A Supramolecular Approach to Structure-Based Design with A Focus on Synthons Hierarchy in Ornithine-Derived Ligands: Review, Synthesis, Experimental and in Silico Studies
Abstract
1. Introduction
1.1. L-Ornithine
1.2. Role of Ornithine in Polyamine Biosynthesis
1.3. Ornithine-Based Inhibitors of Ornithine Decarboxylase Enzyme
1.4. Multi-Target-Directed Ligands: New Anticancer Drug Strategy
1.5. Supramolecular Chemistry as A “New Old” Avenue for Structure-Based Design
1.5.1. Supramolecular Synergy of Small Molecules and Macromolecules
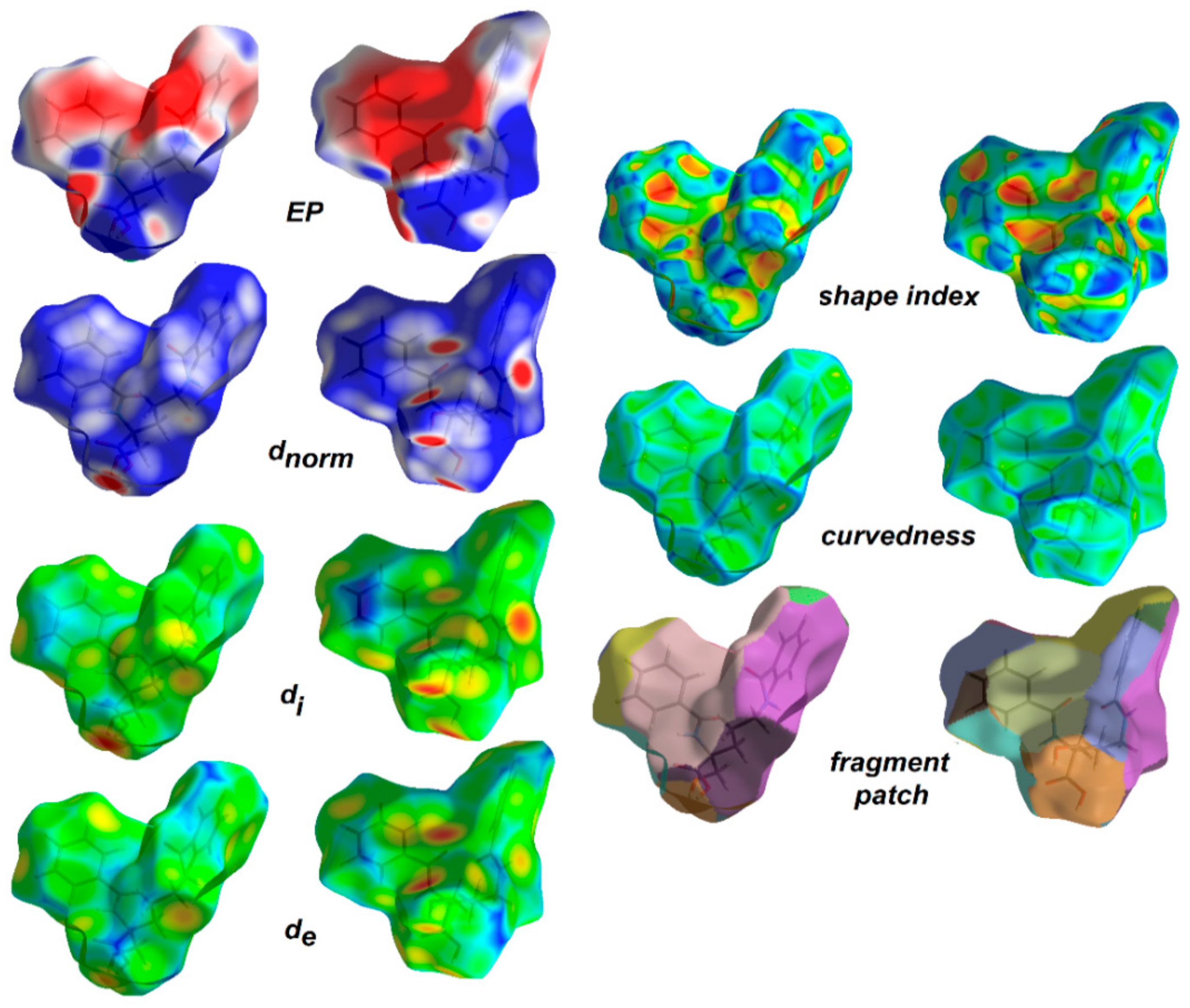
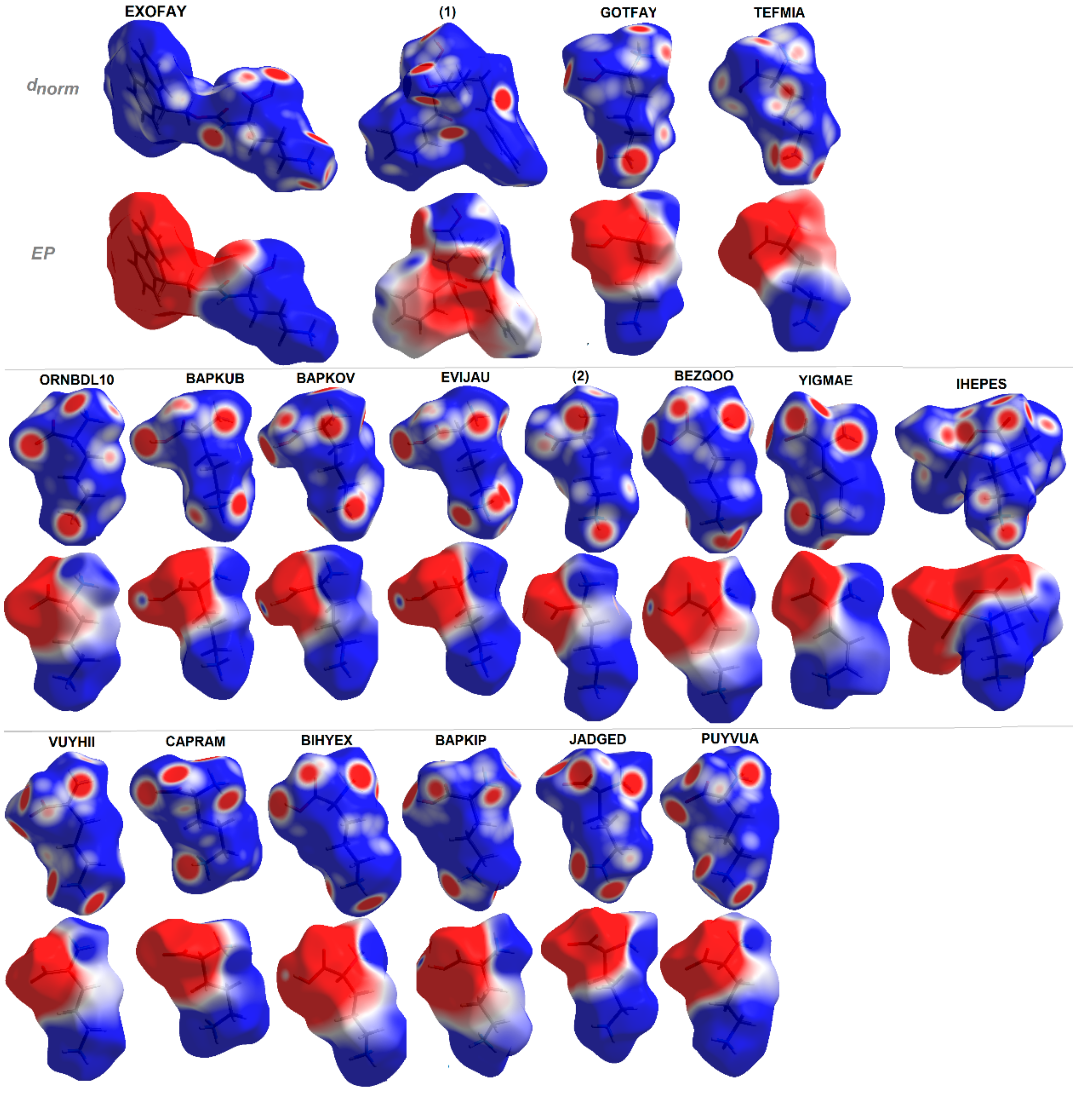
1.5.2. Molecular Hirshfeld Surfaces and Electrostatic Potentials as Useful Tools in Drug Discovery
2. Results and Discussion
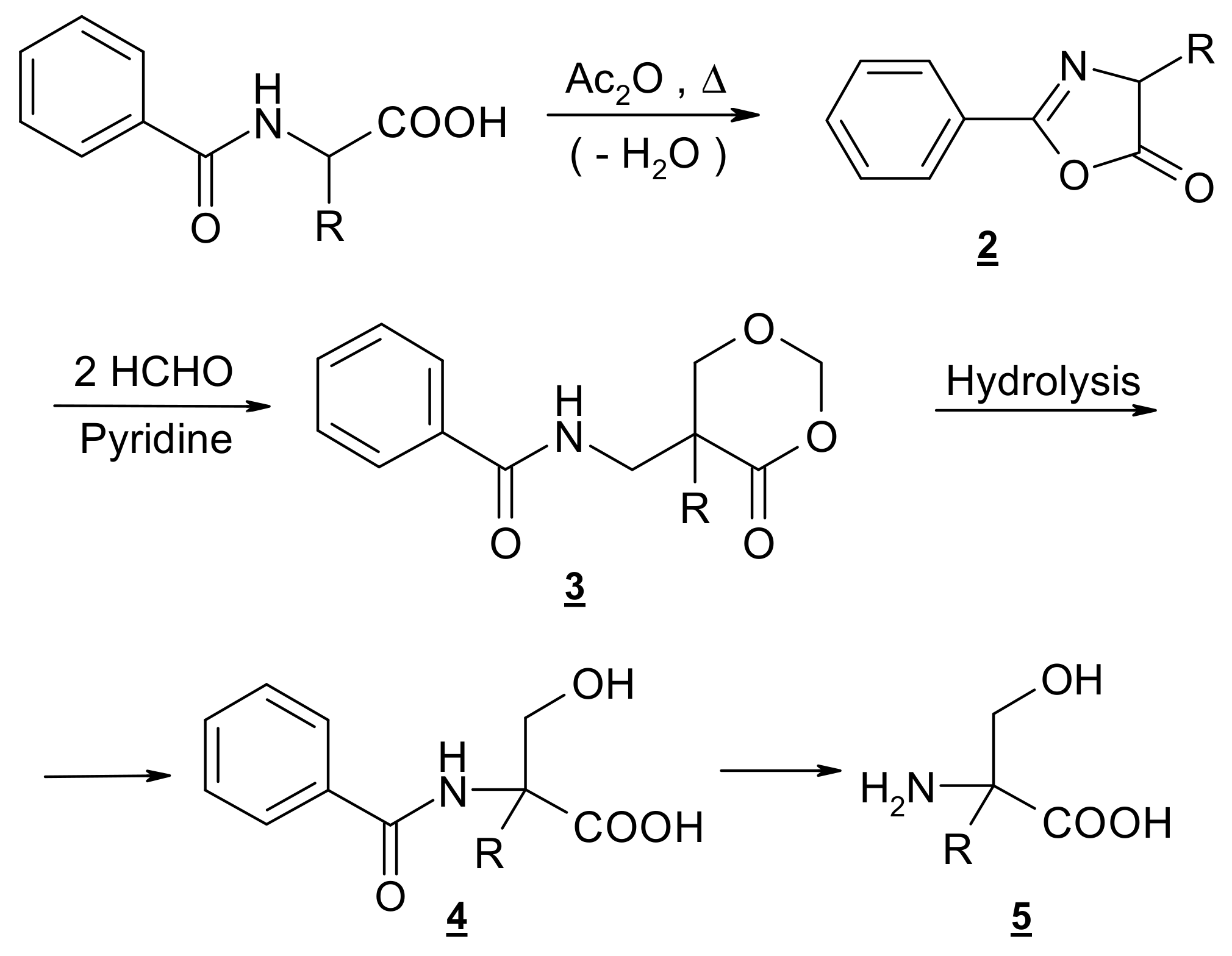
2.1. Synthesis
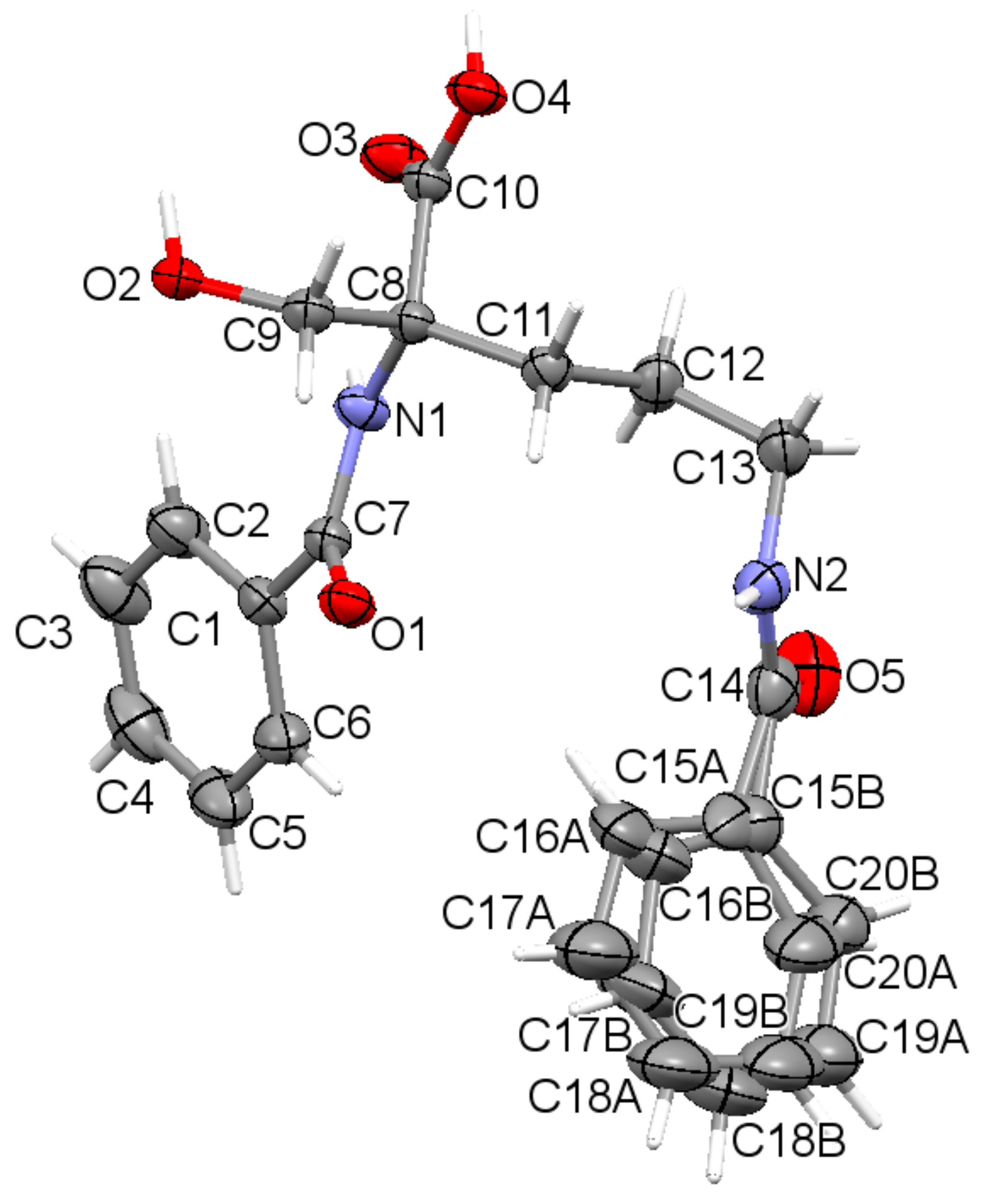
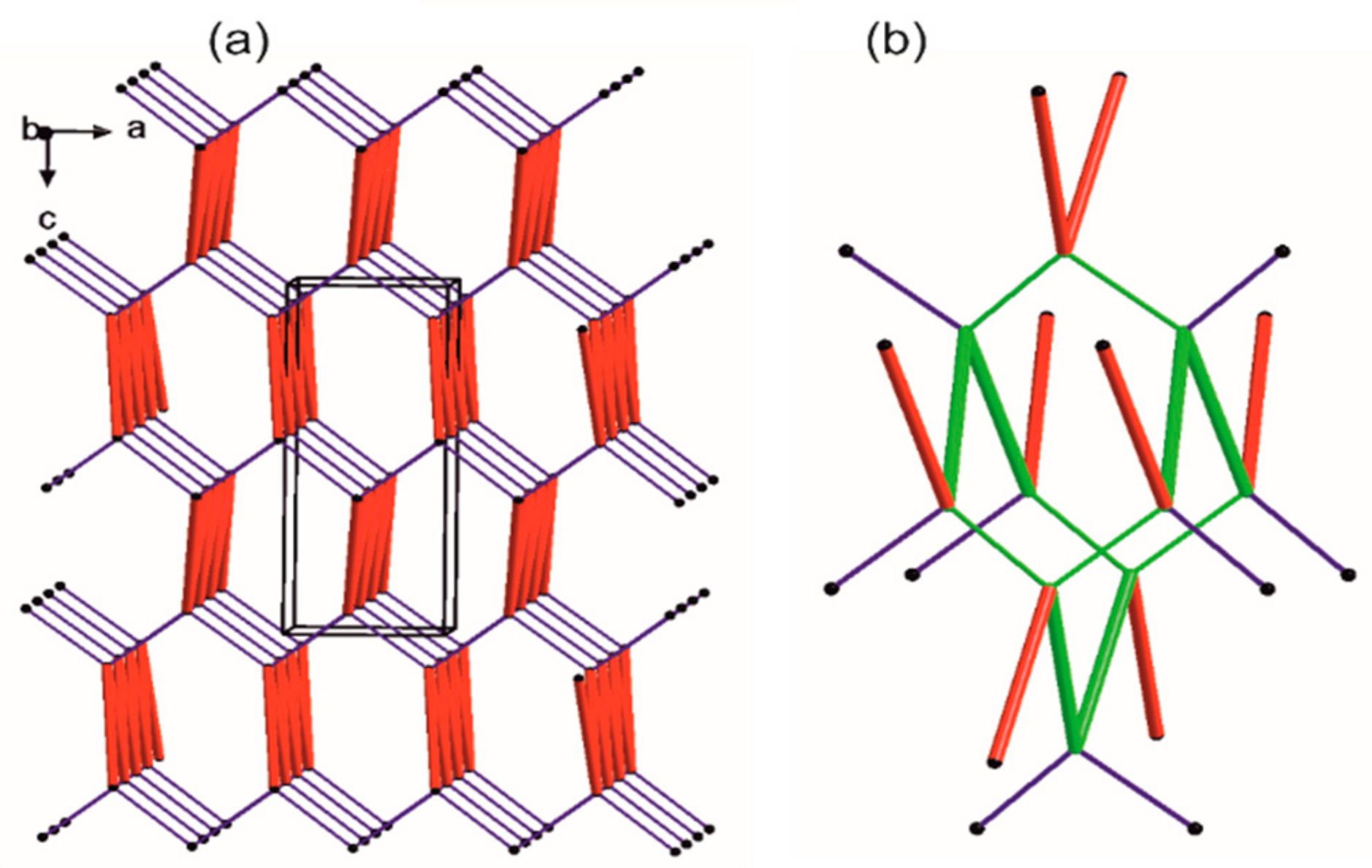
2.2. X-ray Crystal Structure
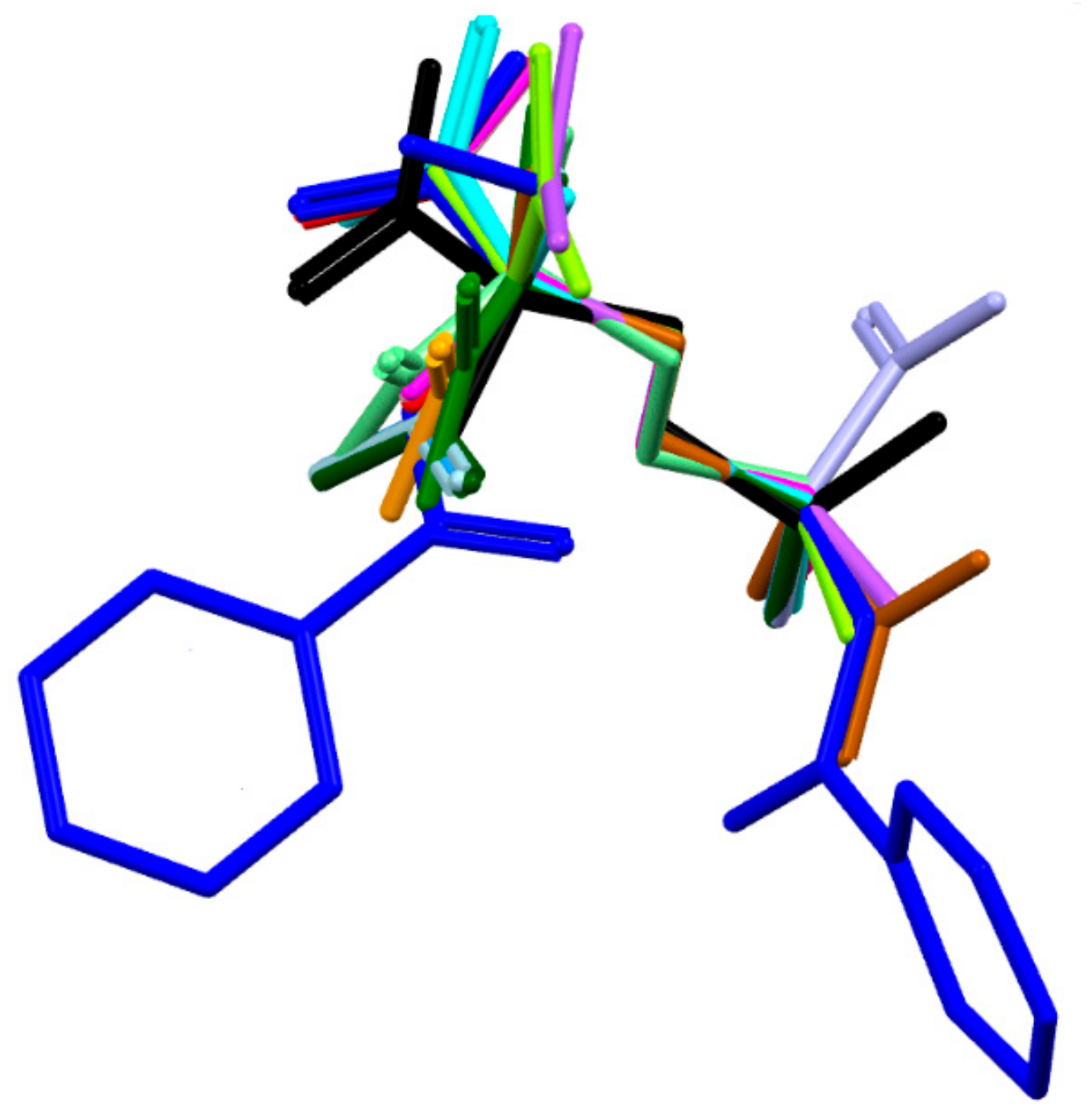
2.3. DFT Study
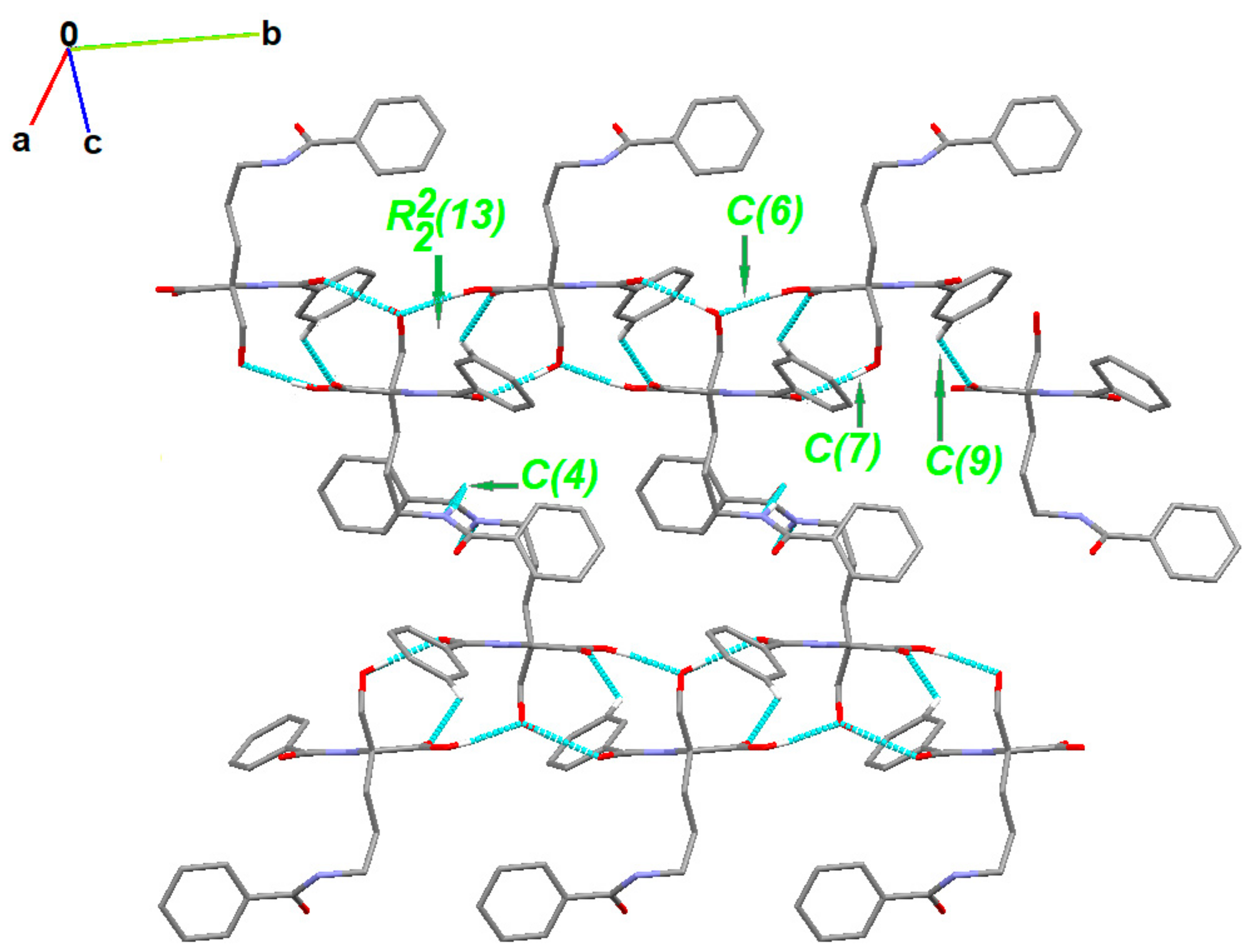
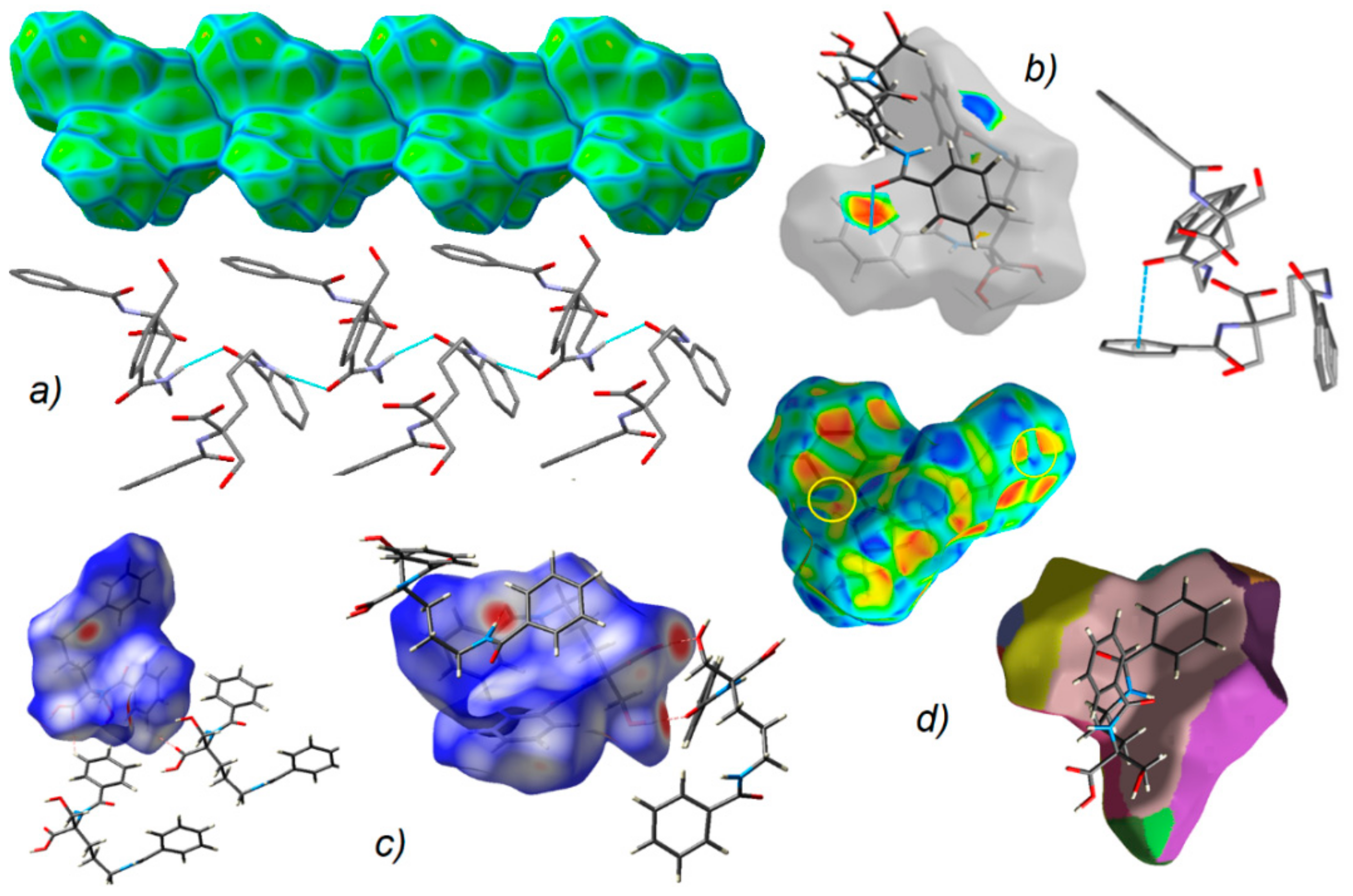
2.4. Supramolecular Commentary
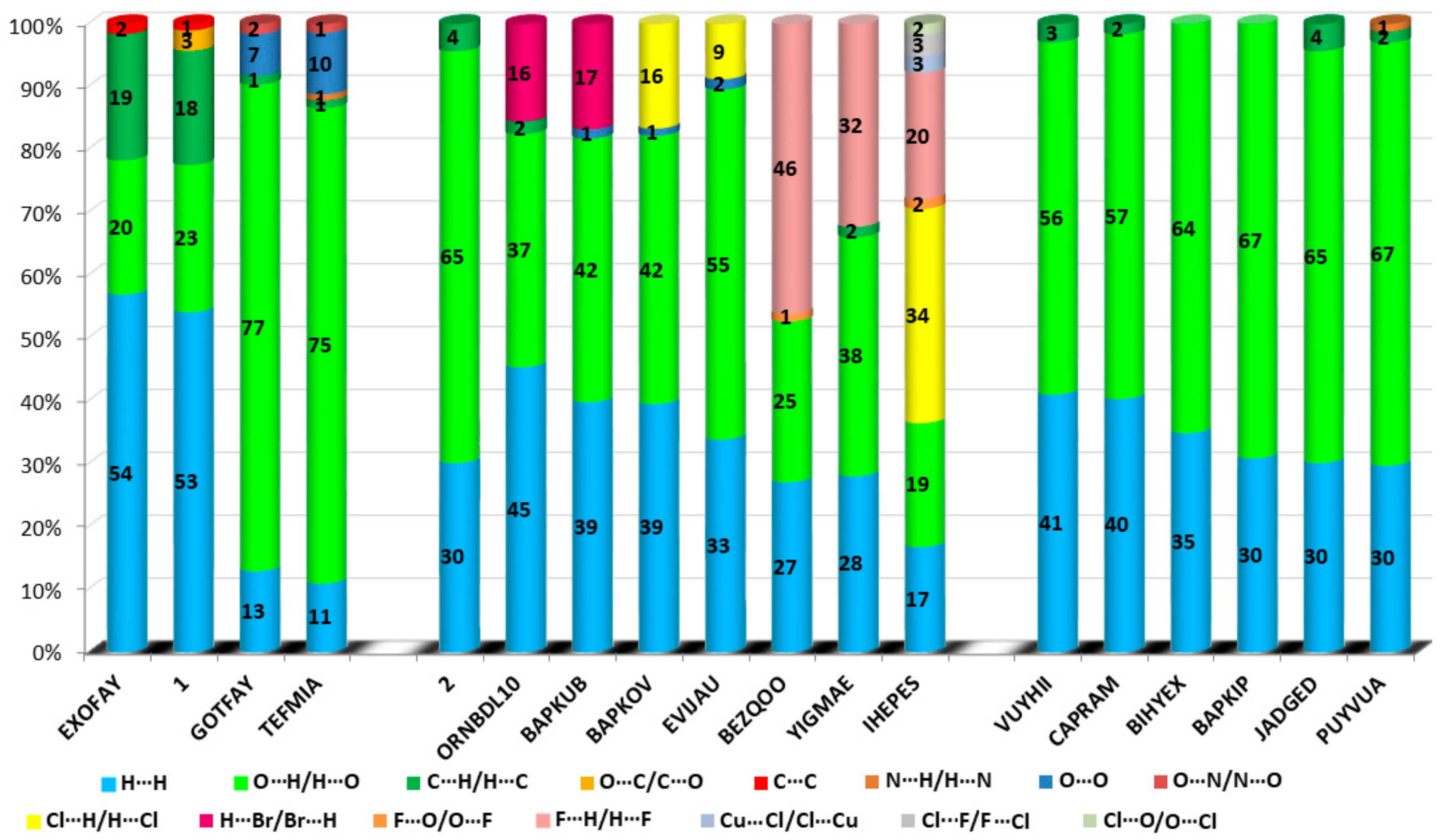
2.5. Hirshfeld Surface Analysis
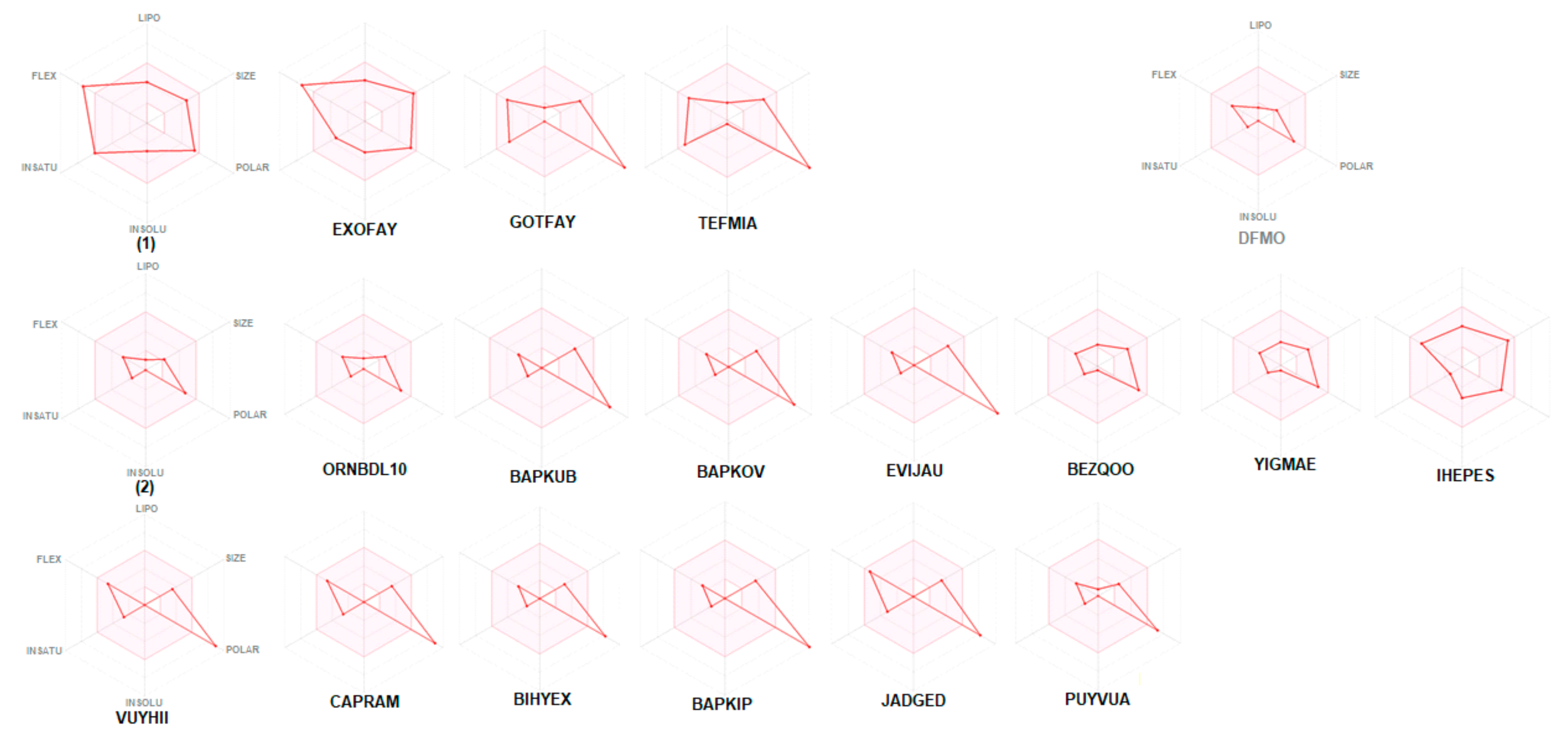
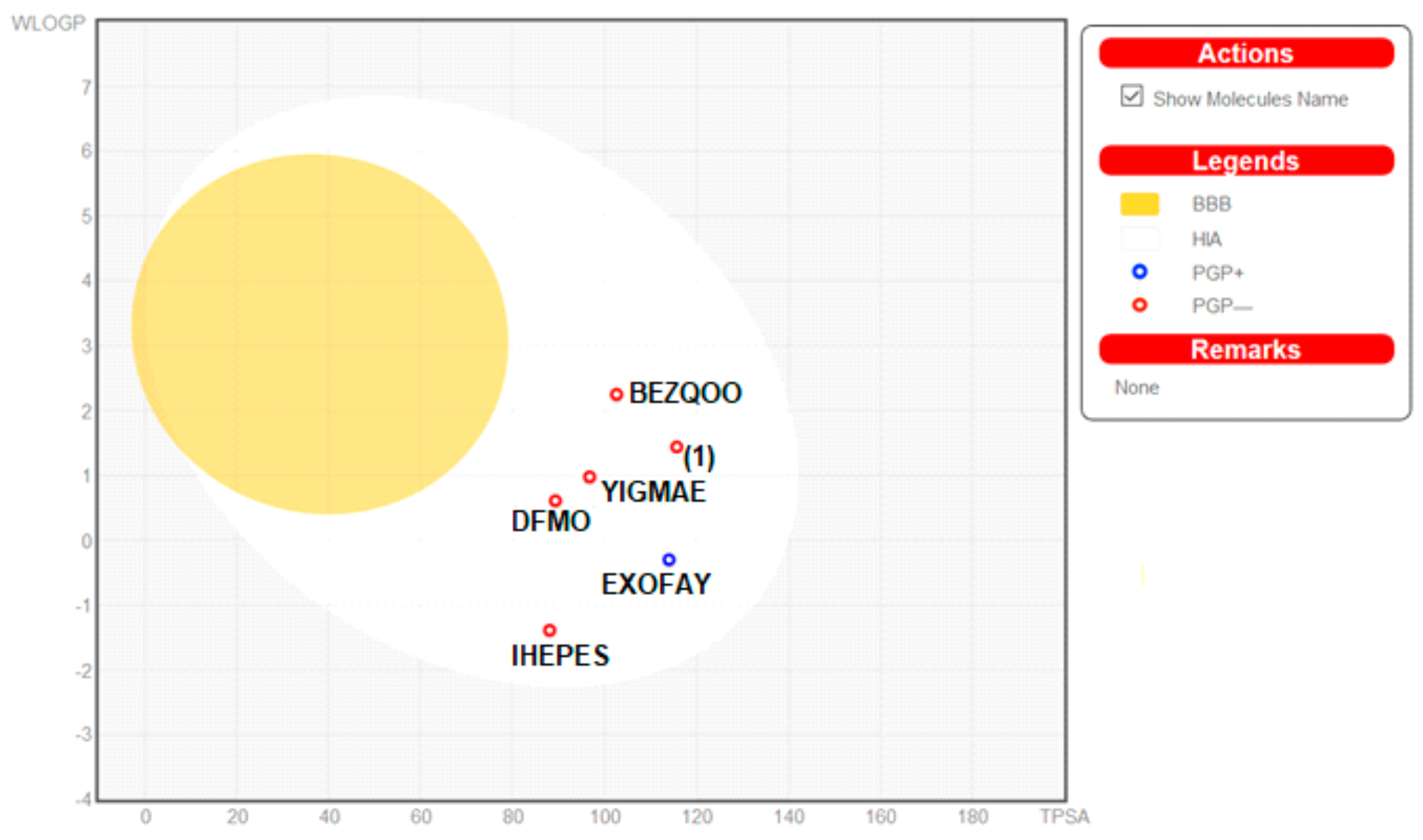
2.6. In Silico ADME Predictions
3. Materials and Methods
