Recent Small-Molecule Inhibitors of the p53–MDM2 Protein–Protein Interaction
Abstract
1. Introduction
2. p53–MDM2 Inhibitors
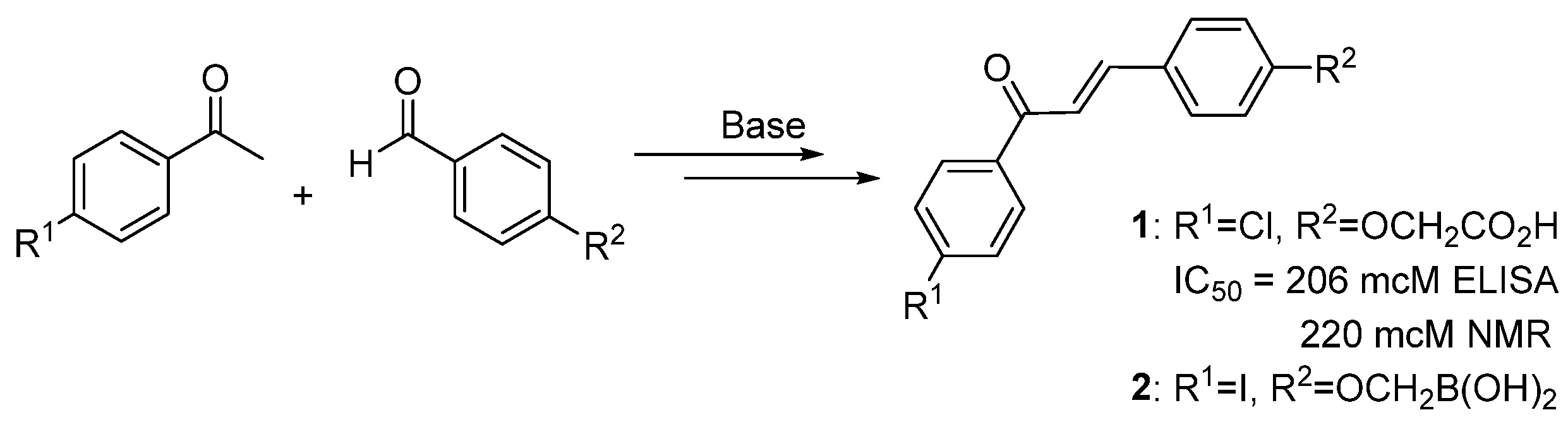
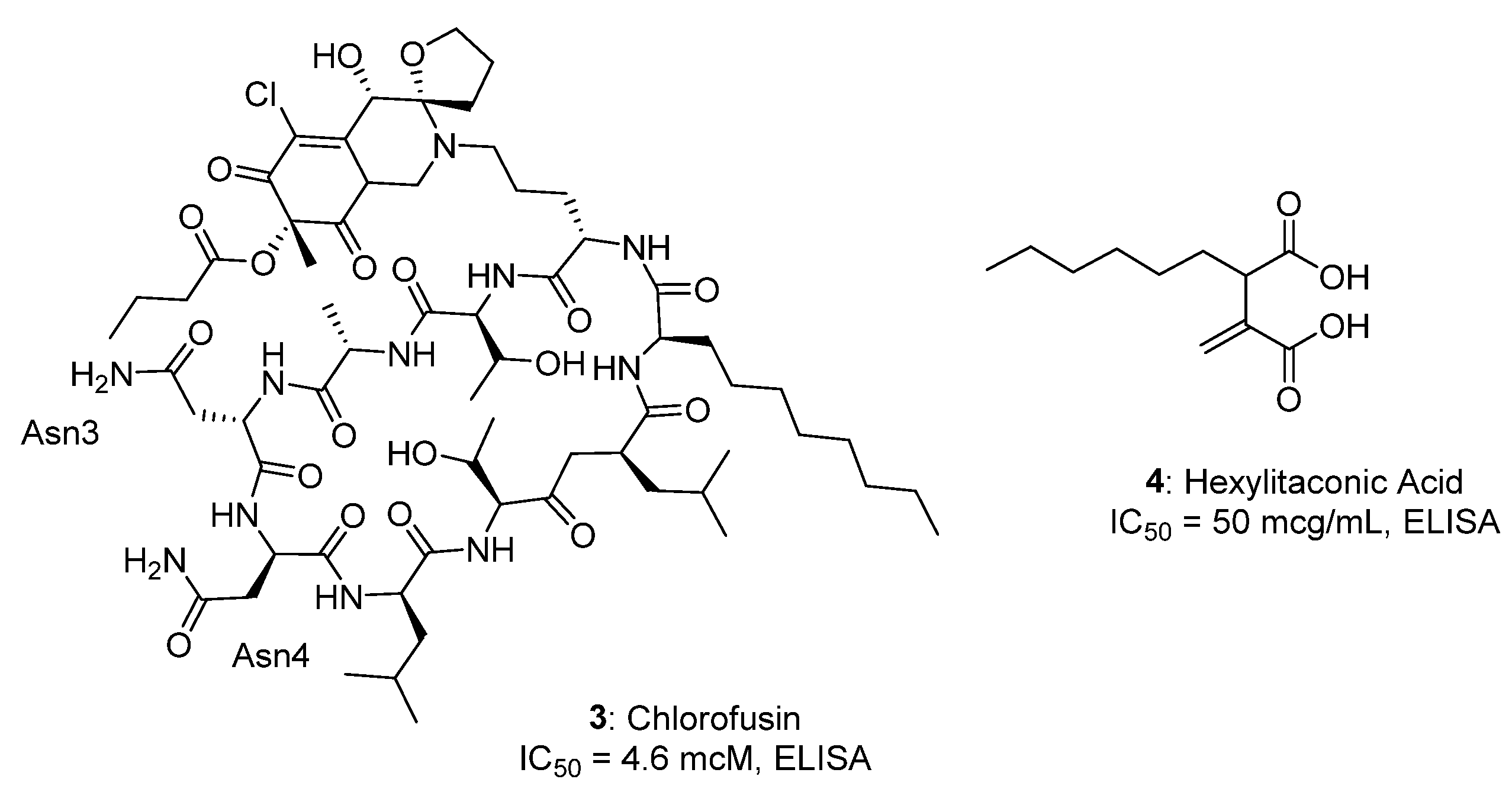
2.1. Natural Compounds
2.2. Nutlin Analogs
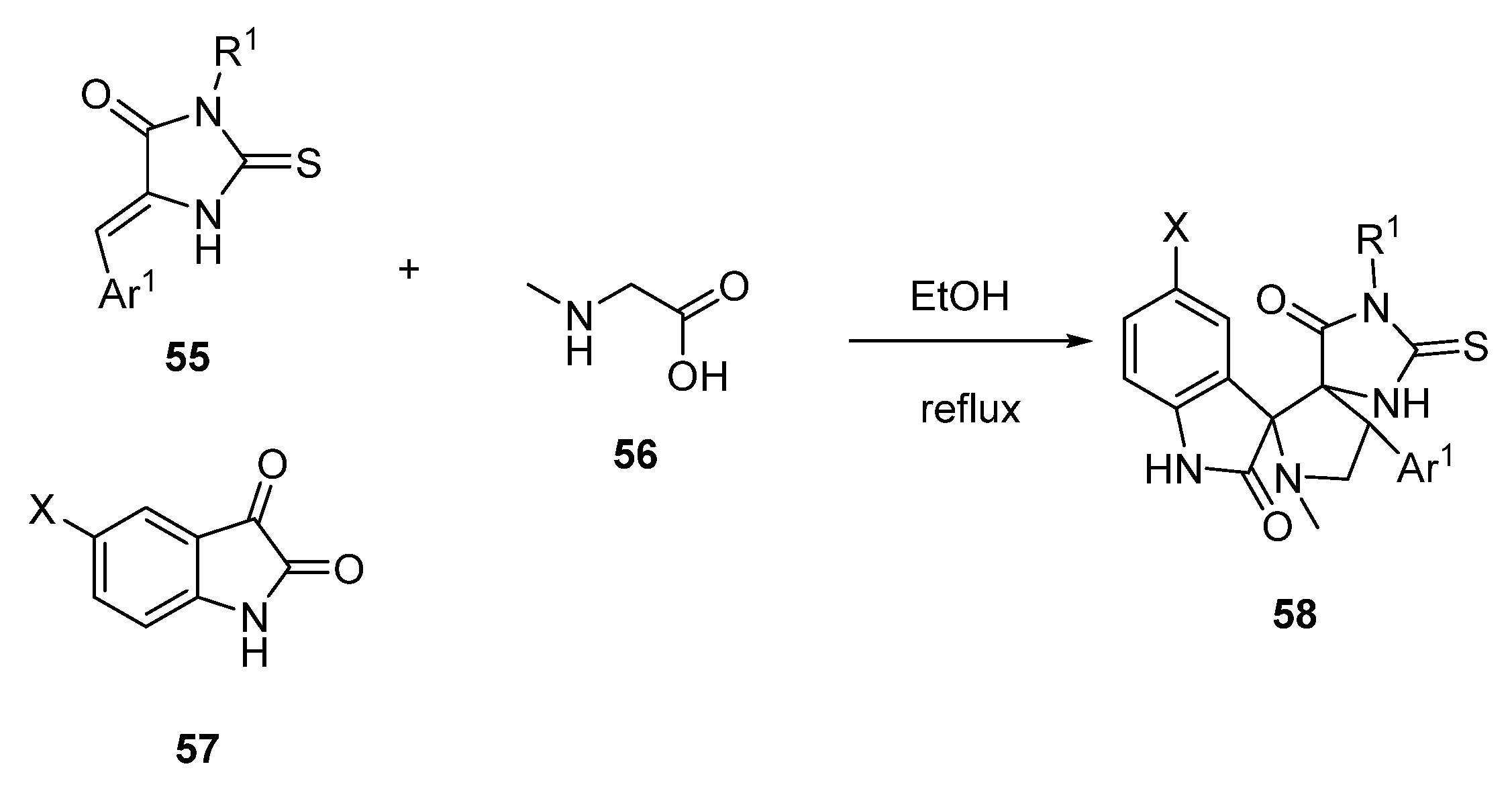
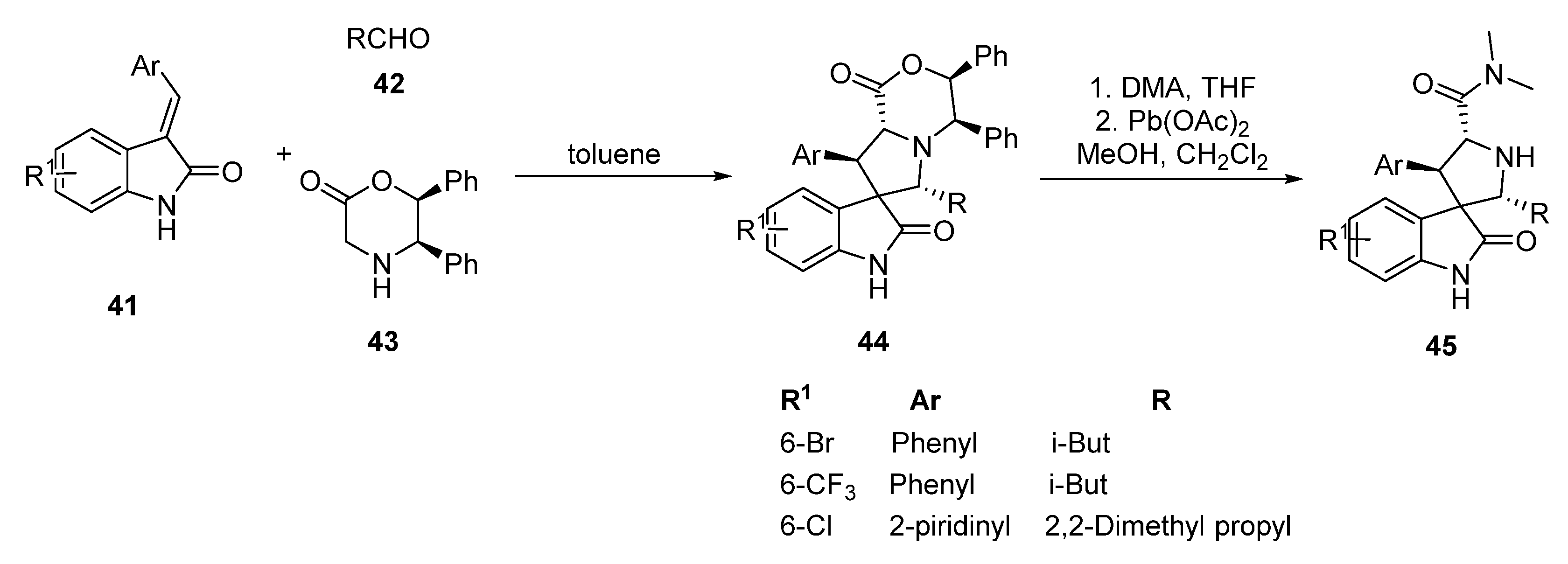
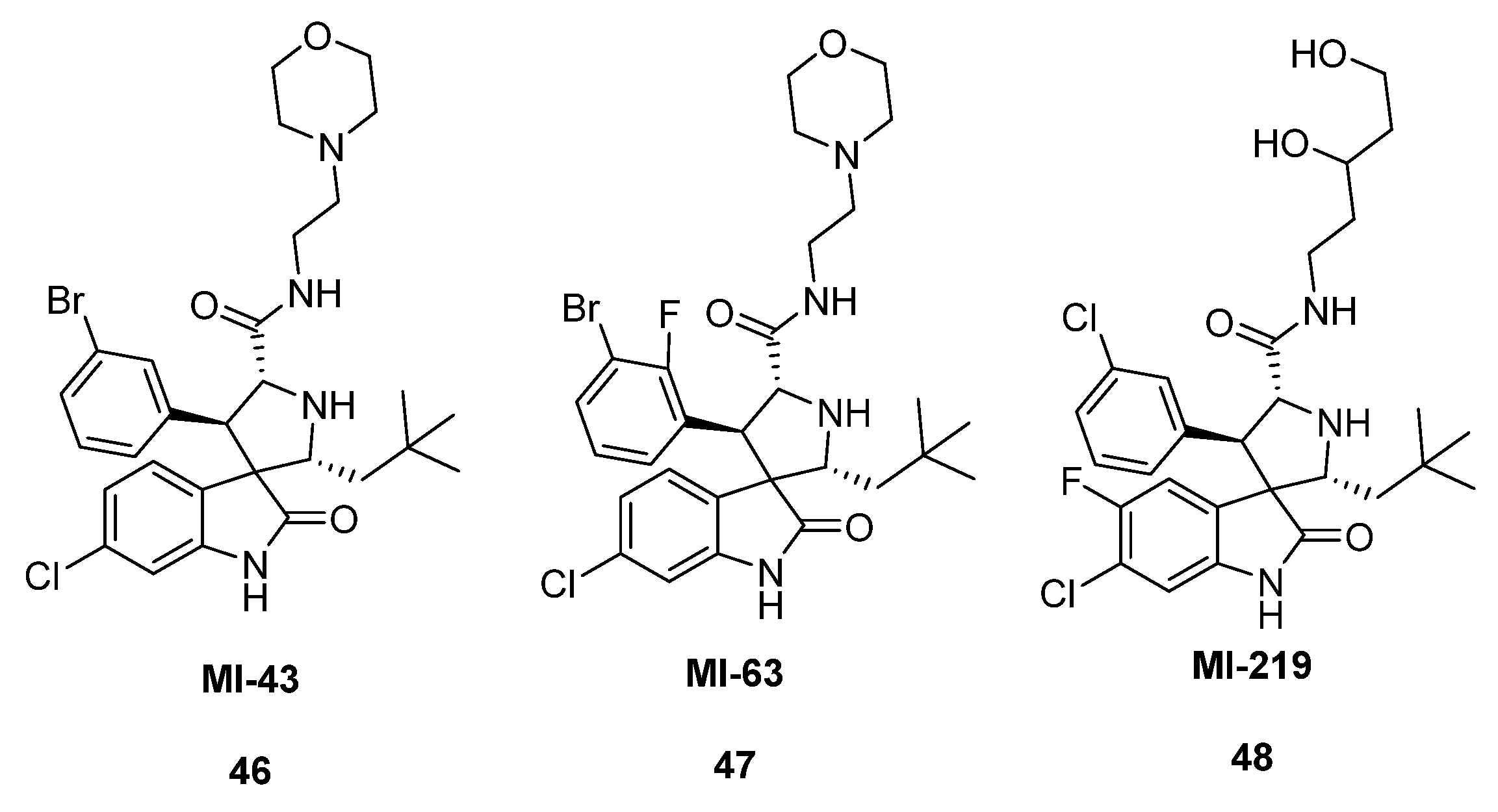
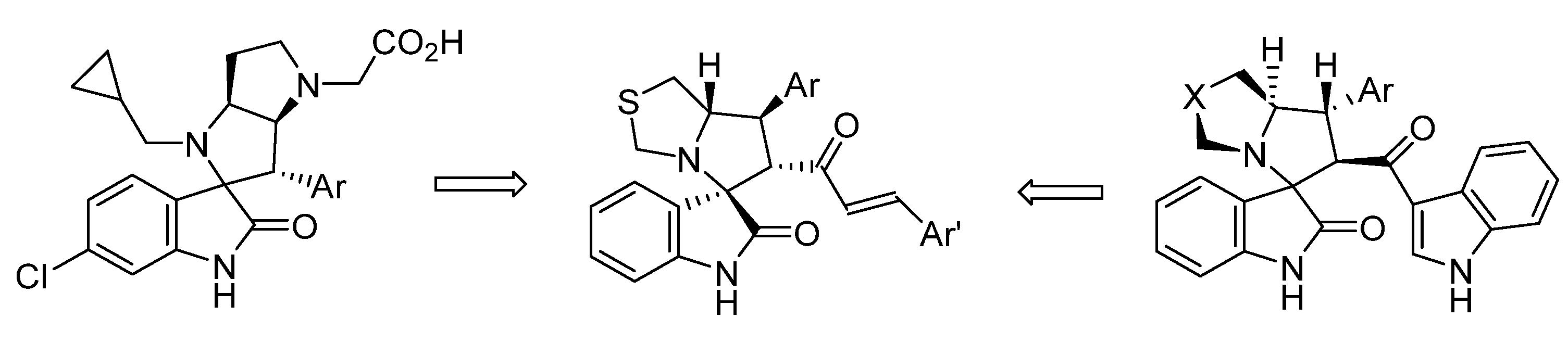
2.3. Spirooxindole Derivatives
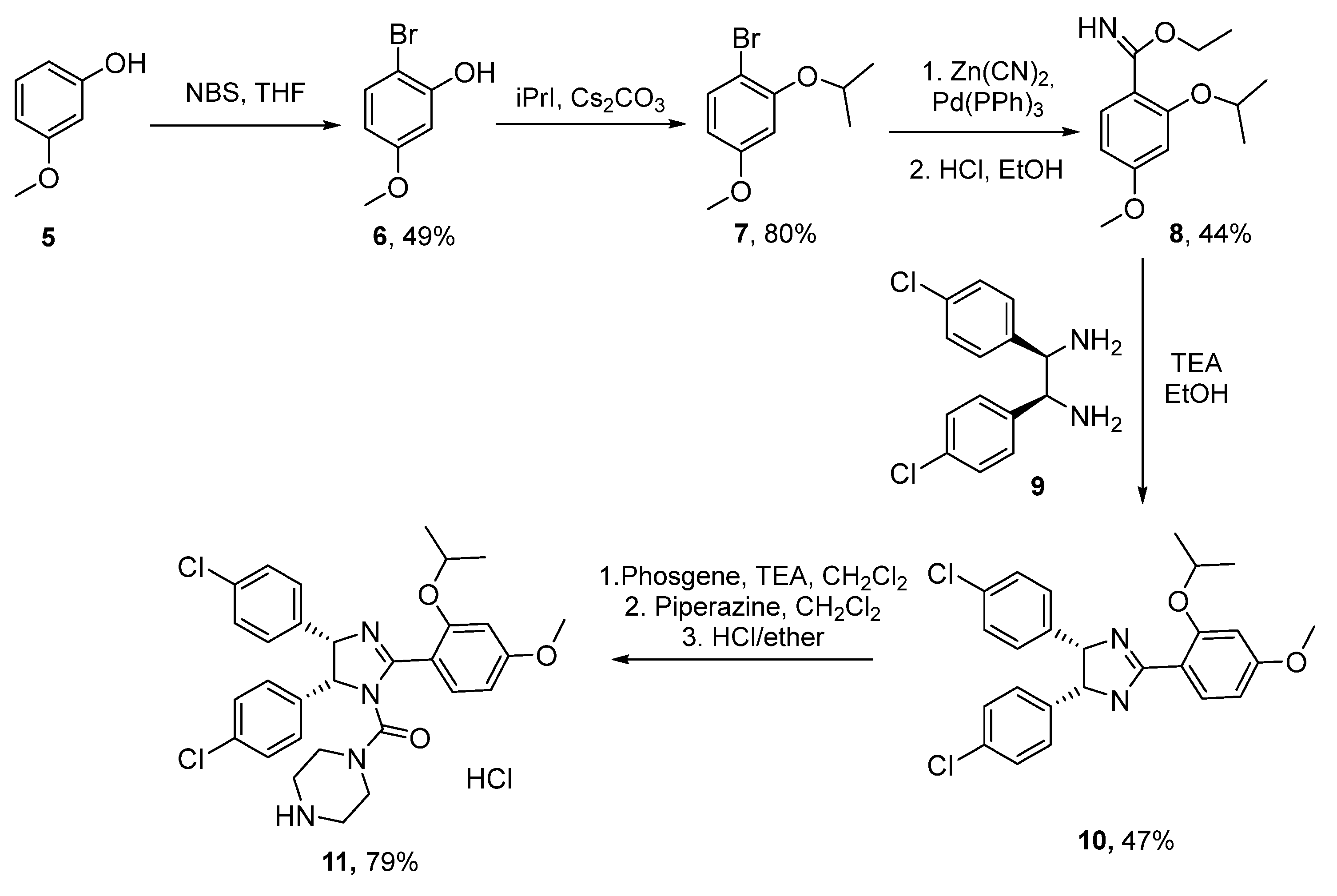
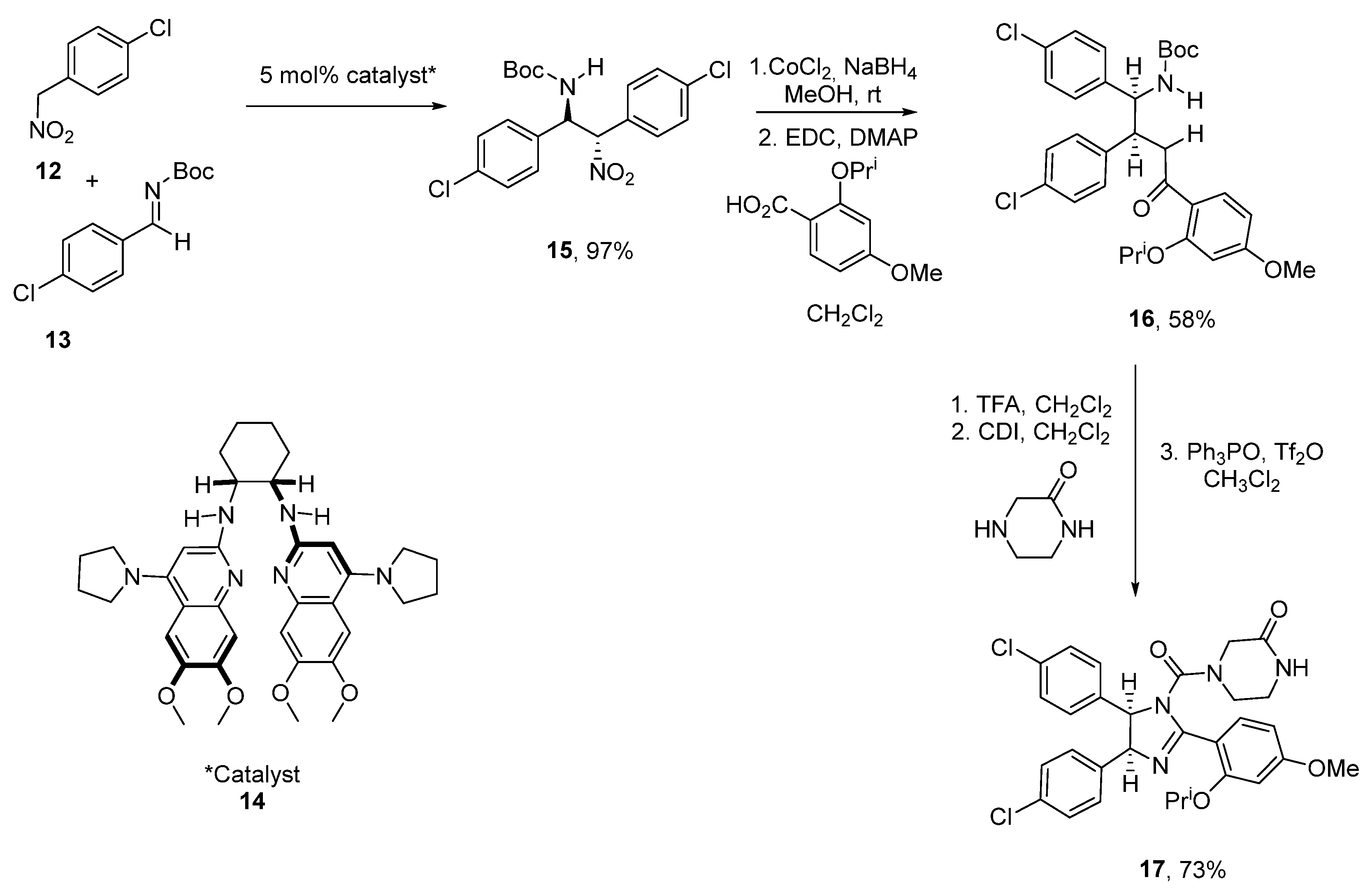
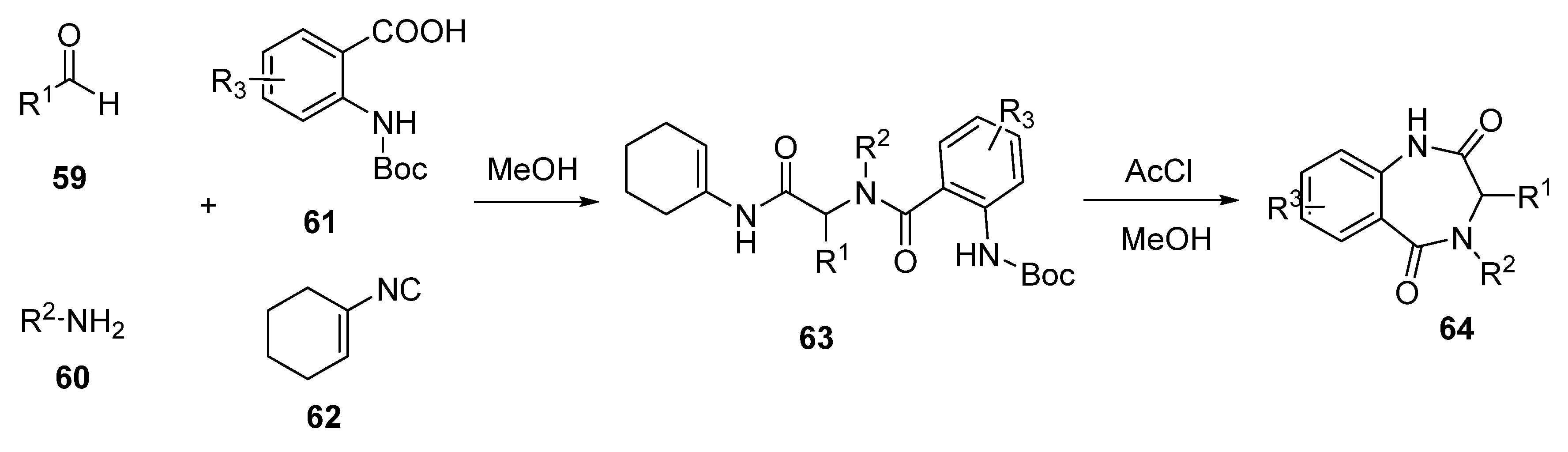
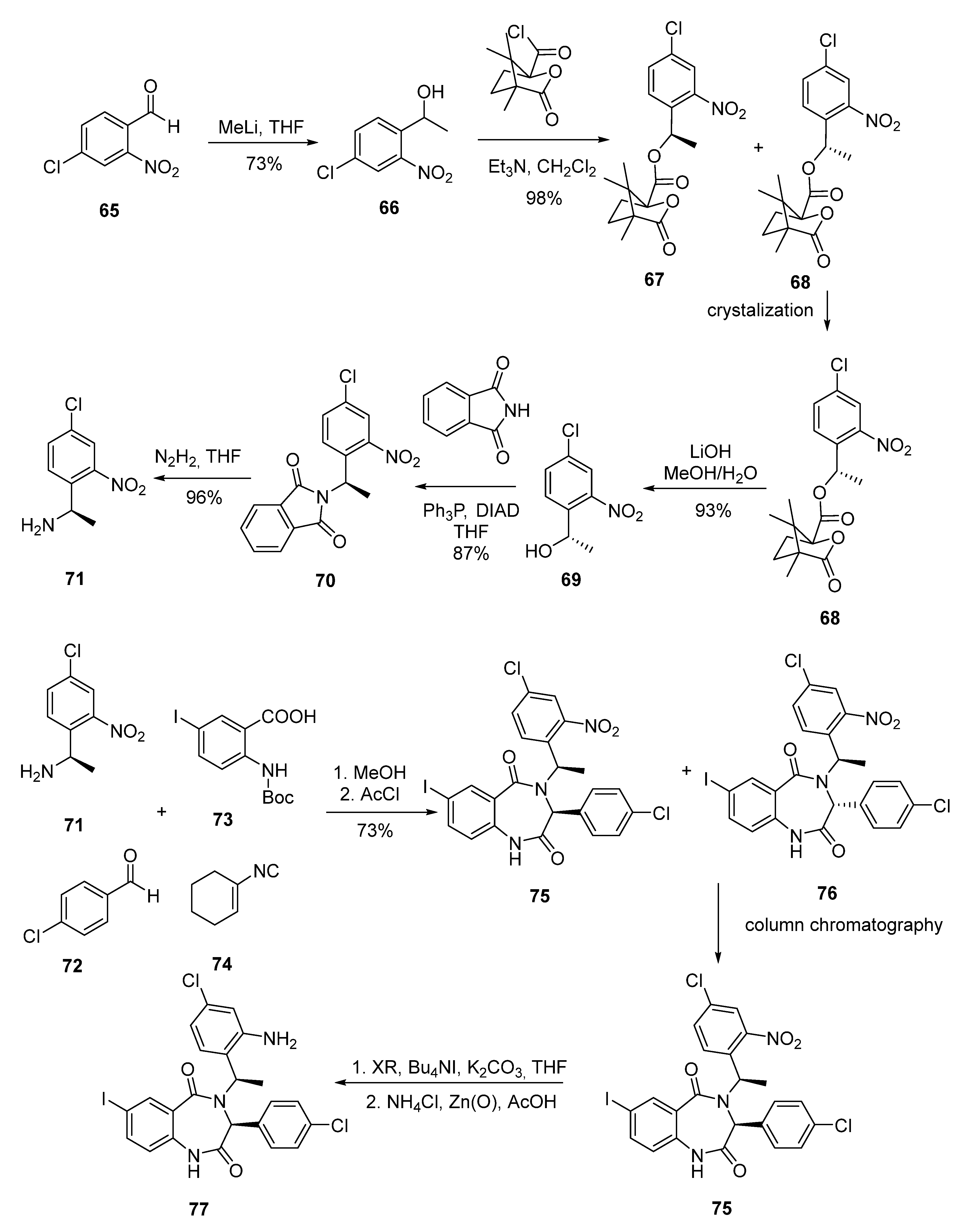
2.4. Benzodiazepine-2,5-diones (BDP)
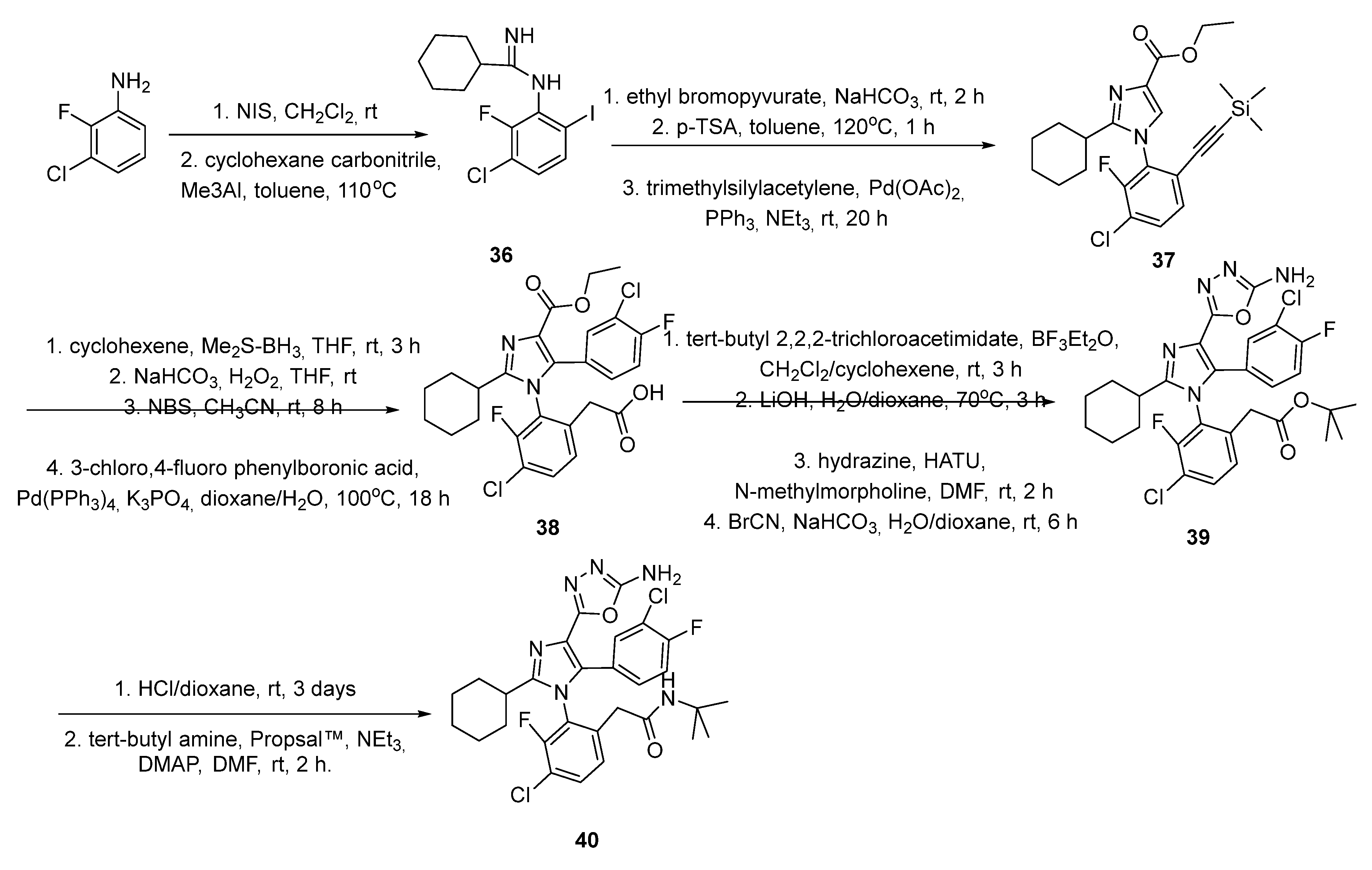
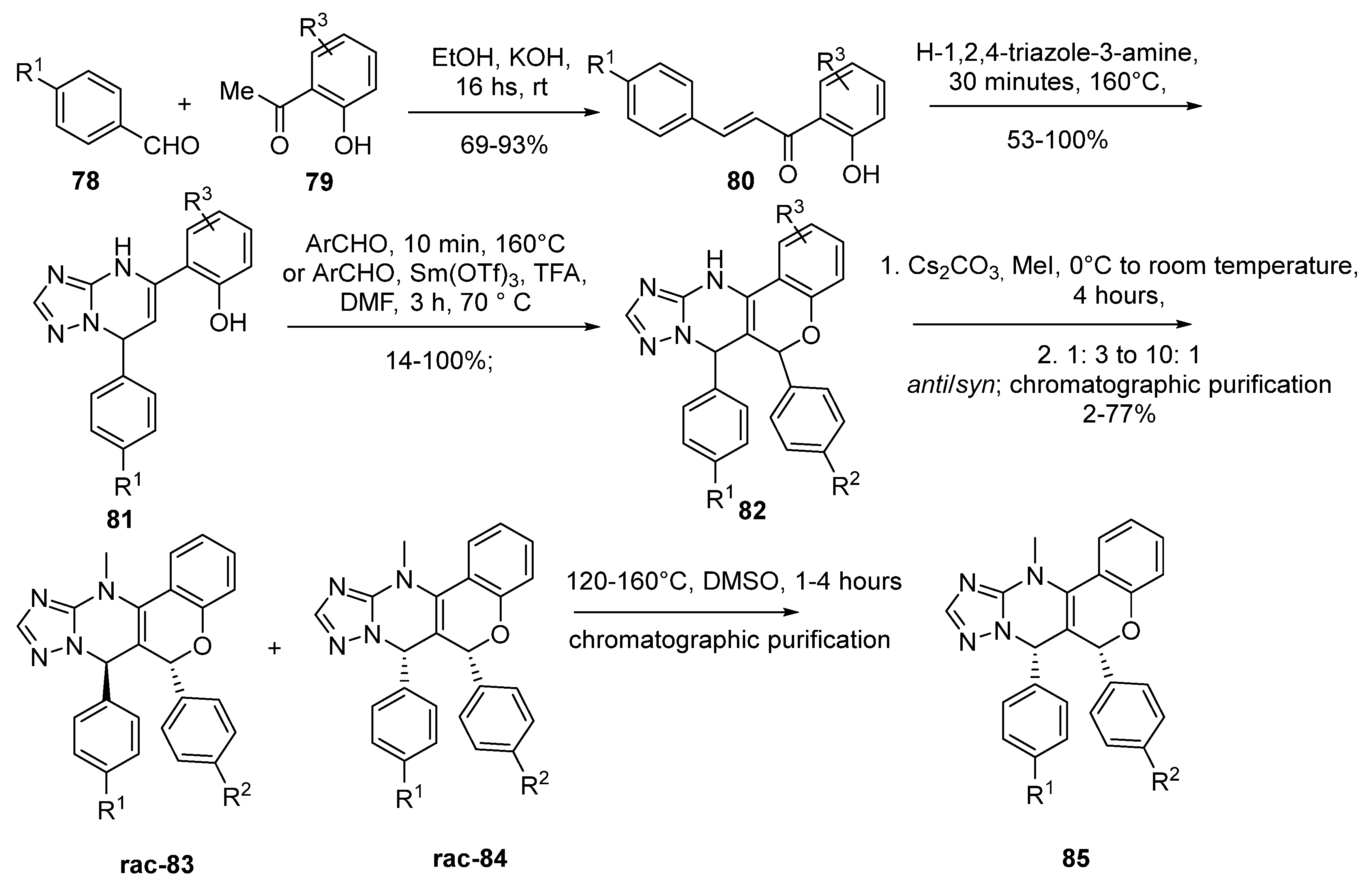
2.5. Chromenotriazole Pyrimidines
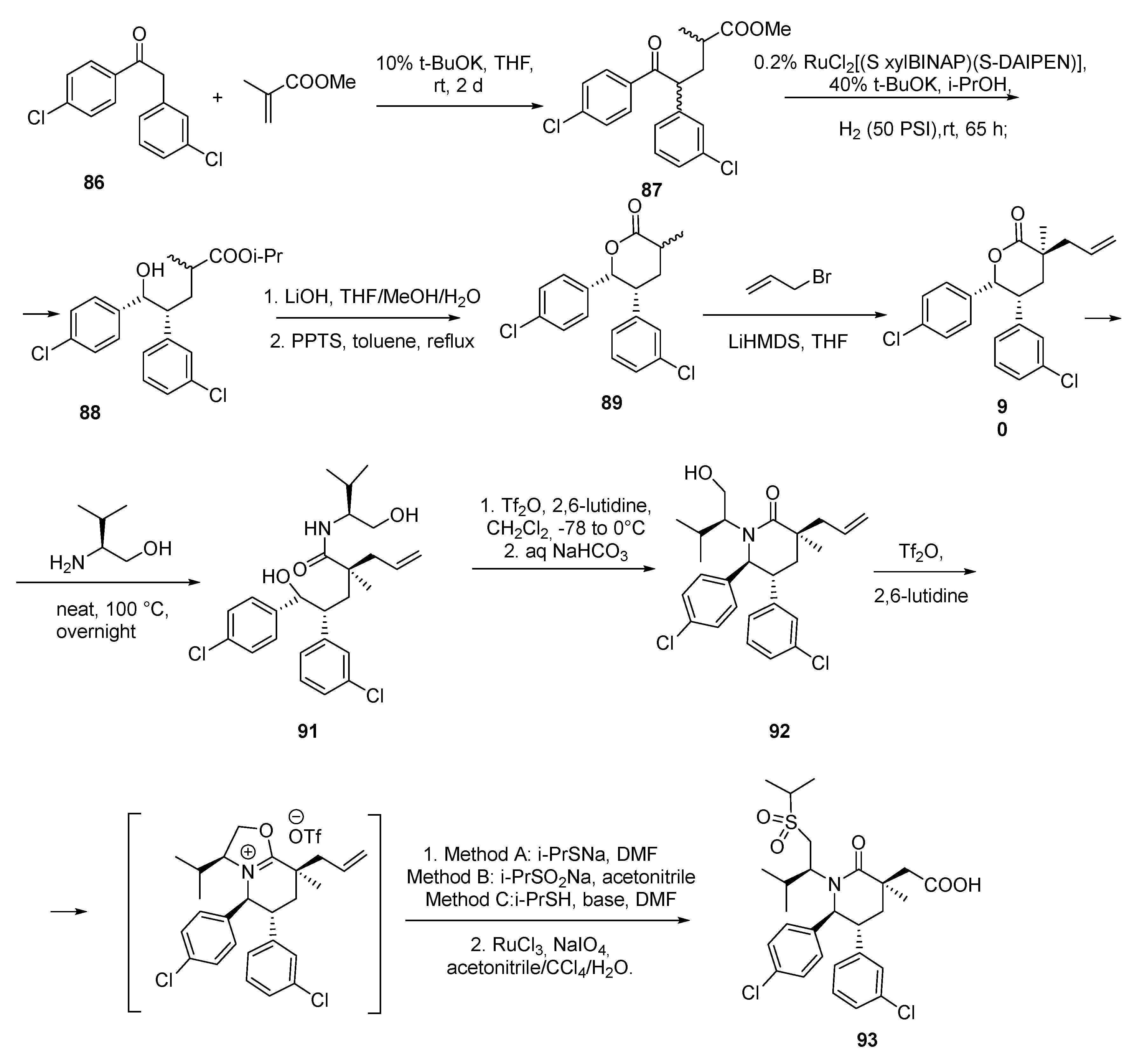
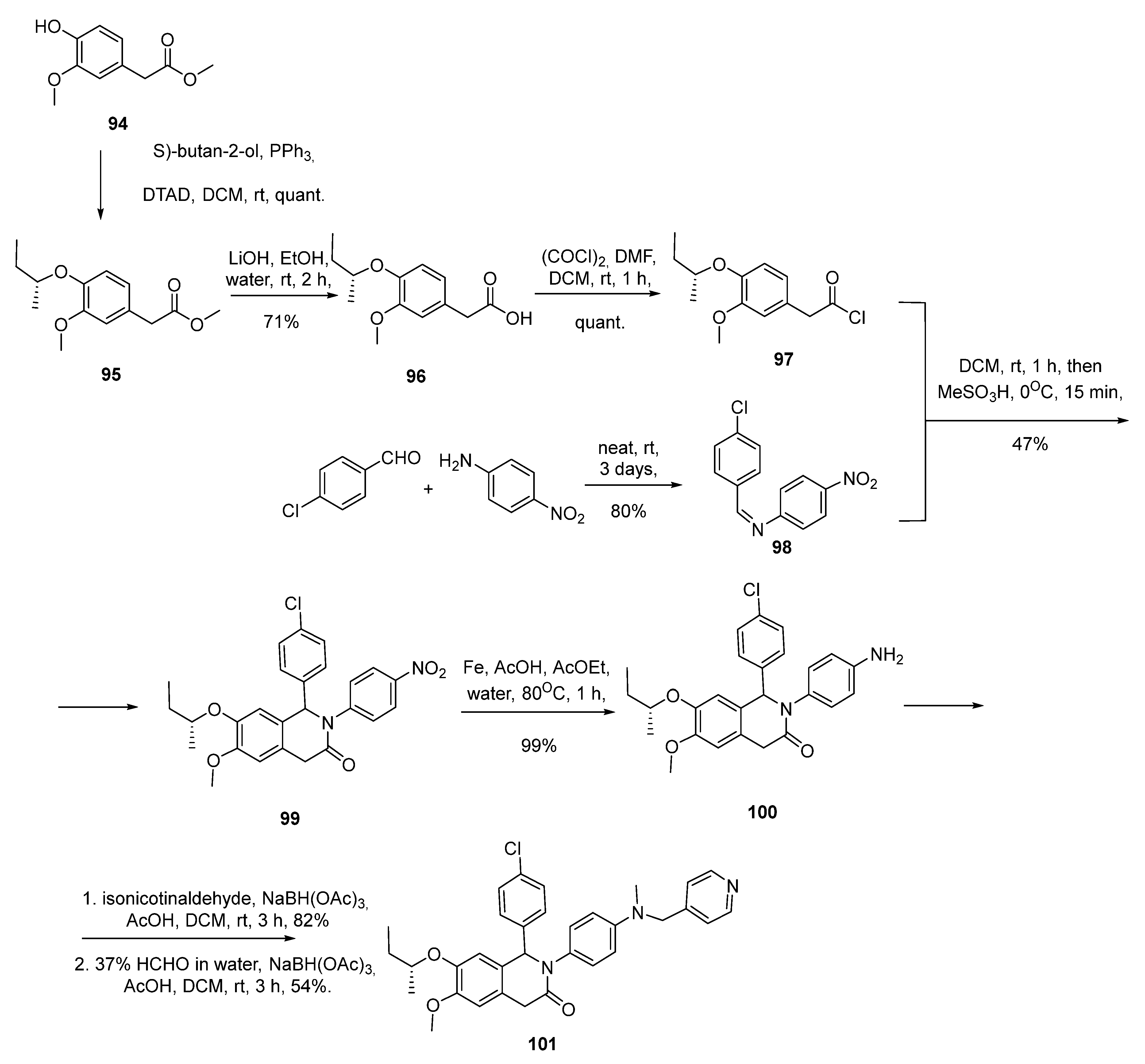
2.6. Piperidinones
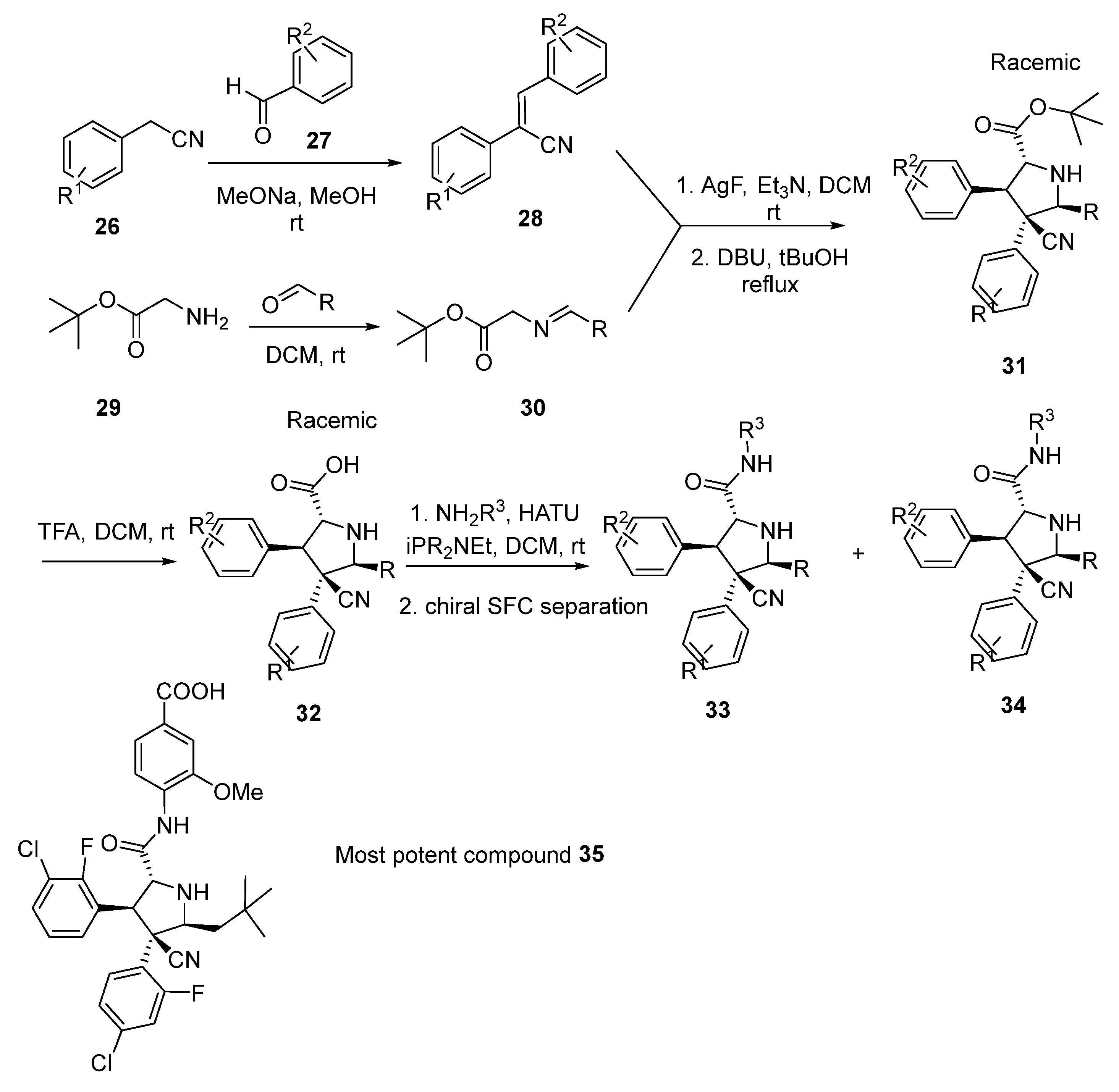
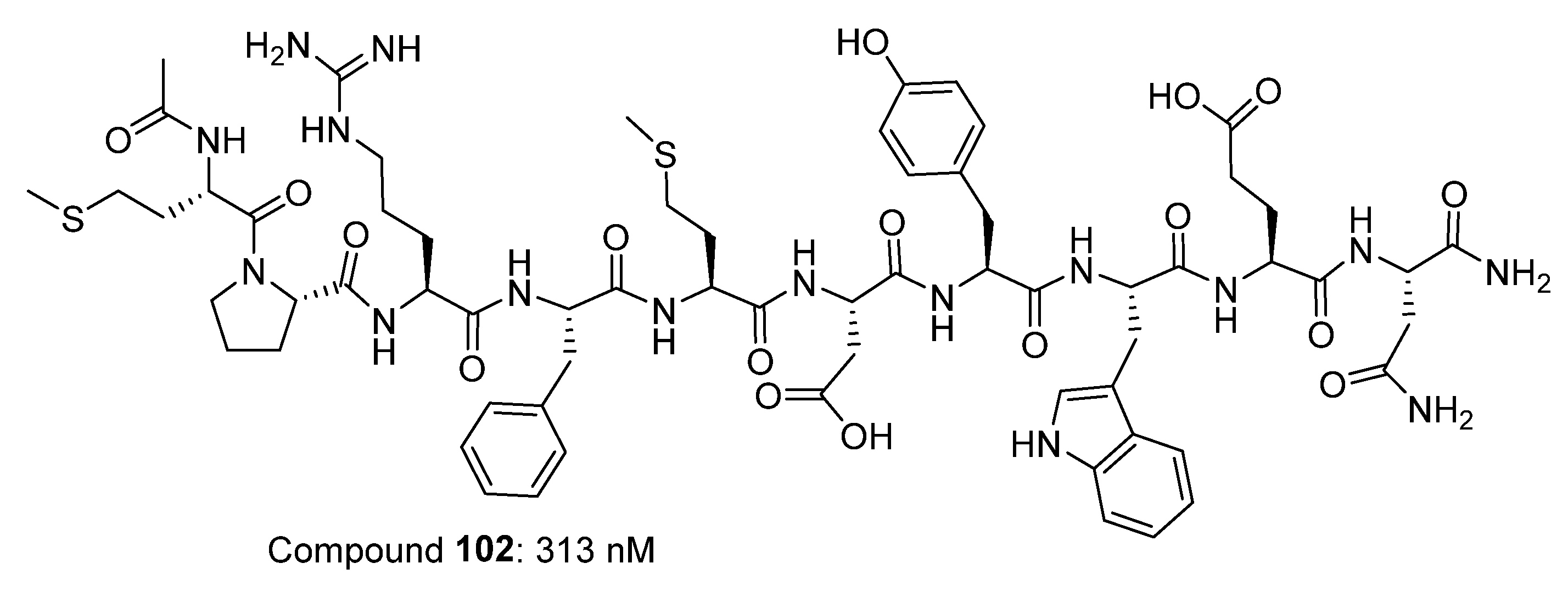
2.7. Peptide-Based Compounds
3. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Sanz, G.; Singh, M.; Peuget, S.; Selivanova, G. Inhibition of p53 inhibitors: Progress, challenges and perspectives. J. Mol. Cell Boil. 2019, 11, 586–599. [Google Scholar] [CrossRef]
- Nayak, S.K.; Khatik, G.L.; Narang, R.; Monga, V.; Chopra, H.K. p53-Mdm2 Interaction Inhibitors as Novel Nongenotoxic Anticancer Agents. Curr. Cancer Drug Targets 2018, 18, 749–772. [Google Scholar] [CrossRef] [PubMed]
- Estrada-Ortiz, N.; Neochoritis, C.G.; Dömling, A. How To Design a Successful P53-MDM2/X Interaction Inhibitor: A Thorough Overview Based on Crystal Structures. ChemMedChem 2015, 11, 757–772. [Google Scholar] [CrossRef] [PubMed]
- Khoury, K.; Dömling, A. p53 Mdm2 Inhibitors. Curr. Pharm. Des. 2012, 18, 4668–4678. [Google Scholar] [CrossRef] [PubMed]
- Popowicz, G.M.; Dömling, A.; Holak, T. The Structure-Based Design of Mdm2/Mdmx-p53 Inhibitors Gets Serious. Angew. Chem. Int. Ed. 2011, 50, 2680–2688. [Google Scholar] [CrossRef]
