Effects of Propolis and Phenolic Acids on Triple-Negative Breast Cancer Cell Lines: Potential Involvement of Epigenetic Mechanisms
Abstract
:1. Introduction
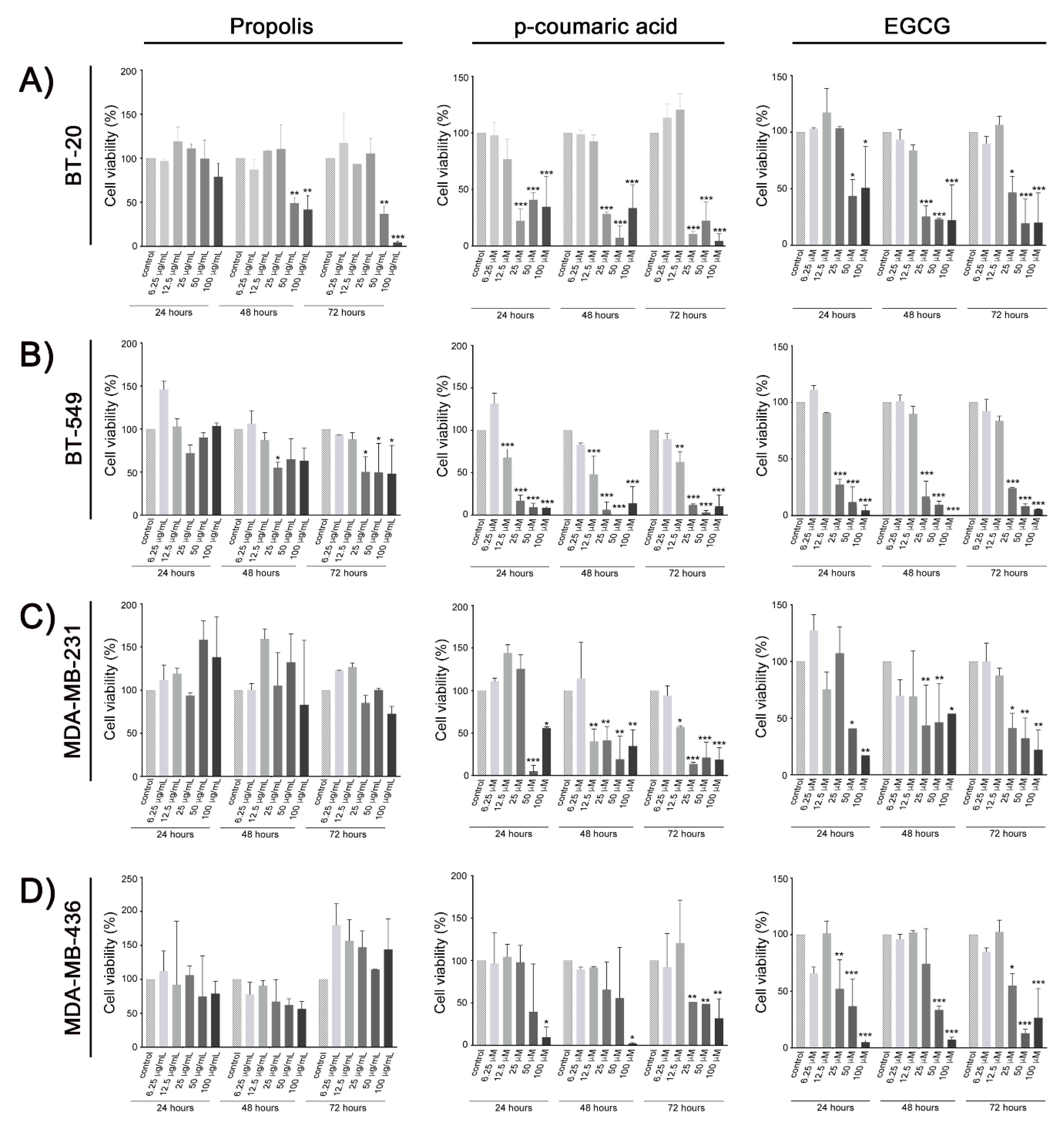
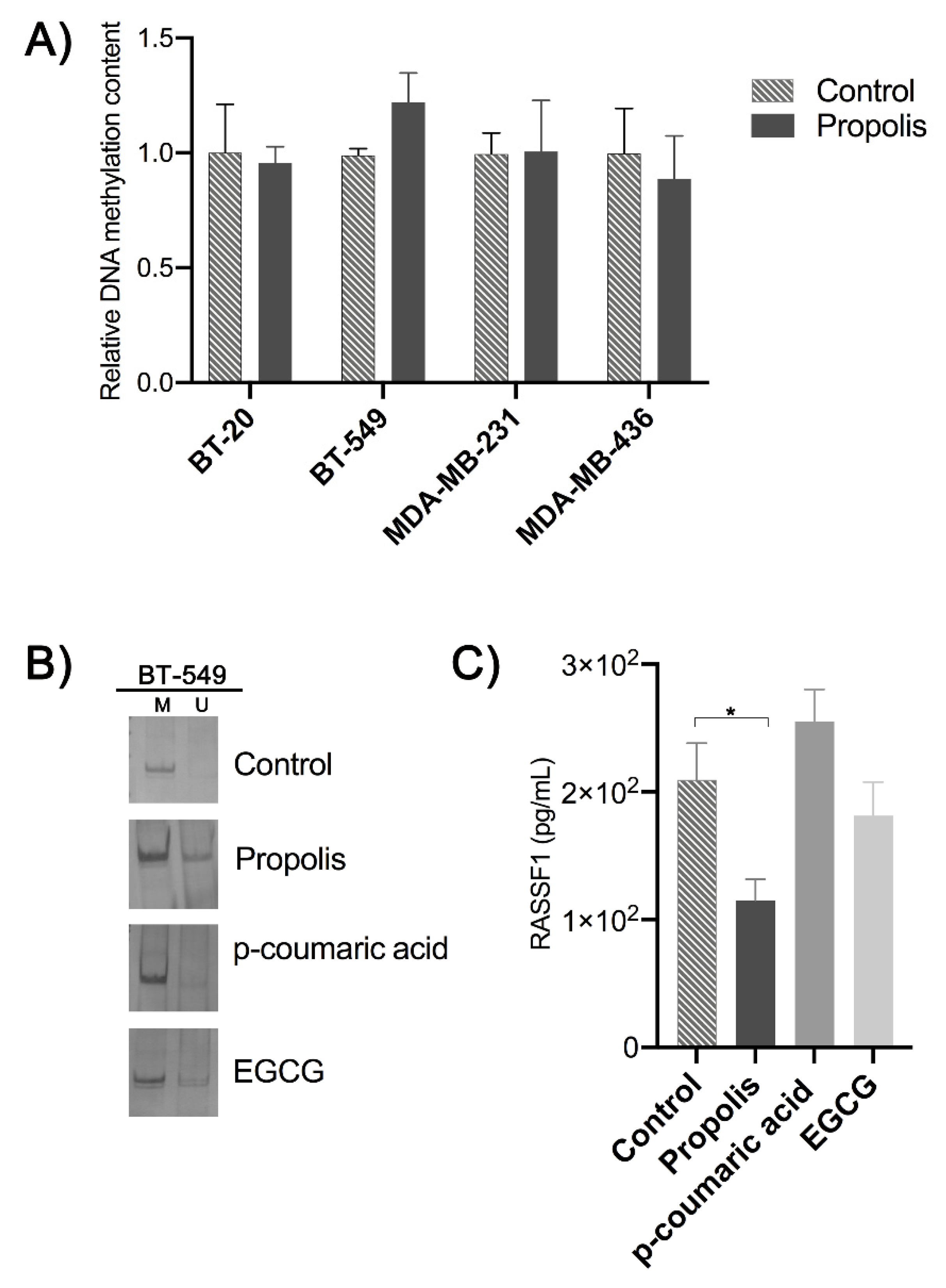
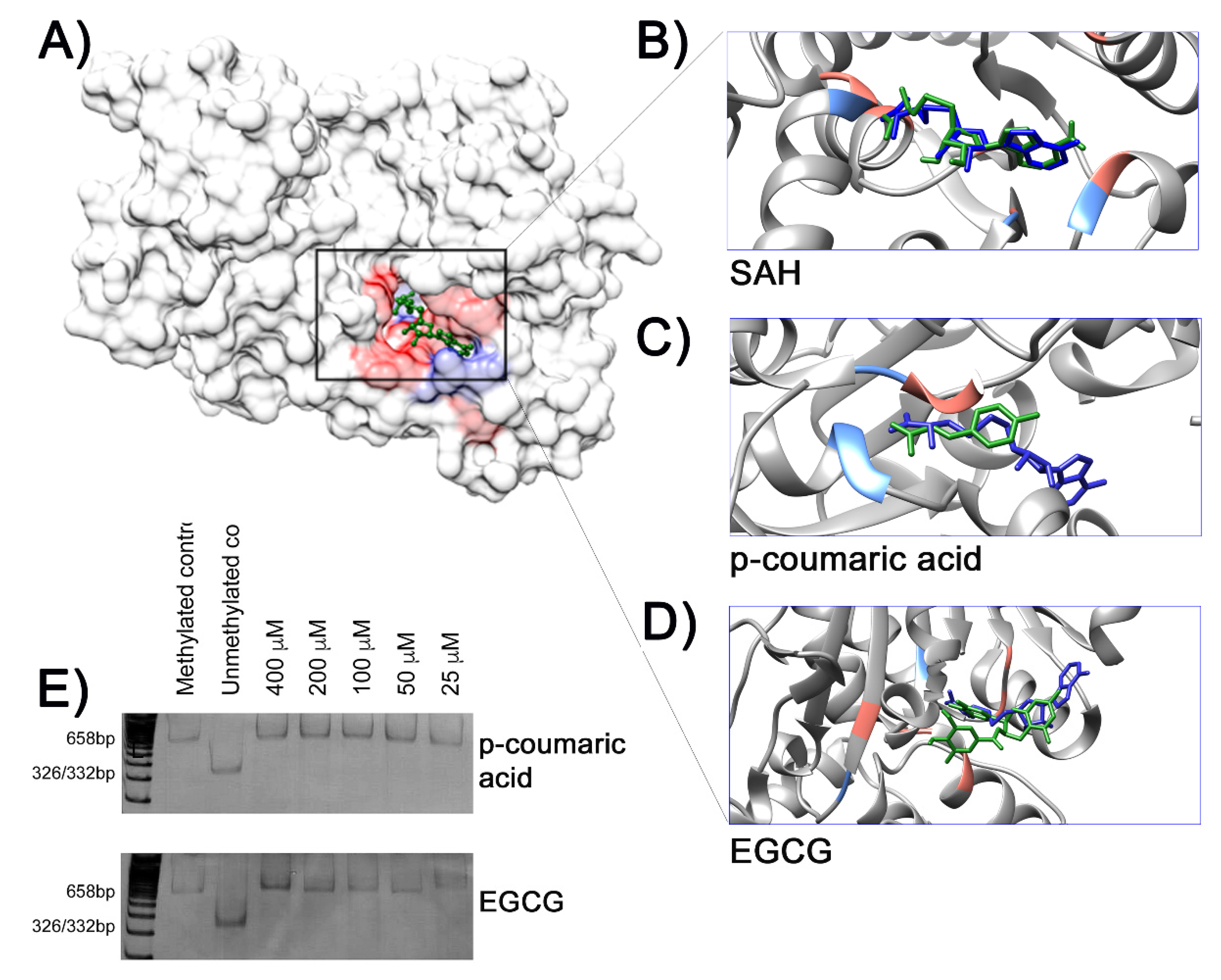
2. Results
3. Discussion
4. Materials and Methods
4.1. Propolis Sample and Chemical Compounds
4.2. Cell Lines and Cell Culture
4.3. Cell Viability Assay
4.4. In itro Treatments and DNA Extraction
4.5. Global DNA Methylation Content
4.6. Methylation-Specific Polymerase Chain Reaction of RASSF1A Promoter
4.7. Expression of the Protein RASSF1
4.8. Molecular Docking
4.9. In Vitro Inhibition of the CpG Methylase M.SssI Assay
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bankova, V. Chemical diversity of propolis and the problem of standardization. J. Ethnopharmacol. 2005, 100, 114–117. [Google Scholar] [CrossRef] [PubMed]
- Bankova, V.; Popova, M.; Trusheva, B. Propolis volatile compounds: Chemical diversity and biological activity: A review. Chem. Cent. J. 2014, 8, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sforcin, J.M. Biological Properties and therapeutic applications of propolis. Phytother Res. 2016, 30, 894–905. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.A.; Amarante, M.K.; Conti, B.J.; Sforcin, J.M. Cytotoxic constituents of propolis inducing anticancer effects: A review. J. Pharm. Pharmacol. 2011, 63, 1378–1386. [Google Scholar] [CrossRef] [PubMed]
- Szliszka, E.; Mertas, A.; Czuba, Z.P.; Król, W. Inhibition of Inflammatory Response by Artepillin C in Activated RAW264.7 Macrophages. Evid. Based Complement. Altern. Med. 2013, 2013, 735176. [Google Scholar] [CrossRef]
- Noronha, V.R.; Araujo, G.S.; Gomes, R.T.; Iwanaga, S.H.; Barbosa, M.C.; Abdo, E.N.; Ferreira e Ferreira, E.; Viana Campos, A.C.; Souza, A.A.; Abreu, S.R.; et al. Mucoadhesive propolis gel for prevention of radiation-induced oral mucositis. Curr. Clin. Pharm. 2014, 9, 359–364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cornara, L.; Biagi, M.; Xiao, J.; Burlando, B. Therapeutic Properties of Bioactive Compounds from Different Honeybee Products. Front. Pharmacol. 2017, 8, 412. [Google Scholar] [CrossRef]