3.1. Crystallization, X-ray Structure Determinations
3.2. Computations
3.2.1. DFT Calculations
3.2.2. H-bond Energies
3.2.3. Hirshfeld Surface Analysis
3.2.4. In Silico ADME Screening
3.3. CSD and RCSB PDB Survey
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ABS | Absorption |
| ADME | absorption, distribution, metabolism and excretion |
| BBB | blood-brain barrier |
| BOILED-Egg | brain or intestinal estimated |
| CNS | central nervous system |
| CPCM | Conductor-like Polarizable Continuum Model |
| CSD | Cambridge Structure Database |
| DFMO | difluoroornithine |
| DFT | Density Functional Theory |
| de | distances from the Hirshfeld surface to the nearest atom outside (external) the surface |
| di | distances from the Hirshfeld surface to the nearest atom inside (internal) the surface |
| dnorm | normalized contact distance |
| EP | molecular electrostatic potential |
| FLEX | flexibility |
| HIA | human gastrointestinal absorption |
| HS | Hirshfeld surface analysis |
| FP | fingerprint plots |
| GI | gastro intestinal absorption |
| INSATU | insaturation |
| INSOLU | insolubility |
| LIPO | lipophilicilty |
| LSAM | Long-Range Synthon Aufbau Module |
| LSHB | the Lippincott and Schroeder H-bond model |
| P-gp | P-glycoprotein |
| POLAR | polarity |