- Vogelstein, B.; Lane, D.P.; Levine, A.J. Surfing the p53 network. Nature 2000, 408, 307–310. [Google Scholar] [CrossRef]
- Levine, A.J. p53, the Cellular Gatekeeper for Growth and Division. Cell 1997, 88, 323–331. [Google Scholar] [CrossRef]
- Wang, Z.; Sun, Y. Targeting p53 for Novel Anticancer Therapy. Transl. Oncol. 2010, 3, 1–12. [Google Scholar] [CrossRef]
- Riley, T.; Sontag, E.; Chen, P.A.; Levine, A. Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Boil. 2008, 9, 402–412. [Google Scholar] [CrossRef]
- Sun, Y. P53 and its downstream proteins as molecular targets of cancer. Mol. Carcinog. 2006, 45, 409–415. [Google Scholar] [CrossRef]
- Campisi, J.; Daddadifagagna, F. Cellular senescence: When bad things happen to good cells. Nat. Rev. Mol. Cell Boil. 2007, 8, 729–740. [Google Scholar] [CrossRef] [PubMed]
- Teodoro, J.; Evans, S.K.; Green, M.R. Inhibition of tumor angiogenesis by p53: A new role for the guardian of the genome. J. Mol. Med. 2007, 85, 1175–1186. [Google Scholar] [CrossRef] [PubMed]
- Levine, B.; Abrams, J.M. p53: The Janus of autophagy? Nat. 2008, 10, 637–639. [Google Scholar] [CrossRef] [PubMed]
- Chumakov, P.M. The p53 protein and its universal functions in a multicellular organism. Adv. Biol. Chem. 2007, 47, 3–52. [Google Scholar]
- Hanahan, U.; Weinberg, R.A. The Hallmarks of Cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Lane, D.P. p53, guardian of the genome. Nature 1992, 358, 15–16. [Google Scholar] [CrossRef]
- Raycroft, L.; Wu, H.; Lozano, G. Transcriptional activation by wild-type but not transforming mutants of the p53 anti-oncogene. Science 1990, 249, 1049–1051. [Google Scholar] [CrossRef]