- Sforcin, J.M.; Bankova, V. Propolis: Is there a potential for the development of new drugs? J. Ethnopharmacol. 2011, 133, 253–260. [Google Scholar] [CrossRef]
- Anjum, S.I.; Ullah, A.; Khan, K.A.; Attaullah, M.; Khan, H.; Ali, H.; Bashir, M.A.; Tahir, M.; Ansari, M.J.; Ghramh, H.A.; et al. Composition and functional properties of propolis (bee glue): A review. Saudi J. Biol. Sci. 2019, 26, 1695–1703. [Google Scholar] [CrossRef]
- Jung, B.I.; Kim, M.S.; Kim, H.A.; Kim, D.; Yang, J.; Her, S.; Song, Y.S. Caffeic acid phenethyl ester, a component of beehive propolis, is a novel selective estrogen receptor modulator. Phytother Res. 2010, 24, 295–300. [Google Scholar] [CrossRef]
- Seda Vatansever, H.; Sorkun, K.; Ismet Deliloğlu Gurhan, S.; Ozdal-Kurt, F.; Turkoz, E.; Gencay, O.; Salih, B. Propolis from Turkey induces apoptosis through activating caspases in human breast carcinoma cell lines. Acta Histochem. 2010, 112, 546–556. [Google Scholar] [CrossRef] [PubMed]
- Ishihara, M.; Naoi, K.; Hashita, M.; Itoh, Y.; Suzui, M. Growth inhibitory activity of ethanol extracts of Chinese and Brazilian propolis in four human colon carcinoma cell lines. Oncol. Rep. 2009, 22, 349–354. [Google Scholar] [PubMed]
- Li, F.; Awale, S.; Tezuka, Y.; Kadota, S. Cytotoxicity of constituents from Mexican propolis against a panel of six different cancer cell lines. Nat. Prod. Commun. 2010, 5, 1601–1606. [Google Scholar] [CrossRef] [Green Version]
- Watabe, M.; Hishikawa, K.; Takayanagi, A.; Shimizu, N.; Nakaki, T. Caffeic acid phenethyl ester induces apoptosis by inhibition of NFkappaB and activation of Fas in human breast cancer MCF-7 cells. J. Biol. Chem. 2004, 279, 6017–6026. [Google Scholar] [CrossRef] [Green Version]
