Protein Engineering of a Pyridoxal-5′-Phosphate-Dependent l-Aspartate-α-Decarboxylase from Tribolium castaneum for β-Alanine Production
Abstract
1. Introduction
2. Results and Discussion
2.1. Protein Engineering of TcPanD
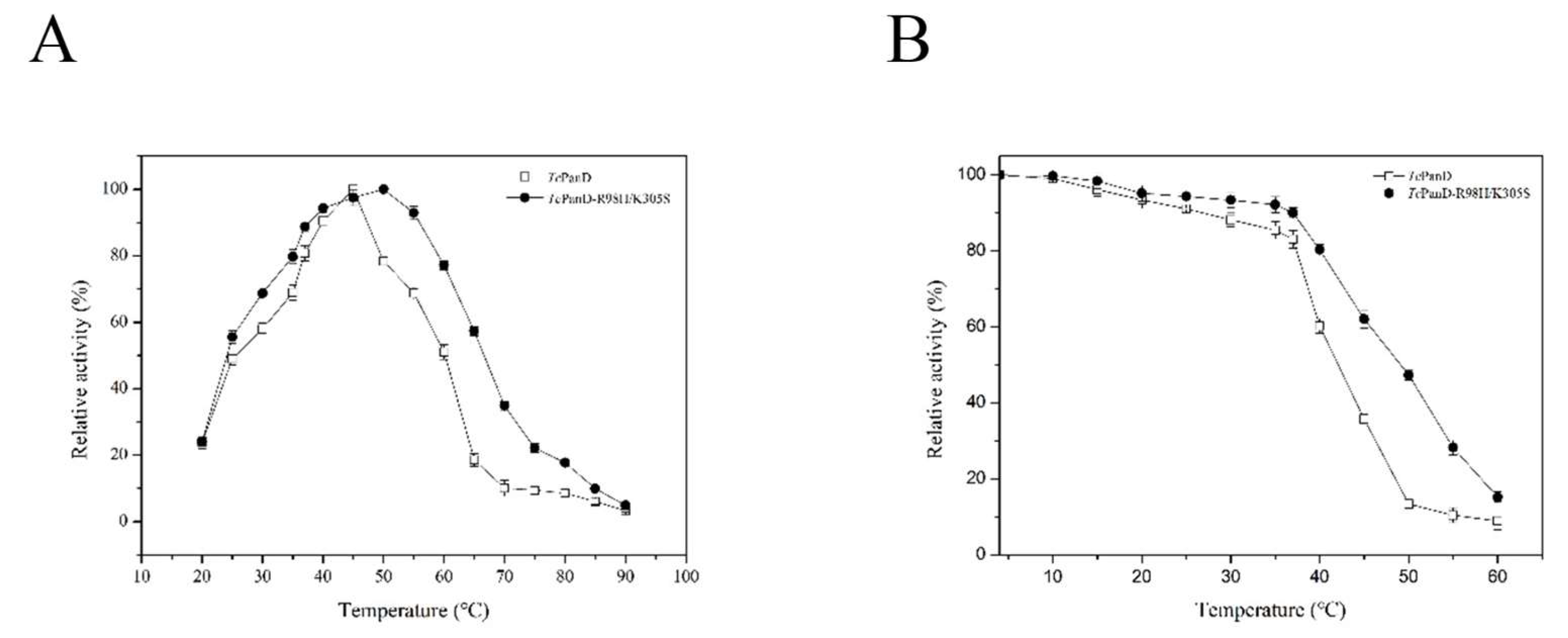
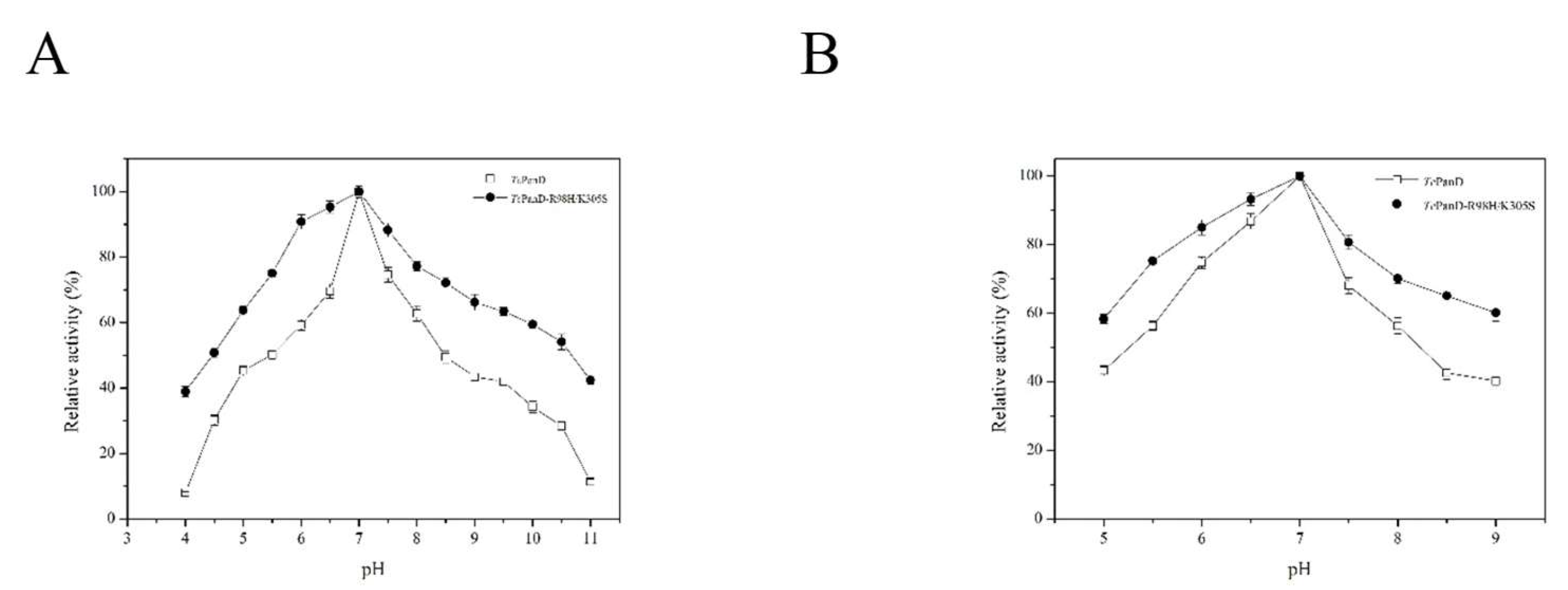
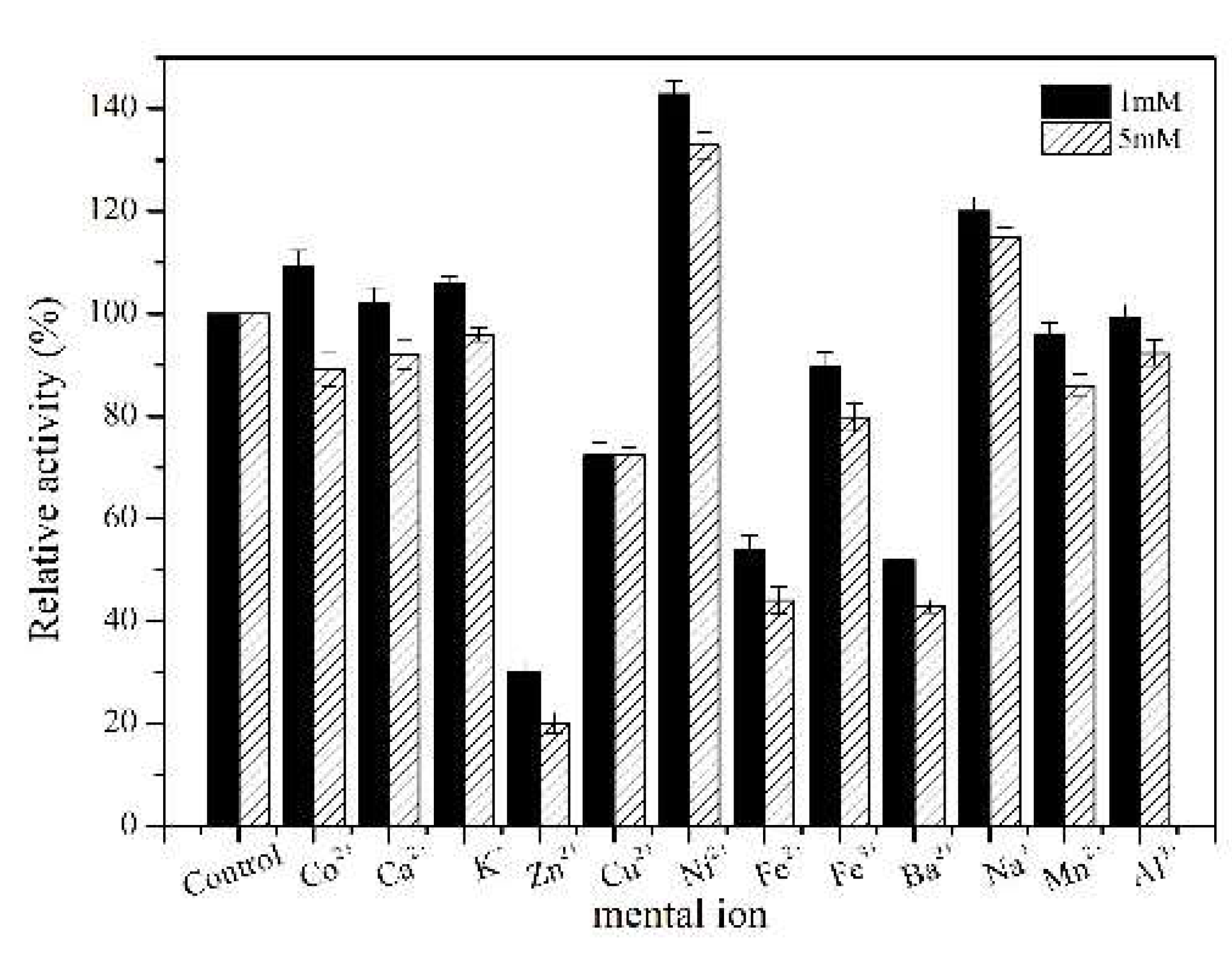
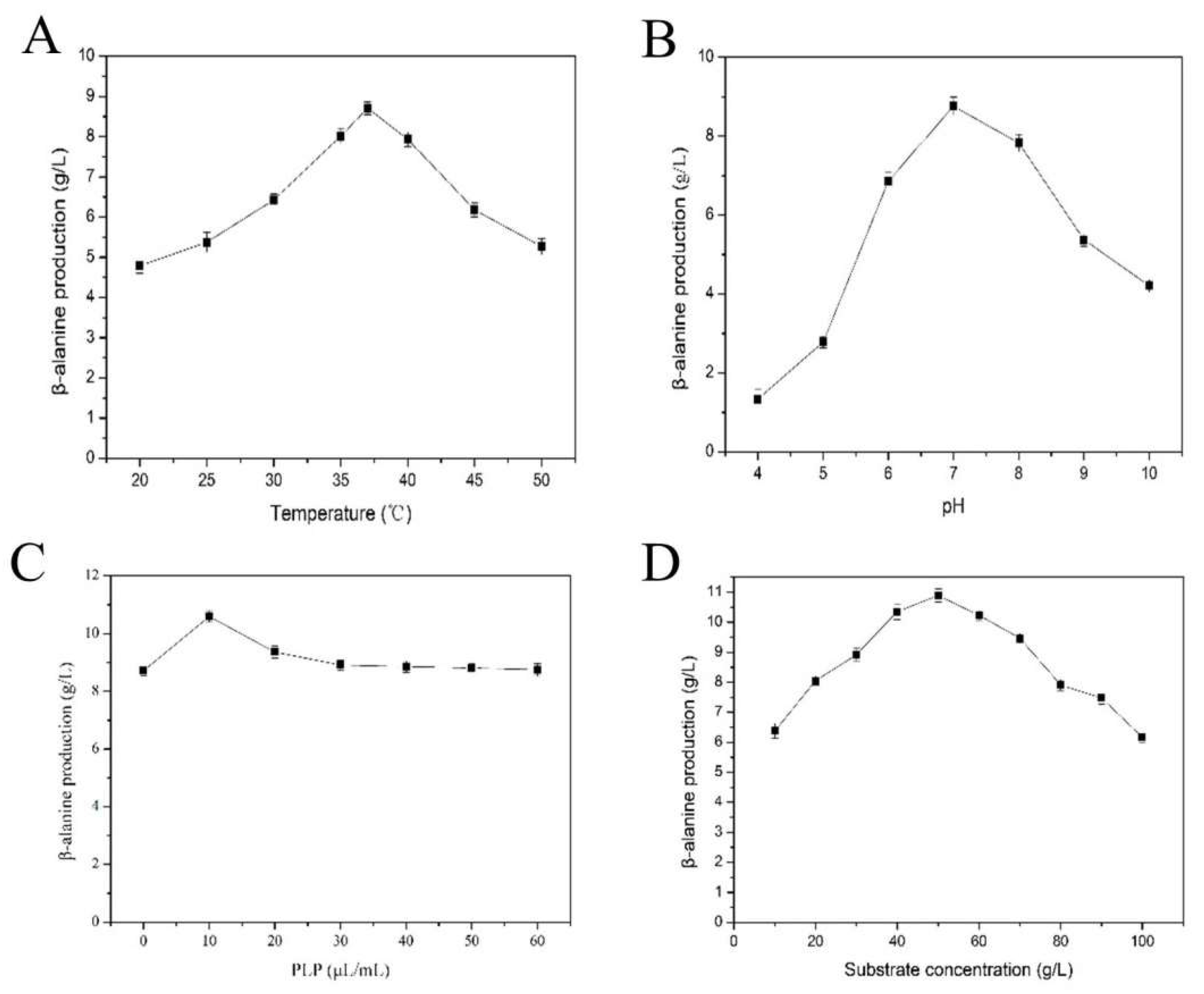
2.2. Purification and Characterization of the TcPanD-R98H/K305S
2.3. β-Alanine Production at a 5-L Scale by the Whole-Cell Catalysis
3. Materials and Methods
3.1. Materials
3.2. Gene Cloning
3.3. Random Mutagenesis and Library Screening
3.4. Site-Directed Saturation Mutation
3.5. Homology Modeling and Molecular Docking
3.6. Protein Expression and Purification
3.7. TcPanD Activity Assay
3.8. The Effect of Coenzyme on the TcPanD Activity
3.9. Temperature Related to Optimum and Stability
3.10. pH Related to Optimum and Stability
3.11. Effects of Different Metal Ions on the TcPanD Activity
3.12. Determination of the Kinetic Parameters
3.13. β-Alanine Production at a 5-L Scale