- Lane, D.P.; Verma, C. Mdm2 in Evolution. Genes Cancer 2012, 3, 320–324. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M.; Snader, K.M. Natural Products as Sources of New Drugs over the Period 1981−2002. J. Nat. Prod. 2003, 66, 1022–1037. [Google Scholar] [CrossRef]
- Maggiolini, M.; Statti, G.; Vivacqua, A.; Gabriele, S.; Rago, V.; Loizzo, M.R.; Menichini, F.; Amdò, S. Estrogenic and antiproliferative activities of isoliquiritigenin in MCF7 breast cancer cells. J. Steroid Biochem. Mol. Boil. 2002, 82, 315–322. [Google Scholar] [CrossRef]
- Neochoritis, C.G.; Miraki, M.K.; Abdelraheem, E.M.M.; Surmiak, E.; Zarganes-Tzitzikas, T.; Łabuzek, B.; Holak, T.; Dömling, A. Design of indole- and MCR-based macrocycles as p53-MDM2 antagonists. Beilstein J. Org. Chem. 2019, 15, 513–520. [Google Scholar] [CrossRef]
- Bowman, M.D.; Jeske, R.C.; Blackwell, H.E. Microwave-Accelerated SPOT-Synthesis on Cellulose Supports. Org. Lett. 2004, 6, 2019–2022. [Google Scholar] [CrossRef]
- Kumar, S.K.; Hager, E.; Pettit, C.; Gurulingappa, H.; Davidson, N.E.; Khan, S.R. Design, Synthesis, and Evaluation of Novel Boronic-Chalcone Derivatives as Antitumor Agents. J. Med. Chem. 2003, 46, 2813–2815. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.-L.; Kuo, P.-L.; Lin, L.-T.; Lin, C.-C. Isoliquiritigenin Inhibits Cell Proliferation and Induces Apoptosis in Human Hepatoma Cells. Planta Medica 2005, 71, 130–134. [Google Scholar] [CrossRef] [PubMed]
- Hegde, V.R.; Silver, J.E.; Patel, M.G.; Gullo, V.P.; Das, P.R.; Puar, M.S. Isolation and Structure of Two Novel Muscarinic Receptor Antagonists. J. Nat. Prod. 1995, 58, 843–847. [Google Scholar] [CrossRef] [PubMed]
- Duncan, S.J.; Cooper, M.A.; Williams, D.H. Binding of an inhibitor of the p53/MDM2 interaction to MDM2. Chem. Commun. 2003, 316–317. [Google Scholar] [CrossRef]
- Tsukamoto, S.; Yoshida, T.; Hosono, H.; Ohta, T.; Yokosawa, H. Hexylitaconic acid: A new inhibitor of p53–HDM2 interaction isolated from a marine-derived fungus, Arthrinium sp. Bioorganic Med. Chem. Lett. 2006, 16, 69–71. [Google Scholar] [CrossRef]