| RCSB PDB | Research Collaboratory fof Structural Bioinformatics |
| RO5 | rule-of-five |
| SC-XRD | single-crystal X-ray diffraction |
| SMILES | simplified molecular-input line-entry system |
| SIB | Swiss Institute of Bioinformatics |
| TPSA | topological polar surface area |
| vdW | van derWaals radii |
References
- Bojarska, J.; Kaczmarek, K.; Zabrocki, J.; Wolf, W.M. Supramolecular Chemistry of Modified Amino Acids and Short Peptides. In Advances in Organic Synthesis; Atta-ur-Rahman, Ed.; Bentham Science Publishers, Ltd.: Sharjah, UAE, 2018; Volume 11, pp. 43–107. [Google Scholar]
- Bojarska, J.; Remko, M.; Wojciechowski, J.M.; Madura, I.D.; Olczak, A.; Kaczmarek, K.; Zabrocki, J.; Wolf, W.M. Supramolecular synthon polymorphism in modified amino acids. Structural, conformational and energy landscapes of N-benzoyl-2`-hydroxy-3-methylisovaline. J. Mol. Struct. 2019, 1190, 11–22. [Google Scholar] [CrossRef]
- Bojarska, J.; Kaczmarek, K.; Zabrocki, J.; Wolf, W.M. Amino Acids: Molecules of life. Int. J. Nutr. Sci. 2019, 4, 1035–1037. [Google Scholar]
- Bojarska, J.; Kaczmarek, K.; Zabrocki, J.; Wolf, W.M. Supramolecular synthons as related to cooperativity in biocomplexes: Towards design and development of oligopeptide-based modern drugs and cosmeceuticals. Biomaterials 2019, 129, 1–27. [Google Scholar]