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ramjee, M.K.; Ramjee, M.K.; Genschel, U.; Genschel, U.; Abell, C.; Abell, C.; Smith, A.G.; Smith, A.G. Escherichia coli L-aspartate-α-decarboxylase: Preprotein processing and observation of reaction intermediates by electrospray mass spectrometry. Biochem. J. 1997, 323, 661–669. [Google Scholar] [CrossRef] [PubMed]
- Sale, C.; Saunders, B.; Harris, R.C. Effect of beta-alanine supplementation on muscle carnosine concentrations and exercise performance. Amino Acids 2010, 39, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Song, C.W.; Lee, S.Y.; Lee, J.; Ko, Y.-S. Metabolic engineering of Escherichia coli for the production of 3-aminopropionic acid. Metab. Eng. 2015, 30, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Artioli, G.G.; Gualano, B.; Smith, A.; Stout, J.; Lancha, A.H. Role of β-alanine supplementation on muscle carnosine and exercise performance. Med. Sci. Sports Exerc. 2010, 42, 1162–1173. [Google Scholar] [PubMed]
- Blancquaert, L.; Everaert, I.; Derave, W. Beta-alanine supplementation, muscle carnosine and exercise performance. Curr. Opin. Clin. Nutr. Metab. Care 2015, 18, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Bug, S.R.; Buc, S.R.; Ford, J.H.; Wise, E.C. An Improved Synthesis of β-Alanine. J. Amer. Chem. Soc. 1945, 67, 92–94. [Google Scholar]