- Vassilev, L.; Vu, B.; Graves, B.; Carvajal, D.; Podlaski, F.; Filipovic, Z.; Kong, N.; Kammlott, U.; Lukacs, C.; Klein, C.; et al. In Vivo Activation of the p53 Pathway by Small-Molecule Antagonists of MDM2. Science 2004, 303, 844–848. [Google Scholar] [CrossRef]
- Wang, S.; Zhao, Y.; Bernard, D.; Aguilar, A.; Kumar, S. Protein-Protein Interactions; Springer: Berlin/Heidelberg, Germany, 2012; pp. 57–79. [Google Scholar]
- Fry, D.C.; Emerson, S.D.; Palme, S.; Vu, B.T.; Liu, C.-M.; Podlaski, F. NMR structure of a complex between MDM2 and a small molecule inhibitor. J. Biomol. NMR 2004, 30, 163–173. [Google Scholar] [CrossRef]
- Davis, T.A.; Johnston, J.N. Catalytic, enantioselective synthesis of stilbene cis-diamines: A concise preparation of (−)-Nutlin-3, a potent p53/MDM2 inhibitor. Chem. Sci. 2011, 2, 1076–1079. [Google Scholar] [CrossRef]
- Miyazaki, M.; Naito, H.; Sugimoto, Y.; Yoshida, K.; Kawato, H.; Okayama, T.; Shimizu, H.; Miyazaki, M.; Kitagawa, M.; Seki, T.; et al. Synthesis and evaluation of novel orally active p53–MDM2 interaction inhibitors. Bioorganic Med. Chem. 2013, 21, 4319–4331. [Google Scholar] [CrossRef] [PubMed]
- Bogen, S.L.; Pan, W.; Gibeau, C.R.; Lahue, B.R.; Ma, Y.; Nair, L.G.; Seigel, E.; Shipps, G.W.; Tian, Y.; Wang, Y.; et al. Discovery of Novel 3,3-Disubstituted Piperidines as Orally Bioavailable, Potent, and Efficacious HDM2-p53 Inhibitors. ACS Med. Chem. Lett. 2016, 7, 324–329. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhao, Y.; Yu, S.; Sun, W.; Liu, L.; Lu, J.; McEachern, N.; Shargary, S.; Bernard, D.; Li, X.; Zhao, T.; et al. A Potent Small-Molecule Inhibitor of the MDM2–p53 Interaction (MI-888) Achieved Complete and Durable Tumor Regression in Mice. J. Med. Chem. 2013, 56, 5553–5561. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhao, Y.; Sun, W.; Kumar, S.; Leopold, L.; Debussche, L.; Barriere, C.; Carry, J.C.; Amaning, K. Spiro-Oxindole MDM2 Antagonists. PCT Patent WO 2012/065022 A2, 18 May 2012. [Google Scholar]