- Kouidhi, B.; Zmantar, T.; Bakhrouf, A. Anti-cariogenic and anti-biofilms activity of Tunisian propolis extract and its potential protective effect against cancer cells proliferation. Anaerobe 2010, 16, 566–571. [Google Scholar] [CrossRef] [PubMed]
- Szliszka, E.; Czuba, Z.P.; Domino, M.; Mazur, B.; Zydowicz, G.; Krol, W. Ethanolic extract of propolis (EEP) enhances the apoptosis- inducing potential of TRAIL in cancer cells. Molecules 2009, 14, 738–754. [Google Scholar] [CrossRef]
- Lee, Y.; Shin, D.H.; Kim, J.H.; Hong, S.; Choi, D.; Kim, Y.J.; Kwak, M.K.; Jung, Y. Caffeic acid phenethyl ester-mediated Nrf2 activation and IkappaB kinase inhibition are involved in NFkappaB inhibitory effect: Structural analysis for NFkappaB inhibition. Eur. J. Pharmacol. 2010, 643, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, I.; Hosoya, T.; Ushida, M.; Kunimasa, K.; Ohta, T.; Kumazawa, S. Nymphaeol-A Isolated from Okinawan Propolis Suppresses Angiogenesis and induces caspase-dependent apoptosis via inactivation of survival signals. Evid Based Complement. Altern. Med. 2013, 2013, 826245. [Google Scholar] [CrossRef] [Green Version]
- Peng, C.Y.; Yang, H.W.; Chu, Y.H.; Chang, Y.C.; Hsieh, M.J.; Chou, M.Y.; Yeh, K.T.; Lin, Y.M.; Yang, S.F.; Lin, C.W. Caffeic Acid phenethyl ester inhibits oral cancer cell metastasis by regulating matrix metalloproteinase-2 and the mitogen-activated protein kinase pathway. Evid. Based Complement. Altern. Med. 2012, 2012, 732578. [Google Scholar] [CrossRef]
- Sulaiman, G.M.; Ad’hiah, A.H.; Al-Sammarrae, K.W.; Bagnati, R.; Frapolli, R.; Bello, E.; Uboldi, S.; Romano, M.; Panini, N.; Scanziani, E.; et al. Assessing the anti-tumour properties of Iraqi propolis in vitro and in vivo. Food Chem. Toxicol. 2012, 50, 1632–1641. [Google Scholar] [CrossRef]
- Liu, Z.; Xie, Z.; Jones, W.; Pavlovicz, R.E.; Liu, S.; Yu, J.; Li, P.K.; Lin, J.; Fuchs, J.R.; Marcucci, G.; et al. Curcumin is a potent DNA hypomethylation agent. Bioorg. Med. Chem Lett. 2009, 19, 706–709. [Google Scholar] [CrossRef]