- Ford, J.H. The alkaline hydrolysis of β-Aminopropionitrile. J. Amer. Chem. Soc. 1945, 67, 876–877. [Google Scholar] [CrossRef]
- Ohara, T.; Sato, T.; Shimizu, N.; Prescher, G.; Schwind, H.; Weiberg, O.; Marten, K.; Greim, H. Acrylic acid and derivatives. Kirk Othmer Encyclop. Chem. Technol. 2000. [Google Scholar] [CrossRef]
- Shen, Y.; Zhao, L.; Li, Y.; Zhang, L.; Shi, G. Synthesis of β-alanine from l-aspartate using l-aspartate-α-decarboxylase from Corynebacterium glutamicum. Biotechnol. Lett. 2014, 36, 1681–1686. [Google Scholar] [CrossRef] [PubMed]
- Van Poelje, P.D.; van Poelje, P.; Snell, E.E.; Snell, E.E. Cloning, sequencing, expression, and site-directed mutagenesis of the gene from clostridium perfringens encoding pyruvoyl-dependent histidine decarboxylase. Biochemistry 1990, 29, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Tomita, H.; Yokooji, Y.; Ishibashi, T.; Imanaka, T.; Atomi, H. An archaeal glutamate decarboxylase homolog functions as an aspartate decarboxylase and is involved in β-alanine and coenzyme A biosynthesis. J. Bacteriol. 2014, 196, 1222–1230. [Google Scholar] [CrossRef] [PubMed]
- Pantaleone David, P.; Fotheringham Ian, G.; Ton Jennifer, L. Process and Composition for Preparing D-Aspartic Acid. U.S. Patent 5,834,259, 28 October 1996. [Google Scholar]