- Vaupel, A.; Bold, G.; De Pover, A.; Stachyra-Valat, T.; Hergovich-Lisztwan, J.; Kallen, J.; Masuya, K.; Furet, P. Tetra-substituted imidazoles as a new class of inhibitors of the p53–MDM2 interaction. Bioorganic Med. Chem. Lett. 2014, 24, 2110–2114. [Google Scholar] [CrossRef] [PubMed]
- Ding, K.; Lu, Y.; Nikolovska-Coleska, Z.; Qiu, S.; Ding, Y.; Gao, W.; Stuckey, J.; Krajewski, K.; Roller, P.P.; Tomita, Y.; et al. Structure-Based Design of Potent Non-Peptide MDM2 Inhibitors. J. Am. Chem. Soc. 2005, 127, 10130–10131. [Google Scholar] [CrossRef] [PubMed]
- Ding, K.; Wang, G.; Deschamps, J.; Parrish, D.A.; Wang, S. Synthesis of Spirooxindoles via Asymmetric 1,3-Dipolar Cycloaddition. Tetrahedron Lett. 2005, 36, 5949–5951. [Google Scholar] [CrossRef]
- Shangary, S.; Ding, K.; Qiu, S.; Nikolovska-Coleska, Z.; Bauer, J.A.; Liu, M.; Wang, G.; Lu, Y.; McEachern, N.; Bernard, D.; et al. Reactivation of p53 by a specific MDM2 antagonist (MI-43) leads to p21-mediated cell cycle arrest and selective cell death in colon cancer. Mol. Cancer Ther. 2008, 7, 1533–1542. [Google Scholar] [CrossRef]
- Shangary, S.; Qin, N.; McEachern, N.; Liu, M.; Miller, R.S.; Qiu, S.; Nikolovska-Coleska, Z.; Ding, K.; Wang, G.; Chen, J.; et al. Temporal activation of p53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proc. Natl. Acad. Sci. USA 2008, 105, 3933–3938. [Google Scholar] [CrossRef]
- Barakat, A.; Islam, M.S.; Ghawas, H.M.; Al-Majid, A.M.; El-Senduny, F.F.; Badria, F.A.; Elshaier, Y.; Ghabbour, H.A. Design and synthesis of new substituted spirooxindoles as potential inhibitors of the MDM2–p53 interaction. Bioorganic Chem. 2019, 86, 598–608. [Google Scholar] [CrossRef]