- Lee, W.J.; Shim, J.Y.; Zhu, B.T. Mechanisms for the inhibition of DNA methyltransferases by tea catechins and bioflavonoids. Mol. Pharm. 2005, 68, 1018–1030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoo, C.B.; Jones, P.A. Epigenetic therapy of cancer: Past, present and future. Nat. Rev. Drug Discov. 2006, 5, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Burbee, D.G.; Forgacs, E.; Zöchbauer-Müller, S.; Shivakumar, L.; Fong, K.; Gao, B.; Randle, D.; Kondo, M.; Virmani, A.; Bader, S.; et al. Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J. Natl. Cancer Inst. 2001, 93, 691–699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, M.A.; Hussain, A.; Sundaram, M.K.; Alalami, U.; Gunasekera, D.; Ramesh, L.; Hamza, A.; Quraishi, U. (-)-Epigallocatechin-3-gallate reverses the expression of various tumor-suppressor genes by inhibiting DNA methyltransferases and histone deacetylases in human cervical cancer cells. Oncol. Rep. 2015, 33, 1976–1984. [Google Scholar] [CrossRef] [Green Version]
- Omene, C.; Kalac, M.; Wu, J.; Marchi, E.; Frenkel, K.; O’Connor, O.A. Propolis and its Active Component, Caffeic Acid Phenethyl Ester (CAPE), Modulate Breast Cancer Therapeutic Targets via an Epigenetically Mediated Mechanism of Action. J. Cancer Sci. Ther. 2013, 5, 334–342. [Google Scholar]
- Shin, E.J.; Jo, S.; Choi, H.K.; Choi, S.; Byun, S.; Lim, T.G. Caffeic Acid Phenethyl Ester Inhibits UV-Induced MMP-1 Expression by Targeting Histone Acetyltransferases in Human Skin. Int. J. Mol. Sci. 2019, 20, 3055. [Google Scholar] [CrossRef] [Green Version]
- Lee, W.J.; Zhu, B.T. Inhibition of DNA methylation by caffeic acid and chlorogenic acid, two common catechol-containing coffee polyphenols. Carcinogenesis 2006, 27, 269–277. [Google Scholar] [CrossRef] [Green Version]
- Grawenda, A.M.; O’Neill, E. Clinical utility of RASSF1A methylation in human malignancies. Br. J. Cancer 2015, 113, 372–381. [Google Scholar] [CrossRef] [Green Version]
- Nedeljković, M.; Damjanović, A. Mechanisms of Chemotherapy Resistance in Triple-Negative Breast Cancer-How We Can Rise to the Challenge. Cells 2019, 8, 957. [Google Scholar] [CrossRef] [Green Version]