- Dusch, N.; Pühler, A.; Kalinowski, J. Expression of the Corynebacterium glutamicum panD gene encoding L- aspartate-α-decarboxylase leads to pantothenate overproduction in Escherichia coli. Appl. Environ. Microbiol. 1999, 65, 1530–1539. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhang, R.; Xu, M.; Zhang, X.; Yang, T.; Liu, F.; Yang, S.; Rao, Z. Glu56Ser mutation improves the enzymatic activity and catalytic stability of Bacillus subtilis L-aspartate α-decarboxylase for an efficient β-alanine production. Proc. Biochem. 2018, 70, 117–123. [Google Scholar] [CrossRef]
- Chopra, S.; Pai, H.; Ranganathan, A. Expression, purification, and biochemical characterization of Mycobacterium tuberculosis aspartate decarboxylase, PanD. Protein Express. Purif. 2002, 25, 533–540. [Google Scholar] [CrossRef]
- Anton, D.L.; Kutny, R. Mechanism of substrate inactivation of Escherichia coli S-adenosylmethionine decarboxylase. Biochemistry 1987, 26, 6444–6447. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-F.; Hess, S.; Pannell, L.K.; Tabor, C.W.; Tabor, H. In vivo Mechanism-based inactivation of S-Adenosylmethionine decarboxylases from Escherichia coli, Salmonella typhimurium, and Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2001, 98, 10578–10583. [Google Scholar] [CrossRef] [PubMed]