- Gollner, A.; Rudolph, D.; Arnhof, H.; Bauer, M.; Blake, S.M.; Boehmelt, G.; Cockroft, X.-L.; Dahmann, G.; Ettmayer, P.; Gerstberger, T.; et al. Discovery of Novel Spiro[3H-indole-3,2′-pyrrolidin]-2(1H)-one Compounds as Chemically Stable and Orally Active Inhibitors of the MDM2–p53 Interaction. J. Med. Chem. 2016, 59, 10147–10162. [Google Scholar] [CrossRef]
- Islam, M.S.; Ghawas, H.M.; El-Senduny, F.F.; Al-Majid, A.M.; Elshaier, Y.; Badria, F.A.; Barakat, A. Synthesis of new thiazolo-pyrrolidine–(spirooxindole) tethered to 3-acylindole as anticancer agents. Bioorganic Chem. 2019, 82, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wolf, S.; Koes, D.; Popowicz, G.M.; Camacho, C.J.; Holak, T.A.; Dömling, A. Exhaustive Fluorine Scanning towards Potent p53-Mdm2 Antagonists. ChemMedChem 2011, 7, 49–52. [Google Scholar] [CrossRef] [PubMed]
- Ivanenkov, Y.A.; Vasilevski, S.V.; Beloglazkina, E.K.; Kukushkin, M.E.; Machulkin, A.; Veselov, M.; Chufarova, N.V.; Chernyaginab, E.S.; Vanzcool, A.S.; Zyk, N.; et al. Design, synthesis and biological evaluation of novel potent MDM2/p53 small-molecule inhibitors. Bioorganic Med. Chem. Lett. 2015, 25, 404–409. [Google Scholar] [CrossRef] [PubMed]
- Beloglazkina, A.A.; Karpov, N.A.; Mefedova, S.R.; Polyakov, V.S.; Skvortsov, D.A.; Kalinina, M.; Tafeenko, V.A.; Majouga, A.G.; Zyk, N.V.; Beloglazkina, E.K. Synthesis of dispirooxindoles containing N-unsubstituted heterocyclic moieties and study of their anticancer activity. Russ. Chem. Bull. 2019, 68, 1006–1013. [Google Scholar] [CrossRef]
- Parks, D.J.; Lafrance, L.V.; Calvo, R.R.; Milkiewicz, K.L.; Gupta, V.; Lattanze, V. 1,4-Benzodiazepine-2,5-diones as small molecule antagonists of the HDM2-p53 interaction discovery and SAR. Bioorg. Med. Chem. Lett. 2005, 15, 765–770. [Google Scholar] [CrossRef]