- Frión-Herrera, Y.; Díaz-García, A.; Ruiz-Fuentes, J.; Rodríguez-Sánchez, H.; Sforcin, J.M. The cytotoxic effects of propolis on breast cancer cells involve PI3K/Akt andERK1/2 pathways, mitochondrial membrane potential, and reactive oxygen species generation. Inflammopharmacology 2019, 27, 1081–1089. [Google Scholar] [CrossRef] [Green Version]
- Farabegoli, F.; Govoni, M.; Spisni, E.; Papi, A. EGFR inhibition by (-)-epigallocatechin-3-gallate and IIF treatments reduces breast cancer cell invasion. Biosci. Rep. 2017, 37, BSR20170168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Negri, A.; Naponelli, V.; Rizzi, F.; Bettuzzi, S. Molecular Targets of Epigallocatechin-Gallate (EGCG): A Special Focus on Signal Transduction and Cancer. Nutrients 2018, 10, 1936. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.H.; Rajamanickam, V.; Nagarajan, S. Supplementation of p-coumaric acid exhibits chemopreventive effect via induction of Nrf2 in a short-term preclinical model of colon cancer. Eur. J. Cancer Prev. 2019, 28, 472–482. [Google Scholar] [CrossRef]
- Roy, N.; Narayanankutty, A.; Nazeem, P.A.; Valsalan, R.; Babu, T.D.; Mathew, D. Plant Phenolics Ferulic Acid and P-Coumaric Acid Inhibit Colorectal Cancer Cell Proliferation through EGFR Down-Regulation. Asian Pac. J. Cancer Prev. 2016, 17, 4019–4023. [Google Scholar]
- Rosa, L.S.; Jordão, N.A.; da Costa Pereira Soares, N.; de Mesquita, J.F.; Monteiro, M.; Teodoro, A.J. Pharmacokinetic, Antiproliferative and Apoptotic Effects of Phenolic Acids in Human Colon Adenocarcinoma Cells Using In Vitro and In Silico Approaches. Molecules 2018, 23, 2569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saenglee, S.; Jogloy, S.; Patanothai, A.; Leid, M.; Senawong, T. Cytotoxic effects of peanut phenolics possessing histone deacetylase inhibitory activity in breast and cervical cancer cell lines. Pharmacol. Rep. 2016, 68, 1102–1110. [Google Scholar] [CrossRef] [PubMed]