- Borodina, I.; Kildegaard, K.R.; Jensen, N.B.; Blicher, T.H.; Maury, J.; Sherstyk, S.; Schneider, K.; Lamosa, P.; Herrgård, M.J.; Rosenstand, I.; et al. Establishing a synthetic pathway for high-level production of 3-hydroxypropionic acid in Saccharomyces cerevisiae via β-alanine. Metab. Eng. 2015, 27, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Pei, W.; Zhang, J.; Deng, S.; Tigu, F.; Li, Y.; Li, Q.; Cai, Z.; Li, Y. Molecular engineering of l-aspartate-α-decarboxylase for improved activity and catalytic stability. Appl. Microbiol. Biotechnol. 2017, 101, 6015–6021. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Huo, L.; Pulley, C.; Liu, A. Decarboxylation mechanisms in biological system. Bioorg. Chem. 2012, 43, 2–14. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |

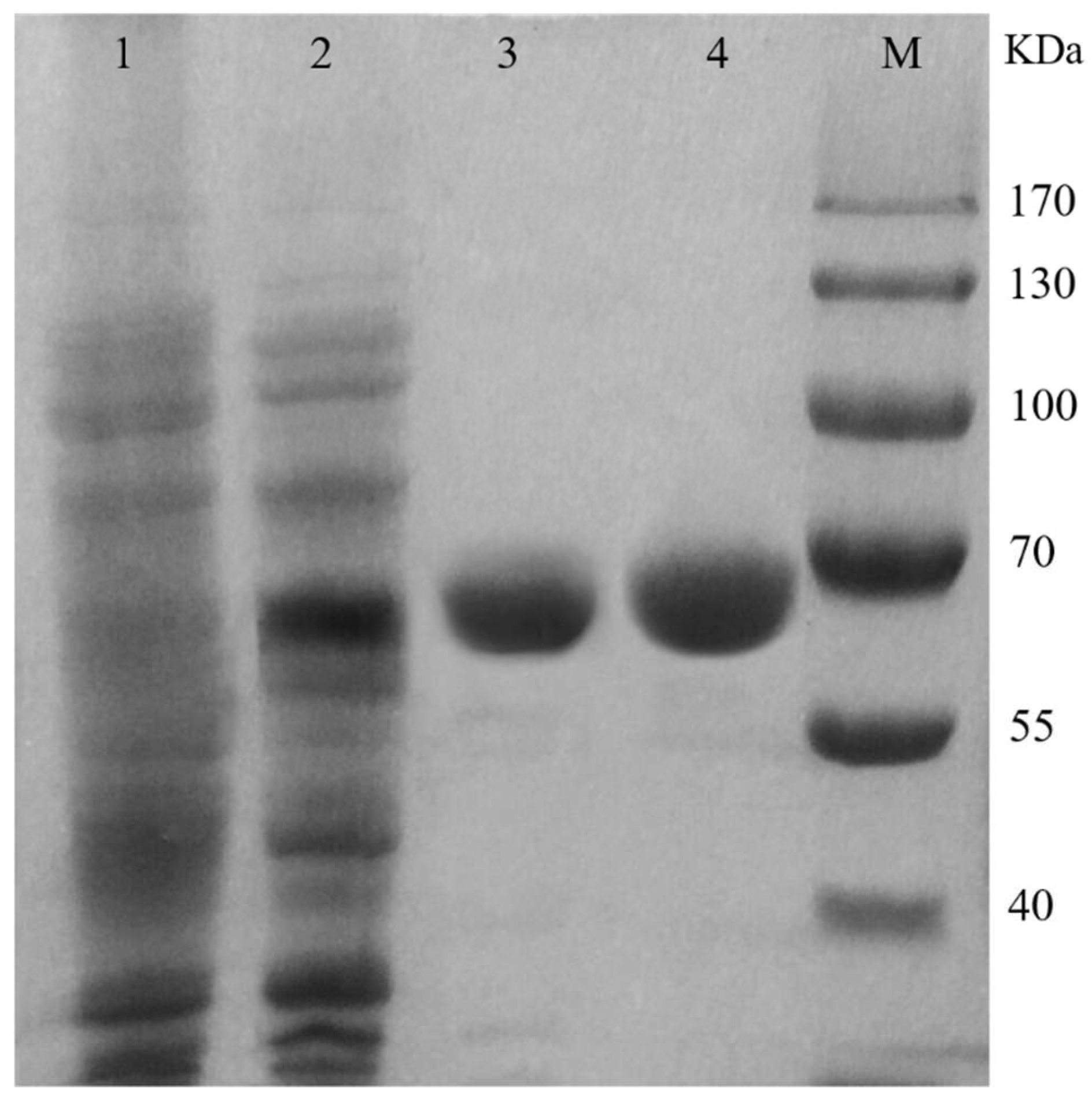

| Purification Steps | Total Protein (mg) | Total Activity (U) | Specific Activity (U/mg) | Fold Purification | Yield (%) | |
|---|---|---|---|---|---|---|
| TcPanD | Crude enzyme | 90.81 | 122.05 | 1.34 | 1 | 100 |