- Marugan, J.J.; Leonard, K.; Raboisson, P.; Gushue, J.M.; Calvo, R.; Koblish, H.K.; Lattanze, J.; Zhao, S.; Cummings, M.D.; Player, M.R.; et al. Enantiomerically pure 1,4-benzodiazepine-2,5-diones as Hdm2 antagonists. Bioorganic Med. Chem. Lett. 2006, 16, 3115–3120. [Google Scholar] [CrossRef]
- Popowicz, G.M.; Czarna, A.; Wolf, S.; Wang, K.; Wang, W.; Dömling, A.; Holak, T. Structures of low molecular weight inhibitors bound to MDMX and MDM2 reveal new approaches for p53-MDMX/MDM2 antagonist drug discovery. Cell Cycle 2010, 9, 1104–1111. [Google Scholar] [CrossRef]
- Huang, Y.; Wolf, S.; Bista, M.; Meireles, L.; Camacho, C.; Holak, T.A.; Dömling, A. 1,4-Thienodiazepine-2,5-diones via MCR (I): Synthesis, virtual space and p53-Mdm2 activity. Chem. Boil. Drug Des. 2010, 76, 116–129. [Google Scholar] [CrossRef]
- Beck, H.P.; DeGraffenreid, M.; Fox, B.; Allen, J.G.; Rew, Y.; Schneider, S.; Saiki, A.; Yu, D.; Oliner, J.D.; Salyers, K.; et al. Improvement of the synthesis and pharmacokinetic properties of chromenotriazolopyrimidine MDM2-p53 protein-protein inhibitors. Bioorganic Med. Chem. Lett. 2011, 21, 2752–2755. [Google Scholar] [CrossRef]
- Sun, D.; Li, Z.; Rew, Y.; Gribble, M.; Bartberger, M.D.; Beck, H.P.; Canon, J.; Chen, A.; Chen, X.; Chow, D.; et al. Discovery of AMG 232, a Potent, Selective, and Orally Bioavailable MDM2–p53 Inhibitor in Clinical Development. J. Med. Chem. 2014, 57, 1454–1472. [Google Scholar] [CrossRef]
- Lucas, B.S.; Fisher, B.; McGee, L.R.; Olson, S.H.; Medina, J.C.; Cheung, E. An Expeditious Synthesis of the MDM2–p53 Inhibitor AM-8553. J. Am. Chem. Soc. 2012, 134, 12855–12860. [Google Scholar] [CrossRef] [PubMed]
- Holzer, P.; Masuya, K.; Furet, P.; Kallen, J.; Valat-Stachyra, T.; Ferretti, S.; Berghausen, J.; Bouisset-Leonard, M.; Buschmann, N.; Pissot-Soldermann, C.; et al. Discovery of a Dihydroisoquinolinone Derivative (NVP-CGM097): A Highly Potent and Selective MDM2 Inhibitor Undergoing Phase 1 Clinical Trials in p53wt Tumors. J. Med. Chem. 2015, 58, 6348–6358. [Google Scholar] [CrossRef] [PubMed]