- Jeltsch, A.; Broche, J.; Bashtrykov, P. Molecular Processes Connecting DNAMethylation Patterns with DNA Methyltransferases and Histone Modifications in Mammalian Genomes. Genes 2018, 9, 566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dawson, M.A.; Kouzarides, T. Cancer epigenetics: From mechanism to therapy. Cell 2012, 150, 12–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okano, M.; Bell, D.W.; Haber, D.A.; Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999, 99, 247–257. [Google Scholar] [CrossRef] [Green Version]
- Jurkowska, R.Z.; Jurkowski, T.P.; Jeltsch, A. Structure and Function of Mammalian DNA Methyltransferases. Chem. Biochem. 2011, 12, 206–222. [Google Scholar] [CrossRef] [PubMed]
- Saldívar-González, F.I.; Gómez-García, A.; Chávez-Ponce de León, D.E.; Sánchez-Cruz, N.; Ruiz-Rios, J.; Pilón-Jiménez, B.A.; Medina-Franco, J.L. Inhibitors of DNA Methyltransferases From Natural Sources: A Computational Perspective. Front. Pharm. 2018, 9, 1144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maldonado-Rojas, W.; Olivero-Verbel, J.; Marrero-Ponce, Y. Computational fishing of new DNA methyltransferase inhibitors from natural products. J. Mol. Graph. Model. 2015, 60, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Dammann, R.H.; Richter, A.M.; Jiménez, A.P.; Woods, M.; Küster, M.; Witharana, C. Impact of Natural Compounds on DNA Methylation Levels of the Tumor Suppressor Gene RASSF1A in Cancer. Int. J. Mol. Sci. 2017, 18, 2160. [Google Scholar] [CrossRef] [Green Version]
- Da Costa Prando, E.; Cavalli, L.R.; Rainho, C.A. Evidence of epigenetic regulation of the tumor suppressor gene cluster flanking RASSF1 in breast cancer cell lines. Epigenetics 2011, 6, 1413–1424. [Google Scholar] [CrossRef] [Green Version]
- Malpeli, G.; Innamorati, G.; Decimo, I.; Bencivenga, M.; Nwabo Kamdje, A.H.; Perris, R.; Bassi, C. Methylation Dynamics of RASSF1A and Its Impact on Cancer. Cancers 2019, 11, 959. [Google Scholar] [CrossRef] [Green Version]