| Ni+ column affinity chromatography | 28.97 | 71.09 | 2.45 | 1.83 | 58.25 | |
| Ultrafiltration concentration | 19.05 | 54.78 | 2.88 | 2.15 | 44.88 | |
| TcPanD-R98H/K305S | Crude enzyme | 120.55 | 306.19 | 2.54 | 1 | 100 |
| Ni+ column affinity chromatography | 33.72 | 164.14 | 4.87 | 1.92 | 53.61 | |
| Ultrafiltration concentration | 18.15 | 127.90 | 7.05 | 2.78 | 41.77 |
| Km (mM) | Vmax (U mg−1) | Kcat (s−1) | Kcat/Km (s−1· mM−1) | |
|---|---|---|---|---|
| TcPanD | 1.35 | 2.88 | 3.12 | 2.31 |
| TcPanD-R98H/K305S | 1.23 | 7.05 | 7.64 | 6.21 |
| Primer Sequence | Restriction Endonuclease | Recognition Sequence | |
|---|---|---|---|
| Forward primer | 5’- TAAGAAGGAGATATACCATGGGCATGCCCGCTACCG-3’ | Nco Ι | CCATGG |
| Reverse primer | 5’- GTGGTGGTGGTGGTGCTCGAGTAAGTCCGAGCCAAGACG-3’ | Xho Ι | CTCGAG |
| Primer Sequence | Altered Bases | |
|---|---|---|
| 98-For | 5’-GAACCGGAGGAACTTNNNCGCTTGATGGATTTT-3’ | NNN |
| 98-Rev | 5’-AAAATCCATCAAGCGNNNAAGTTCCTCCGGTTC-3’ | |
| 305-For | 5’-TTCGATCCCATTGAGNNNATCGCCGACGTGTGT-3’ | |
| 305-Rev | 5’-ACACACGTCGGCGATNNNCTCAATGGGATCGAA-3’ | |
| 451-For | 5’-GGTTTCGAAATGGTANNNGCCGAGCCCGAATAT-3’ | |