- Gessier, F.; Kallen, J.; Jacoby, E.; Chène, P.; Stachyra-Valat, T.; Ruetz, S.; Jeay, S.; Holzer, P.; Masuya, K.; Furet, P. Discovery of dihydroisoquinolinone derivatives as novel inhibitors of the p53–MDM2 interaction with a distinct binding mode. Bioorganic Med. Chem. Lett. 2015, 25, 3621–3625. [Google Scholar] [CrossRef] [PubMed]
- Chène, P.; Fuchs, J.; Bohn, J.; García-Echeverría, C.; Furet, P.; Fabbro, D. A small synthetic peptide, which inhibits the p53-hdm2 interaction, stimulates the p53 pathway in tumour cell lines 1 1Edited by A. R. Fersht. J. Mol. Boil. 2000, 299, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Böttger, A.; Böttger, V.; Sparks, A.; Liu, W.-L.; Howard, S.F.; Lane, D.P. Design of a synthetic Mdm2-binding mini protein that activates the p53 response in vivo. Curr. Boil. 1997, 7, 860–869. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beloglazkina, A.; Zyk, N.; Majouga, A.; Beloglazkina, E. Recent Small-Molecule Inhibitors of the p53–MDM2 Protein–Protein Interaction. Molecules 2020, 25, 1211. https://doi.org/10.3390/molecules25051211
Beloglazkina A, Zyk N, Majouga A, Beloglazkina E. Recent Small-Molecule Inhibitors of the p53–MDM2 Protein–Protein Interaction. Molecules. 2020; 25(5):1211. https://doi.org/10.3390/molecules25051211
Chicago/Turabian StyleBeloglazkina, Anastasia, Nikolai Zyk, Alexander Majouga, and Elena Beloglazkina. 2020. "Recent Small-Molecule Inhibitors of the p53–MDM2 Protein–Protein Interaction" Molecules 25, no. 5: 1211. https://doi.org/10.3390/molecules25051211
APA StyleBeloglazkina, A., Zyk, N., Majouga, A., & Beloglazkina, E. (2020). Recent Small-Molecule Inhibitors of the p53–MDM2 Protein–Protein Interaction. Molecules, 25(5), 1211. https://doi.org/10.3390/molecules25051211