- Bojarska, J.; Remko, M.; Kaczmarek, K.; Zabrocki, J.; Wolf, W.M. New synthons in supramolecular chemistry of short biologically active peptides. In Proceedings of the 32nd European Crystallographic Meeting, Vienna, Austria, 18–20 August 2019; p. MS35-P33, (soon in Supplement of Acta Cryst. A). [Google Scholar]
- Bojarska, J.; Kaczmarek, K.; Zabrocki, J.; Wolf, W. Novel biologically important supramolecular synthons in short peptides. In Proceedings of the World Peptide Congress, Osaka, Japan, 24–25 October 2019. [Google Scholar]
- Bojarska, J.; Kaczmarek, K.; Zabrocki, J.; Wolf, W. Supramolecular synthon concept targeting biomolecules. In Proceedings of the World Structural and Molecular Biology Conference, Rome, Italy, 26–28 November 2018. [Google Scholar]
- Rosowsky, A.; Bader, H.; Cucchi, C.A.; Moran, R.G.; Kohler, W.; Freisheim, J.H. Methotrexate analogues. N-delta-acyl-N-alpha-(4-amino-4-deoxypteroyl)-L-ornithine derivatives: Synthesis and in vitro antitumor activity. J. Med. Chem. 1988, 1, 1332–1337. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Spackman, P.; Yu, L.-J.; Morton, C.J.; Parker, M.W.; Bond, C.S.; Spackman, M.A.; Jayatilaka, D.; Thomas, S. Bridging Crystal Engineering and Drug Discovery by Utilizing Intermolecular Interactions and Molecular Shapes in Crystals. Angew. Chem. 2019, 131, 16936–16940. [Google Scholar] [CrossRef]