| 451-Rev | 5’-ATATTCGGGCTCGGCNNNTACCATTTCGAAACC-3’ |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, X.-J.; Huang, C.-Y.; Xu, X.-D.; Chen, H.; Liang, M.-J.; Xu, Z.-X.; Xu, H.-X.; Wang, Z. Protein Engineering of a Pyridoxal-5′-Phosphate-Dependent l-Aspartate-α-Decarboxylase from Tribolium castaneum for β-Alanine Production. Molecules 2020, 25, 1280. https://doi.org/10.3390/molecules25061280
Yu X-J, Huang C-Y, Xu X-D, Chen H, Liang M-J, Xu Z-X, Xu H-X, Wang Z. Protein Engineering of a Pyridoxal-5′-Phosphate-Dependent l-Aspartate-α-Decarboxylase from Tribolium castaneum for β-Alanine Production. Molecules. 2020; 25(6):1280. https://doi.org/10.3390/molecules25061280
Chicago/Turabian StyleYu, Xin-Jun, Chang-Yi Huang, Xiao-Dan Xu, Hong Chen, Miao-Jie Liang, Zhe-Xian Xu, Hui-Xia Xu, and Zhao Wang. 2020. "Protein Engineering of a Pyridoxal-5′-Phosphate-Dependent l-Aspartate-α-Decarboxylase from Tribolium castaneum for β-Alanine Production" Molecules 25, no. 6: 1280. https://doi.org/10.3390/molecules25061280
APA StyleYu, X.-J., Huang, C.-Y., Xu, X.-D., Chen, H., Liang, M.-J., Xu, Z.-X., Xu, H.-X., & Wang, Z. (2020). Protein Engineering of a Pyridoxal-5′-Phosphate-Dependent l-Aspartate-α-Decarboxylase from Tribolium castaneum for β-Alanine Production. Molecules, 25(6), 1280. https://doi.org/10.3390/molecules25061280