- Morel, D.; Jeffery, D.; Aspeslagh, S.; Almouzni, G.; Postel-Vinay, S. Combining epigenetic drugs with other therapies for solid tumours-past lessons and future promise. Nat. Rev. Clin. Oncol. 2020, 17, 91–107. [Google Scholar] [CrossRef]
- Da Silva, L.M.; Frión-Herrera, Y.; Bartolomeu, A.R.; Gorgulho, C.M.; Sforcin, J.M. Mechanisms involved in the cytotoxic action of Brazilian propolis and caffeic acid against HEp-2 cells and modulation of P-glycoprotein activity. J. Pharm. Pharm. 2017, 69, 1625–1633. [Google Scholar] [CrossRef]
- Conti, B.J.; Bankova, V.; Sforcin, J.M. Chemical Composition of the Same Brazilian Propolis Sample Analyzed in 1997 and in 2012: No Freezing Effect. Nat. Prod. Commun. 2015, 10, 1279–1280. [Google Scholar] [CrossRef] [Green Version]
- Calanca, N.; Paschoal, A.P.; Munhoz, É.P.; Galindo, L.T.; Barbosa, B.M.; Caldeira, J.R.F.; Oliveira, R.A.; Cavalli, L.R.; Rogatto, S.R.; Rainho, C.A. The long non-coding RNA ANRASSF1 in the regulation of alternative protein-coding transcripts RASSF1A and RASSF1C in human breast cancer cells: Implications to epigenetic therapy. Epigenetics 2019, 14, 741–750. [Google Scholar] [CrossRef]
- Negraes, P.D.; Favaro, F.P.; Camargo, J.L.; Oliveira, M.L.; Goldberg, J.; Rainho, C.A.; Salvadori, D.M. DNA methylation patterns in bladder cancer and washing cellsediments: A perspective for tumor recurrence detection. BMC Cancer 2008, 8, 238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera-A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Brueckner, B.; Garcia Boy, R.; Siedlecki, P.; Musch, T.; Kliem, H.C.; Zielenkiewicz, P.; Suhai, S.; Wiessler, M.; Lyko, F. Epigenetic reactivation of tumor suppressor genes by a novel small-molecule inhibitor of human DNA methyltransferases. Cancer Res. 2005, 65, 6305–6311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Sample Availability: Samples of the compounds are not available from the authors. |
| Ligand | CID | 2D Molecular Structures * | Binding Energy (Kcal/mol) | Max RMSD ** | Hydrophobic Contacts | Hydrogen Bonds |