- Van Dun, S.; Ottmann, C.; Milroy, L.-G.; Brunsveld, L. Supramolecular Chemistry Targeting Proteins. J. Am. Chem. Soc. 2017, 139, 13960–13968. [Google Scholar] [CrossRef]
- Jin, X.; Zhu, L.; Xue, B.; Zhu, X.; Yan, D. Supramolecular nanoscale drug-delivery system with ordered structure. Natl. Sci. Rev. 2019, 6, 1128–1137. [Google Scholar] [CrossRef]
- Fahmy, S.A.; Brüßler, J.; Alawak, M.; El-Sayed, M.; Bakowsky, U.; Shoeib, T. Chemotherapy Based on Supramolecular Chemistry: A Promising Strategy in Cancer Therapy. Pharm. 2019, 11, 292. [Google Scholar] [CrossRef]
- Bulusu, G.; Desiraju, G.R. Strong and Weak Hydrogen Bonds in Protein–Ligand Recognition. J. Indian Inst. Sci. 2019, 100, 31–41. [Google Scholar] [CrossRef]
- Desiraju, G.R. Supramolecular Synthons in Crystal Engineering—A New Organic Synthesis. Angew. Chem. Int. Ed. 1995, 34, 2311–2327. [Google Scholar] [CrossRef]
- Murherjee, A. Building upon supramolecular synthons: Some aspects of crystal engineering. Cryst. Growth & Des. 2015, 15, 3076–3085. [Google Scholar]
- SIB Swiss Institute of Bioinformatics Members The SIB Swiss Institute of Bioinformatics’ resources: Focus on curated databases. Nucleic Acids Res. 2015, 44, D27–D37.
