b,c,d |
| 3f (4-Cl) | 0.87 ± 0.04 b,c,d | 0.91 ± 0.03 b,c |
| 3g (3,4-diOCH3) | 0.62 ± 0.05 g,h,i,j | 0.66 ± 0.05 f,g,h |
| 3h (4-N(CH3)2) | 0.41 ± 0.05 m | 0.43 ± 0.05 l,m |
| 3i (3-NO2) | 0.52 ± 0.05 h,i,j,k,l,m | 0.54 ± 0.05 h,i,j,k,l,m |
| 3j (4-F) | 0.89 ± 0.03 b,c | 0.91 ± 0.03 b,c |
| 3k (imidazole) | 0.46 ± 0.05 k,l,m | 0.48 ± 0.05 j,k,l,m |
| 3l (2-OCH3, 4-NO2) | 0.74 ± 0.05 e,f,g | 0.77 ± 0.04 d,e,f |
| 3m (2-OH, 5-NO2) | 0.59 ± 0.05 h,i,j,k | 0.64 ± 0.05 f,g,h,i |
| 3n (4-OH) | 0.50 ± 0.05 i,j,k,l,m | 0.52 ± 0.05 h,i,j,k,l,m |
| Temephos | 0.99 ± 0.01 a | 1.00 ± 0.00 a |
| Acetone | 0.01 ± 0.01 m | 0.04 ± 0.02 n |
| Entry | Solubility Level | Blood–Brain Barrier Penetration (BBB) Level | CYP450 Inhibition | Hepatotoxicity | Intestinal Absorption Level | Plasma Protein Binding | AlogP | PSA 2D |
|---|---|---|---|---|---|---|---|---|
| Temephos | 2 | 0 | - | + | 0 | + | 5.656 | 53.58 |
| 3a (4-OCH3) | 3 | 2 | - | + | 0 | + | 2.432 | 51.851 |
| 3b (4-CN) | 3 | 2 | - | + | 0 | + | 2.327 | 65.856 |
| 3c (3-OH) | 3 | 2 | - | + | 0 | + | 2.206 | 63.736 |
| 3d (4-OCF3) | 1 | 1 | - | + | 0 | + | 4.568 | 51.851 |
| 3e (3-I) | 2 | 1 | - | + | 0 | + | 3.026 | 42.921 |
| 3f (4-Cl) | 2 | 1 | - | + | 0 | + | 3.113 | 42.921 |
| 3g (3,4-diOCH3) | 3 | 2 | - | + | 0 | + | 2.415 | 60.781 |
| 3h (4-N(CH3)2) | 3 | 2 | - | + | 0 | + | 2.610 | 46.273 |
| 3i (3-NO2) | 3 | 3 | - | + | 0 | + | 2.343 | 85.744 |
| 3j (4-F) | 3 | 2 | - | + | 0 | + | 2.654 | 42.921 |
| 3k (imidazole) | 4 | 3 | - | + | 0 | - | 0.632 | 69.237 |
| 3l (2-OCH3, 4-NO2) | 3 | 2 | - | + | 0 | + | 2.326 | 94.674 |
| 3m (2-OH, 5-NO2) | 3 | 3 | - | + | 0 | + | 2.101 | 106.559 |
| 3n (4-OH) | 3 | 2 | - | + | 0 | + | 2.206 | 63.736 |
| Entry | Larvicidal Activity | Stereoisomer | Binding Energy (kJ/mol) | Residues Interaction | |
|---|---|---|---|---|---|
| H-Bond (Interacting Atom) | Hydrophobic (π–π) | ||||
| Native ligand | - | - | −46.68 | Ser 280, Tyr 282 | Gly 279, Trp 285, Tyr 489 |
| Temephos | - | - | 0 | - | - |
| 3a (4-OCH3) | 0.77 | R | −93.21 | Trp 245 (NH), Tyr 489 (NHCO) | Trp 245, His 600 |
| S | −114.50 | Gly 279 (NHCO), Ser 280 (NHCO) Tyr 489 (NH) | Tyr 282, Trp 245, Tyr 489 | ||
| 3b (4-CN) | 0.86 | R | −98.03 | Ser 283 (NHCO), Tyr 489 (NH) Tyr 291 (CN) | Trp 245, Tyr 282, Asp 233 (ion-pi) |
| S | −81.74 | Trp 245 (NH), Tyr 291 (CN) Gly 278 (CN), Tyr 282 (NHCO) Tyr 489 (NHCO) | Trp 245 | ||
| 3c (3-OH) | 0.51 | R | −99.16 | Ser 289 (NHCO), Tyr 291 (OH) Glu 359 (OH) | Trp 245, Tyr 489 |
| S | −89.42 | Ser 280 (OH), Tyr 282 (OH) Ser 283 (NHCO), Tyr 289 (NH) | Trp 245, Tyr 489 | ||
| 3d (4-OCF3) | 0.91 | R | −110.82 | Gly 278 (CF3), Tyr 291 (CF3) Tyr 489 (NH) | Trp 245, Tyr 282, Asp 233 (ion-pi), Glu 359 (F), Gly 278 (F) |
| S | −106.47 | Ser 283 (NHCO) Tyr 489 (NH) | Trp 245, Tyr 282, Tyr 489 | ||
| 3e (3-I) | 0.82 | R | −105.04 | Tyr 489 (NH) | Trp 245, Tyr 282, Tyr 489, Asn 246 (I) |
| S | −75.88 | Trp 245 (NH) | Trp 245 | ||
| 3f (4-Cl) | 0.87 | R | −108.19 | Ser 283 (NHCO) Tyr 489 (NH) | Tyr 282, Trp 245, Asp 233 (ion-pi) |
| S | −81.59 | Trp 245 (NH), Tyr 282 (NHCO) Tyr 489 (NHCO), Glu 359 (Cl) | Tyr 245 | ||
| 3g (3,4-diOCH3) | 0.62 | R | −95.90 | Ser 283 (NHCO) Tyr 489 (NH) | Trp 245, Tyr 282, Tyr 489 |