|---|---|---|---|---|---|---|
| S-adenosyl-homocysteine (SAH) | 439155 | 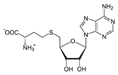 | −8.3 | 4.547 | Phe1145, Leu1151, Glu1168, Cys1191, Leu1247, Ala1579, Val1580 | Met1169, Asp1190, Ans1578 |
| caffeic acid | 689043 | 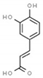 | −6.7 | 6.809 | Ser1146, Gly1147, Cys1148, Asn1578, Ala1579 | Gly1149, Gly1150, Leu1151, Val1580 |
| hydrocinnamic acid | 107 |  | −5.2 | 7.170 | Ser1146, Gly1147, Asn1578, Gly1223 | Leu1151, Val1580 |
| p-coumaric acid | 637542 | 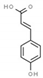 | −6.0 | 6.049 | Gly1147, Asn1578, Ala1579 | Gly1149, Gly1150, Leu1151, Val1580 |
| (−)-epigallocatchin−3-gallate | 65064 | 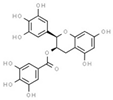 | −10.4 | 0.1564 | Arg1312, Asn1578, Val1580, Gly1223, Gly1147, Phe1145 | Glu1266, Arg1310 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Assumpção, J.H.M.; Takeda, A.A.S.; Sforcin, J.M.; Rainho, C.A. Effects of Propolis and Phenolic Acids on Triple-Negative Breast Cancer Cell Lines: Potential Involvement of Epigenetic Mechanisms. Molecules 2020, 25, 1289. https://doi.org/10.3390/molecules25061289
Assumpção JHM, Takeda AAS, Sforcin JM, Rainho CA. Effects of Propolis and Phenolic Acids on Triple-Negative Breast Cancer Cell Lines: Potential Involvement of Epigenetic Mechanisms. Molecules. 2020; 25(6):1289. https://doi.org/10.3390/molecules25061289
Chicago/Turabian StyleAssumpção, João Henrique Maia, Agnes Alessandra Sekijima Takeda, José Maurício Sforcin, and Cláudia Aparecida Rainho. 2020. "Effects of Propolis and Phenolic Acids on Triple-Negative Breast Cancer Cell Lines: Potential Involvement of Epigenetic Mechanisms" Molecules 25, no. 6: 1289. https://doi.org/10.3390/molecules25061289
APA StyleAssumpção, J. H. M., Takeda, A. A. S., Sforcin, J. M., & Rainho, C. A. (2020). Effects of Propolis and Phenolic Acids on Triple-Negative Breast Cancer Cell Lines: Potential Involvement of Epigenetic Mechanisms. Molecules, 25(6), 1289. https://doi.org/10.3390/molecules25061289