- Daina, A.; Michielin, O.; Zoete, V. iLOGP: A Simple, Robust, and Efficient Description of n-Octanol/Water Partition Coefficient for Drug Design Using the GB/SA Approach. J. Chem. Inf. Model. 2014, 54, 3284–3301. [Google Scholar] [CrossRef]
- Daina, A.; Zoete, V. A BOILED-Egg To Predict Gastrointestinal Absorption and Brain Penetration of Small Molecules. Chem. Med. Chem. 2016, 11, 1117–1121. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Daina, A.; Blatter, M.-C.; Gerritsen, V.B.; Palagi, P.M.; Marek, D.; Xenarios, I.; Schwede, T.; Michielin, O.; Zoete, V. Drug Design Workshop: A Web-Based Educational Tool To Introduce Computer-Aided Drug Design to the General Public. J. Chem. Educ. 2017, 94, 335–344. [Google Scholar] [CrossRef]
- Banerjee, P.; Eckert, O.A.; Schrey, A.K.; Preissner, R. ProTox-II: A webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018, 46, W257–W263. [Google Scholar] [CrossRef]
- Drwal, M.N.; Banerjee, P.; Dunkel, M.; Wettig, M.R.; Preissner, R. ProTox: A webserver for the in silico predicition of rodent oral toxicity. Nucleic Acid Res 2014, 42, W53–W58. [Google Scholar] [CrossRef]
- Jiang, L.-Y.; Ling, C.; Zhang, Y.-Y.; Liu, J.-Z. Metabolic evolution of Corynebacterium glutamicum for increased production of L-ornithine. BMC Biotechnol. 2013, 13, 47. [Google Scholar] [CrossRef]
- Qian, Z.-G.; Xia, X.-X.; Lee, S.Y. Metabolic engineering ofEscherichia colifor the production of putrescine, a four carbon diamine. Biotechnol. Bioeng. 2009, 104, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Schneider, J.; Eberhardt, D.; Wendisch, V.F. Improving putrescine production by Corynebacterium glutamicum by fine-tuning ornithine transcarbomolyase activity using a plasmid addiction system. Appl. Microbiol. Biotechnol. 2012, 95, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.; Kaan, H.Y.K.; Zheng, X.; Tang, X.; He, Y.; Tan, Q.V.; Zhang, N.; Song, H. Structural basis of Ornithine Decarboxylase inactivation and accelerated degradation by polyamine sensor Antizyme1. Sci. Rep. 2015, 5, 14738. [Google Scholar] [CrossRef] [PubMed]
- Ginguay, A.; Cynober, L.; Curis, E.; Nicolis, I. Ornithine Aminotransferase, an Important Glutamate-Metabolizing Enzyme at the Crossroads of Multiple Metabolic Pathways. Biology 2017, 6, 18. [Google Scholar] [CrossRef] [PubMed]
- Andersson, L.; Blomberg, L.; Flegel, M.; Lepsa, L.; Nilsson, B.; Verlander, M. Large-scale synthesis of peptides. Biopolym. 2000, 55, 227–250. [Google Scholar] [CrossRef]
- Syud, E.A.; Stanger, H.E.; Gellman, S.H. Rules for antiparallel ß-sheet design: D-Pro-Gly is superior to 1-Asn-Gly for ß-hairpin nucleation. J. Am. Chem. Soc. 1998, 123, 8667–8677. [Google Scholar] [CrossRef]
- Salvatore, F.; Cimino, F.; D’Ayello-Caracciolo, M.; Cittadini, D. Mechanism of the protection by l-ornithine-l-aspartate mixture and by l-arginine in ammonia intoxication. Arch. Biochem. Biophys. 1964, 107, 499–503. [Google Scholar] [CrossRef]
- Shi, H.P.; Fishel, R.S.; Efron, D.T.; Williams, J.Z.; Fishel, M.H.; Barbul, A. Effect of supplemental ornithine on wound healing. J. Surg. Res. 2002, 106, 299–302. [Google Scholar] [CrossRef]
- Bucci, L.; Hickson, J.F.; Pivarnik, J.M.; Wolinsky, I.; McMahon, J.C.; Turner, S.D. Ornithine ingestion and growth hormone release in bodybuilders. Nutr. Res. 1990, 10, 239–245. [Google Scholar] [CrossRef]
- Harada, D.; Nagamachi, S.; Aso, K.; Ikeda, K.; Takahashi, Y.; Furuse, M. Oral administration of l-ornithine increases the content of both collagen constituting amino acids and polyamines in mouse skin. Biochem. Biophys. Res. Commun. 2019, 512, 712–715. [Google Scholar] [CrossRef]
- Miyake, M.; Kirisako, T.; Kokubo, T.; Miura, Y.; Morishita, K.; Okamura, H.; Tsuda, A. Randomised controlled trial of the effects of L-ornithine on stress markers and sleep quality in healthy workers. Nutr. J. 2014, 13, 53. [Google Scholar] [CrossRef] [PubMed]
- Demura, S.; Morishita, K.; Yamada, T.; Yamaji, S.; Komatsu, M. Effect of L-ornithine hydrochloride ingestion on intermittent maximal anaerobic cycle ergometer performance and fatigue recovery after exercise. Graefe’s Arch. Clin. Exp. Ophthalmol. 2011, 111, 2837–2843. [Google Scholar] [CrossRef] [PubMed]
- Sugino, T.; Shirai, T.; Kajimoto, Y.; Kajimoto, O. l-Ornithine supplementation attenuates physical fatigue in healthy volunteers by modulating lipid and amino acid metabolism. Nutr. Res. 2008, 28, 738–743. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Falla, T.J. Cosmeceuticals and peptides. Clin. Dermatol. 2009, 27, 485–494. [Google Scholar] [CrossRef]
- Ito, N.; Saito, H.; Seki, S.; Ueda, F.; Asada, T. Effects of Composite Supplement Containing Astaxanthin and Sesamin on Cognitive Functions in People with Mild Cognitive Impairment: A Randomized, Double-Blind, Placebo-Controlled Trial. J. Alzheimer’s Dis. 2018, 62, 1767–1775. [Google Scholar] [CrossRef]
- Vong, L.B.; Ibayashi, Y.; Lee, Y.; Ngo, D.-N.; Nishikawa, Y.; Nagasaki, Y. Poly(ornithine)-based self-assembling drug for recovery of hyperammonemia and damage in acute liver injury. J. Control. Release 2019, 310, 74–81. [Google Scholar] [CrossRef]
- Bachrach, U.; Weinstein, A. Effect of Aliphatic Polyamines on Growth and Macromolecular Syntheses in Bacteria. J. Gen. Microbiol. 1970, 60, 159–165. [Google Scholar] [CrossRef]
- Russell, D.H. The roles of the polyamines, putrescine, spermidine, and spermine in normal and malignant tissues. Life Sci. 1973, 13, 1635–1647. [Google Scholar] [CrossRef]
- Somani, R.R.; Rai, P.R.; Kandpile, P.S. Ornithine Decarboxylase Inhibition: A Strategy to Combat Various Diseases. Mini-Reviews Med. Chem. 2018, 18, 1008–1021. [Google Scholar] [CrossRef]
- Pegg, A.E. Regulation of Ornithine Decarboxylase. J. Boil. Chem. 2006, 281, 14529–14532. [Google Scholar] [CrossRef]
- Valdés-Santiago, L.; Ruiz-Herrera, J. Stress and polyamine metabolism in fungi. Front. Chem. 2014, 1, 42. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.; Dey, A.; Gupta, B. Plant polyamines in abiotic stress responses. Acta Physiol. Plant. 2013, 35, 2015–2036. [Google Scholar] [CrossRef]
- El-Sayed, A.S.; George, N.M.; Yassin, M.A.; Alaidaroos, B.A.; Bolbol, A.A.; Mohamed, M.S.; Rady, A.M.; Azize, S.W.; Zayed, R.A.; Sitohy, M.Z. Purification and Characterization of Ornithine Decarboxylase from Aspergillus terreus; Kinetics of Inhibition by Various Inhibitors. Mol. 2019, 24, 2756. [Google Scholar] [CrossRef] [PubMed]
- Bey, P.; Van Dorseelar, V.; Mamont, P.; Jung, M.; Tardif, C. Analogues of ornithine as inhibitors of ornithine decarboxylase. New deductions concerning the topography of the enzyme`s active site. J. Med. Chem. 1978, 21, 50–55. [Google Scholar] [CrossRef]
- Meyskens, F.L.; Gerner, E.W. Development of difluoromethylornithine (DFMO) as a chemoprevention agent. Clin. Cancer Res. 1999, 5, 945–951. [Google Scholar]
- Alexiou, G.A.; Lianos, G.D.; Ragos, V.; Galani, V.; Kyritsis, A.P. Difluoromethylornithine in cancer: New advances. Futur. Oncol. 2017, 13, 809–819. [Google Scholar] [CrossRef]
- Hukkeri, S.; Ambrosio, A.B. In-silico Identification of Candidate Inhibitory Ligands against Ornithine Decarboxylase Enzyme for Human Sleeping Sickness Causing Trypanosoma brucei. J. Biomol. Res. Ther. 2017, 6, 1–7. [Google Scholar]
- Enríquez, M.M.M.; Alcántara-Farfán, V.; Faisal, J.L.A.; Trujillo-Ferrara, J.G.; Rodriguez-Paez, L.; Vargas-Ramírez, A.L. N-ω-chloroacetyl-L-ornithine, a new competitive inhibitor of ornithine decarboxylase, induces selective growth inhibition and cytotoxicity on human cancer cells versus normal cells. J. Enzym. Inhib. Med. Chem. 2014, 30, 345–353. [Google Scholar] [CrossRef]
- Vargas-Ramírez, A.L.; Enríquez, M.M.M.; Cordero-Rodríguez, N.I.; Ruiz-Cuello, T.; Faisal, J.L.A.; Trujillo-Ferrara, J.G.; Alcántara-Farfán, V.; Rodriguez-Paez, L. N-ω-chloroacetyl-L-ornithine has in-vitro activity against cancer cell lines and in-vivo activity against ascitic and solid tumors. Anti-Cancer Drugs 2016, 27, 508–518. [Google Scholar] [CrossRef]
- El Bissati, K.; Redel, H.; Ting, L.-M.; Lykins, J.D.; McPhillie, M.J.; Upadhya, R.; Woster, P.M.; Yarlett, N.; Kim, K.; Weiss, L.M. Novel Synthetic Polyamines Have Potent Antimalarial Activities in vitro and in vivo by Decreasing Intracellular Spermidine and Spermine Concentrations. Front. Microbiol. 2019, 9, 9. [Google Scholar] [CrossRef]