| S | −91.29 | Trp 245 (NH), Tyr 282 (NHCO) Tyr 489 (NHCO) | Tyr 245 | ||
| 3h (4-N(CH3)2) | 0.41 | R | −92.98 | Tyr 489 (NH) | Trp 245, Tyr 282, Tyr 489 |
| S | −91.01 | Ser 283 (NHCO) Tyr 489 (NH) | Trp 245, Tyr 489 | ||
| 3i (3-NO2) | 0.52 | R | −108.38 | Asp 233 (NO2), Ser 283 (NHCO) Tyr 489 (NH) | Trp 245, Tyr 282, Tyr 489 |
| S | −131.24 | Ser 280 (NHCO) Tyr 489 (NH) | Trp 245, Tyr 282 | ||
| 3j (4-F) | 0.89 | R | −101.15 | Ser 283 (NHCO) Tyr 489 (NH) | Trp 245, Tyr 489 |
| S | −76.85 | Tyr 291 (NHCO) Tyr 489 (NH) | Trp 245, Tyr 489 | ||
| 3k (imidazole) | 0.46 | R | −91.64 | Ser 283 (NHCO) Tyr 489 (NH) | Trp 245 |
| S | −80.14 | Ser 283 (NHCO) Tyr 489 (NH) | Trp 245, Tyr 489 | ||
| 3l (2-OCH3, 4-NO2) | 0.74 | R | −120.40 | Gly 279 (NHCO), Ser 289 (NHCO) Ser 289 (OCH3) | Trp 245, Tyr 489, His 600, Glu 359 (ion-pi) |
| S | −89.15 | Ser 283 (NHCO) Tyr 489 (NH) | Trp 245, Tyr 282, Tyr 489, Tyr 493 (ion-pi) | ||
| 3m (2-OH, 5-NO2) | 0.59 | R | −127.01 | Ser 283 (NHCO), Tyr 489 (NH) Tyr 489 (OH) | Trp 245, Tyr 282, Tyr 489 |
| S | −113.14 | Trp 245 (NH) Ser 283 (OH) | Trp 245, Glu 359 (ion-pi), His 600 (ion-pi) | ||
| 3n (4-OH) | 0.50 | R | −91.36 | Ser 280 (NHCO), Tyr 291 (OH) Glu 359 (OH) | Trp 245, Tyr 282, Tyr 489 |
| S | −121.58 | Ser 280 (NHCO), Tyr 291 (OH) Glu 359 (OH) | Trp 245 | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Venugopala, K.N.; Ramachandra, P.; Tratrat, C.; Gleiser, R.M.; Bhandary, S.; Chopra, D.; Morsy, M.A.; Aldhubiab, B.E.; Attimarad, M.; Nair, A.B.; et al. Larvicidal Activities of 2-Aryl-2,3-Dihydroquinazolin -4-ones against Malaria Vector Anopheles arabiensis, In Silico ADMET Prediction and Molecular Target Investigation. Molecules 2020, 25, 1316. https://doi.org/10.3390/molecules25061316
Venugopala KN, Ramachandra P, Tratrat C, Gleiser RM, Bhandary S, Chopra D, Morsy MA, Aldhubiab BE, Attimarad M, Nair AB, et al. Larvicidal Activities of 2-Aryl-2,3-Dihydroquinazolin -4-ones against Malaria Vector Anopheles arabiensis, In Silico ADMET Prediction and Molecular Target Investigation. Molecules. 2020; 25(6):1316. https://doi.org/10.3390/molecules25061316
Chicago/Turabian StyleVenugopala, Katharigatta N., Pushpalatha Ramachandra, Christophe Tratrat, Raquel M. Gleiser, Subhrajyoti Bhandary, Deepak Chopra, Mohamed A. Morsy, Bandar E. Aldhubiab, Mahesh Attimarad, Anroop B. Nair, and et al. 2020. "Larvicidal Activities of 2-Aryl-2,3-Dihydroquinazolin -4-ones against Malaria Vector Anopheles arabiensis, In Silico ADMET Prediction and Molecular Target Investigation" Molecules 25, no. 6: 1316. https://doi.org/10.3390/molecules25061316
APA StyleVenugopala, K. N., Ramachandra, P., Tratrat, C., Gleiser, R. M., Bhandary, S., Chopra, D., Morsy, M. A., Aldhubiab, B. E., Attimarad, M., Nair, A. B., Sreeharsha, N., Venugopala, R., Deb, P. K., Chandrashekharappa, S., Khalil, H. E., Alwassil, O. I., Abed, S. N., Bataineh, Y. A., Palenge, R., ... Mohanlall, V. (2020). Larvicidal Activities of 2-Aryl-2,3-Dihydroquinazolin -4-ones against Malaria Vector Anopheles arabiensis, In Silico ADMET Prediction and Molecular Target Investigation. Molecules, 25(6), 1316. https://doi.org/10.3390/molecules25061316