- Kim, H.I.; Schultz, C.R.; Buras, A.L.; Friedman, E.; Fedorko, A.; Seamon, L.; Chandramouli, G.V.R.; Maxwell, G.L.; Bachmann, A.; Risinger, J. Ornithine decarboxylase as a therapeutic target for endometrial cancer. PLoS ONE 2017, 12, e0189044. [Google Scholar] [CrossRef] [PubMed]
- Levin, V.A.; Ictech, S.E.; Hess, K.R. Clinical importance of eflornithine (α-difluoromethylornithine) for the treatment of malignant gliomas. CNS Oncol. 2018, 7, CNS16. [Google Scholar] [CrossRef] [PubMed]
- Sierra, J.C.; Suarez, G.; Piazuelo, M.B.; Luis, P.B.; Baker, D.R.; Romero-Gallo, J.; Barry, D.P.; Schneider, C.; Morgan, D.R.; Peek, R.M.; et al. Alpha-difluoromethylornithine reduces gastric carcinogenesis by causing mutations in Heliobacter pyroli cagY. PNAS 2019, 116, 5077–5085. [Google Scholar] [CrossRef] [PubMed]
- Sholler, G.L.S.; Ferguson, W.; Bergendahl, G.; Bond, J.P.; Neville, K.; Eslin, D.; Brown, V.; Roberts, W.; Wada, R.K.; Oesterheld, J.; et al. Maintenance DFMO Increases Survival in High Risk Neuroblastoma. Sci. Rep. 2018, 8, 14445. [Google Scholar] [CrossRef] [PubMed]
- Logiudice, N.; Le, L.; Abuan, I.; Leizorek, Y.; Roberts, S. Alpha-Difluoromethylornithine, an Irreversible Inhibitor of Polyamine Biosynthesis, as a Therapeutic Strategy against Hyperproliferative and Infectious Diseases. Med Sci. 2018, 6, 12. [Google Scholar] [CrossRef]
- Obaleye, J.A.; Tella, A.C.; Osunniran, W.A.; Simon, N.; Omojasola, P. Synthesis, Characterization, Crystal Structure and Antimicrobial Evaluation of a Novel –M–X–M–X– Type Infinite Chain 1D Cu(II) Complex with Eflornithine Hydrochloride Hydrate as Ligand. J. Inorg. Organomet. Polym. Mater. 2014, 24, 827–835. [Google Scholar] [CrossRef]
- Trujillo-Ferrara, J.G. An approach to anti-cancer therapy with multi-targeting ligands. J. Cancer Sci. Ther. 2014, 6, 10. [Google Scholar]
- Luna-Rojas, L.; Avila-Trejo, A.M.; Farfán, V.A.; Rodríguez-Páez, L.; Pastor-Alonso, M.O.; Aguilar-Faisal, J.L.; Pastor-Alonso, O.; Faisal, J.L.A. Antiviral activity of N-ω-Chloroacetyl-L-Ornithine on in vitro replication of Chikungunya virus. bioRxiv 2019, arXiv:745455. [Google Scholar]
- Raghavendra, N.M.; Pingili, D.; Kadasi, S.; Mettu, A.; Prasad, S. Dual or multi-targeting inhibitors: The next generation anticancer agents. Eur. J. Med. Chem. 2018, 143, 1277–1300. [Google Scholar] [CrossRef]
- Ramsay, R.R.; Nikolić, M.P.; Nikolic, K.; Uliassi, E.; Bolognesi, M.L. A perspective on multi-target drug discovery and design for complex diseases. Clin. Transl. Med. 2018, 7, 3. [Google Scholar] [CrossRef]
- Singh, H.; Kinarivala, N.; Sharma, S. Multi-Targeting Anticancer Agents: Rational Approaches, Synthetic Routes and Structure Activity Relationship. Anti-Cancer Agents Med. Chem. 2019, 19, 842–874. [Google Scholar] [CrossRef] [PubMed]
- Alcaro, S.; Bolognesi, M.L.; García-Sosa, A.T.; Rapposelli, S. Editorial: Multi-Target-Directed Ligands (MTDL) as Challenging Research Tools in Drug Discovery: From Design to Pharmacological Evaluation. Front. Chem. 2019, 7, 71. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.K.; Gemma, S. Structure-Based Design of Drugs and Other Bioactive Molecules; Wiley: Hoboken, NJ, USA, 2014. [Google Scholar]
- Batool, M.; Ahmad, B.; Choi, S. A Structure-Based Drug Discovery Paradigm. Int. J. Mol. Sci. 2019, 20, 2783. [Google Scholar] [CrossRef] [PubMed]
- Bissantz, C.; Kuhn, B.; Stahl, M. A medicinal chemists`s guide to molecular interactions. J. Med. Chem. Perspective. 2010, 53, 5061–5084. [Google Scholar] [CrossRef] [PubMed]
- Vepuri, S.B.; Anbazhagan, S.; Divya, D.; Padmini, D. A review on supramolecular chemistry in drug design and formulation research. Indonesian J. Pharm. 2013, 24, 131–150. [Google Scholar]
- Sarkhel, S.; Desiraju, G.R. N-H. O, O-H. O and C-H. O Hydrogen Bonds in Protein–Ligand Complexes: Strong and Weak Interactions in Molecular Recognition. Proteins 2004, 54, 247–259. [Google Scholar] [CrossRef]
- Panigrahi, S.K. Strong and weak hydrogen bonds in protein-ligand complexes of kinases: A comparative study. Amino Acids 2008, 34, 617–633. [Google Scholar] [CrossRef]
- Armstrong, D.R.; Berrisford, J.M.; Conroy, M.J.; Gutmanas, A.; Anyango, S.; Choundhary, P.; Clark, A.R.; Dana, J.M.; Deshpande, M.; Dunlop, R. PDBe: Improved fundability of macromolecular structure data in the PDB. Nucleic Acids Res. 2020, D1, D335–D343. [Google Scholar] [CrossRef]
- Burley, S.K.; Berman, H.M.; Bhikadiya, C.; Bi, C.; Chen, L.; Di Costanzo, L.; Christie, C.; Dalenberg, K.; Duarte, J.M.; Dutta, S.; et al. RCSB Protein Data Bank: Biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2018, 47, D464–D474. [Google Scholar] [CrossRef]
- Allen, F.H. The Cambridge Structural Database: A quarter of a million crystal structures and rising. Acta Crystallogr. Sect. B Struct. Sci. 2002, 58, 380–388. [Google Scholar] [CrossRef]
- Groom, C.; Bruno, I.; Lightfoot, M.; Ward, S.C. The Cambridge Structural Database. Acta Crystallogr. Sect. B Struct. Sci. Cryst. Eng. Mater. 2016, 72, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.; Wiggin, S.; Stanzione, F. New insights and innovation from a million crystal structures in the Cambridge Structural Database. Struct. Dyn. 2019, 6, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Vologzhanina, A.V. Intermolecular Interactions in Functional Crystalline Materials: From Data to Knowledge. Cryst. 2019, 9, 478–528. [Google Scholar] [CrossRef]
- Groom, C.; Cole, J.C. The use of small-molecule structures to complement protein-ligand crystal structures in drug discovery. Acta Crystallogr. Sect. D Struct. Boil. 2017, 73, 240–245. [Google Scholar] [CrossRef] [PubMed]
- Kaminski, Z.J.; Leplawy, M.T.; Zabrocki, J. α-Hydroxymethylation of α-Amino-acids. Synth. 1973, 1973, 792–793. [Google Scholar] [CrossRef]
- Kaminski, Z.; Leplawy, M.T. Preparation of N-acyl-α-alkylerines by selective cleavage of 5-acylamino-5-alkyl-4-oxo-1,3-dioxanes. Synthesis 1974, 292–293. [Google Scholar] [CrossRef]
- Ito, Y.; Ma, Y. Peptides. In Proceedings of the Encyclopedia of Chromatography, 3rd ed.; (Print Version); Informa UK: Colchester, UK, 2009; p. 323. [Google Scholar]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. GAUSSIAN09. Revision D.01; Gaussian Inc.: Wallingford, CT, USA, 2013. [Google Scholar]
- Parr, R.G.; Wang, W. Density-Functional Theory of Atoms and Molecules; Oxford University Press: New York, NY, USA, 1994. [Google Scholar]
- Neumann, R.; Nobes, R.H.; Handy, N.C. Exchange functionals and potentials. Mol. Physics 1996, 87, 1–36. [Google Scholar] [CrossRef]
- Bickelhaupt, F.M.; Baerends, E.J. Kohn-Sham Density Functional Theory: Predicting and Understanding Chemistry. Rev. Comp. Ch. 2007, 15, 1–86. [Google Scholar]
- Zhao, Y.; Truhlar, D.G. The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: Two new functionals and systematic testing of four M06 functionals and 12 other functionals. Theor. Chem. Accounts 2008, 119, 525. [Google Scholar] [CrossRef]
- Hehre, W.J. Ab initio molecular orbital theory. Accounts Chem. Res. 1976, 9, 399–406. [Google Scholar] [CrossRef]
- Klamt, A.; Schüürmann, G. COSMO: A new approach to dielectric screening in solvents with explicit expressions for the screening energy and its gradient. J. Chem. Soc. Perkin Trans. 2 1993, 2, 799–805. [Google Scholar] [CrossRef]
- Barone, V.; Cossi, M. Quantum Calculation of Molecular Energies and Energy Gradients in Solution by a Conductor Solvent Model. J. Phys. Chem. A 1998, 102, 1995–2001. [Google Scholar] [CrossRef]
- Cossi, M.; Rega, N.; Scalmani, G.; Barone, V. Energies structures, and electronic properties of molecules in solution with the C-PCM salvation model. J. Comp. Chem. 2003, 24, 669–681. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, J.; Davis, R.E.; Shimoni, L.; Chang, N.-L. Patterns in Hydrogen Bonding: Functionality and Graph Set Analysis in Crystals. Angew. Chem. Int. Ed. 1995, 34, 1555–1573. [Google Scholar] [CrossRef]
- Etter, M.C. Encoding and decoding hydrogen-bond patterns of organic compounds. Accounts Chem. Res. 1990, 23, 120–126. [Google Scholar] [CrossRef]
- Etter, M.C. Hydrogen bonds as design elements in organic chemistry. J. Phys. Chem. 1991, 95, 4601–4610. [Google Scholar] [CrossRef]
- Ganguly, P.; Desiraju, G.R. Long-range synthon Aufbau modules (LSAM) in crystal structures: Systematic changes in C6H6−nFn(0 ≤ n ≤ 6) fluorobenzenes. Cryst. Eng. Comm. 2010, 12, 817–833. [Google Scholar] [CrossRef]
- Wells, A.F.; Pearce, P.; Shlichta, P.J. Three Dimensional Nets and Polyhedra and Structure in Nature is a Strategy for Design. Phys. Today 1979, 32, 51. [Google Scholar] [CrossRef]
- Brylinski, M. Aromatic interactions at the ligand-protein interface: Implications for the development of docking scoring functions. Chem. Boil. Drug Des. 2017, 91, 380–390. [Google Scholar] [CrossRef]
- Mazur, L.; Koziol, A.E.; Rzeszotarska, B.; Masiukiewicz, E. Molecular and crystal structure of Nα-(9-fluorenyl)-methoxycarbonyl-L-ornithine hydrochloride diethyl ether solvate. Int. J. Pept. Res. Ther. 2002, 9, 255–260. [Google Scholar] [CrossRef]
- Balaprabhakaran, S.; Chandrasekaran, J.; Babu, B.; Thirumurugan, R.; Anitha, K. Synthesis, crystal growth and physiochemical characterization of organic NLO crystal: L-ornithinium dipicrate (LODP). Spectrochim. Acta Part A: Mol. Biomol. Spectrosc. 2015, 136, 700–706. [Google Scholar] [CrossRef] [PubMed]
- Nagata, H.; In, Y.; Tomoo, K.; Doi, M.; Ishida, T.; Wakahara, A. Structural Feature and Molecular Interaction of Basic Amino Acid-Picric Acid Complexes by X-Ray Crystal Analyses. Chem. Pharm. Bull. 1995, 43, 1836–1843. [Google Scholar] [CrossRef]
- Jackson, L.K.; Goldsmith, E.J.; Phillips, M.A. X-ray structure of trypanosome brucei ornithine decarboxylase bound to D- ornithine and to G418: Insights into substrate binding and ODC conformational flexibility. J. Biol. Chem. 2003, 278, 22037–22043. [Google Scholar] [CrossRef] [PubMed]
- Grishin, N.V.; Osterman, A.L.; Brooks, H.B.; Phillips, M.A.; Goldsmith, E.J. X-ray structure of ornithine decarboxylase from Trypanosoma brucei: The native structure and the structure in complex with alpha-difluoromethylornithine. Biochem. 1999, 38, 15174–15184. [Google Scholar] [CrossRef]
- Grabowski, S.J. Noncovalent Interactions in Crystal Structures: Quantifying Cooperativity in Hydrogen and Halogen Bonds. In Intermolecular Interactions in Crystals: Fundamentals of Crystal Engineering; Novoa, J.J., Ed.; Royal Society in Chemistry: London, UK, 2008; Volume 4, pp. 673–718. [Google Scholar]
- Mahadevi, A.S.; Narahari, S. Cooperativity in noncovalent ineractions. Chem. Rev. 2016, 116, 2775–2825. [Google Scholar] [CrossRef]
- Spackman, M.; Jayatilaka, D. Hirshfeld surface analysis. Cryst. Eng.Comm. 2009, 11, 19–32. [Google Scholar] [CrossRef]
- Spackman, M.A.; McKinnon, J.J. Fingerprint intermolecular interactions in molecular crystals. Cryst. Eng. Comm. 2002, 4, 378–392. [Google Scholar] [CrossRef]
- Fleck, M.; Ghazaryan, V.V.; Petrosyan, A.M. Amino acid hexafluorosilicates–an overview. Z. Kristallogr. 2013, 228, 240–249. [Google Scholar] [CrossRef]
- Ghazaryan, V.; Fleck, M.; Petrosyan, A.M. Salts of amino acids with hexafluorosilicate anion. J. Cryst. Growth 2013, 362, 162–166. [Google Scholar] [CrossRef]
- Kalyanaraman, A.R.; Srinivasan, R. The crystal structure of DL-ornithine hydrobromide. Acta Cryst. B. 1971, 27, 1420–1427. [Google Scholar] [CrossRef]
- Ghazaryan, V.V.; Fleck, M.; Petrosyan, A.M. Mixed salts of amino acids: New analogs of the di-L-ornithinum(2+) chloride nitrate sulfate crystal. J. Crystallization Phys. Chem. 2: 7–16. J. Cryst. Phys. Chem. 2011, 2, 7–16. [Google Scholar]
- Ramaswamy, S.; Sridhar, B.; Ramakrishnan, V.; Rajaram, R.K. Bis( DL -methioninium) sulfate. Acta Cryst. Sect. E. 2004, 60, 768–770. [Google Scholar] [CrossRef]
- Corpinot, M.K.; Bučar, D.-K. A Practical Guide to the Design of Molecular Crystals. Cryst. Growth Des. 2018, 19, 1426–1453. [Google Scholar] [CrossRef]
- Krämer, S.D.; Wunderli-Allenspach, H. Physicochemical properties in pharmacokinetic lead optimization. Il Farm. 2001, 56, 145–148. [Google Scholar] [CrossRef]
- Neervannan, S. Preclinical formulations for discovery and toxicology: Physicochemical challenges. Expert Opin. Drug Metab. Toxicol. 2006, 2, 715–731. [Google Scholar] [CrossRef] [PubMed]
- Sliwoski, G.; Kothiwale, S.; Meiler, J.; Lowe, E.W. Computational methods in drug discovery. Pharmacol. Rev. 2013, 66, 334–395. [Google Scholar] [CrossRef]
- Müller, J.; Martins, A.; Csábi, J.; Fenyvesi, F.; Könczöl, Á.; Hunyadi, A.; Balogh, G.T. BBB penetration-targeting physicochemical lead selection: Ecdysteroids as chemo-sensitizers against CNS tumors. Eur. J. Pharm. Sci. 2017, 96, 571–577. [Google Scholar] [CrossRef]
- Doogue, M.; Polasek, T.M. The ABCD of clinical pharmacokinetics. Ther. Adv. Drug Saf. 2013, 4, 5–7. [Google Scholar] [CrossRef]
- Lipinski, C.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Delivery Rev. 1997, 23, 3–26. [Google Scholar] [CrossRef]
- Ghose, A.K.; Viswanadhan, V.N.; Wendoloski, J J. Prediction of Hydrophobic (Lipophilic) Properties of Small Organic Molecules Using Fragmental Methods: An Analysis of ALOGP and CLOGP Methods. J. Phys. Chem. A. 1998, 102, 3762–3772. [Google Scholar] [CrossRef]
- Ghose, A.K.; Viswanadhan, V.N.; Wendoloski, J.J. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J. Comb. Chem. 1999, 1, 55–68. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.-Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular Properties That Influence the Oral Bioavailability of Drug Candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef] [PubMed]
- Egan, W.J.; Lauri, G. Prediction of intestinal permeability. Adv. Drug Deliv. Rev. 2002, 54, 273–289. [Google Scholar] [CrossRef]
- Egan, W.J.; Merz, K.M.; Baldwin, J.J. Prediction of drug absorption using multivariate statistics. J. Med. Chem. 2000, 43, 3867–3877. [Google Scholar] [CrossRef] [PubMed]
- Muegge, I.; Heald, S.L.; Brittelli, D. Simple selection criteria for drug-like chemical matter. J. Med. Chem. 2001, 44, 1841–1846. [Google Scholar] [CrossRef]
- Azam, F.; Madi, A.M.; Ali, H.I. Molecular Docking and Prediction of Pharmacokinetic Properties of Dual Mechanism Drugs that Block MAO-B and Adenosine A2A Receptors for the Treatment of Parkinson’s Disease. J. Young Pharm. 2012, 4, 184–192. [Google Scholar] [CrossRef]
- Zhao, M.Y.; Abraham, M.H.; Le, J.; Hersey, A.; Luscombe, C.N.; Beck, G.; Sherborne, B. Rate-limited steps of human oral absorption and QSAR studies. Pharm. Res. 2002, 19, 1446–1457. [Google Scholar]
- Querlex, B. Computational Biophysics of the Skin; Jenny Stanford Publishing: New York, NY, USA, 2016; pp. 1–558. [Google Scholar]
- Delaney, J. ESOL: Estimating Aqueous Solubility Directly from Molecular Structure. J. Chem. Inf. Comput. Sci. 2004, 44, 1000–1005. [Google Scholar] [CrossRef]
- Pires, D.; Blundell, T.L.; Ascher, D.B. pkCSM: Predicting Small-Molecule Pharmacokinetic and Toxicity Properties Using Graph-Based Signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Spek, A.L. Structure validation in chemical crystallography. Acta Crystallogr. Sect. D Boil. Crystallogr. 2009, 65, 148–155. [Google Scholar] [CrossRef]
- Farrugia, L.J. WinGX and ORTEP for Windows: An update. J. Appl. Crystallogr. 2012, 45, 849–854. [Google Scholar] [CrossRef]
- Macrae, C.; Bruno, I.; Chisholm, J.A.; Edgington, P.R.; McCabe, P.; Pidcock, E.; Rodriguez-Monge, L.; Taylor, R.; Van De Streek, J.; Wood, P.A. Mercury CSD 2.0– new features for the visualization and investigation of crystal structures. J. Appl. Crystallogr. 2008, 41, 466–470. [Google Scholar] [CrossRef]
- Rigaku, O.D. CrysAlis PRO; Rigaku Oxford Diffraction Ltd.: Yarnton, UK, 2018. [Google Scholar]
- Becke, A.D. A new mixing of Hartree-Fock and local density – functional theories. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar] [CrossRef]
- Dennington, R.; Keith, T.; Millam, J. GaussView, version 5; Semichem, Inc.: Shawnee Mission, KS, USA, 2009. [Google Scholar]
- Lippincott, E.R.; Schroeder, R. One-Dimensional Model of the Hydrogen Bond. J. Chem. Phys. 1955, 23, 1099. [Google Scholar] [CrossRef]
- Schroeder, R.; Lippincott, E.R. Potential Function Model of Hydrogen Bonds. II. J. Phys. Chem. 1957, 61, 921–928. [Google Scholar] [CrossRef]
- Gilli, P.; Gilli, G. LSHB. A Computer Program for Performing Lippincott and Schroeder HB Calculations; University of Ferrara: Ferrara, Italy, 1992. [Google Scholar]
- McKinnon, J.; Jayatilaka, D.; Spackman, M. Towards quantitative analysis of intermolecular interactions with Hirshfeld surfaces. Chem. Commun. 2007, 3814–3816. [Google Scholar] [CrossRef]
- Turner, M.J.; McKinnon, J.J.; Wolff, S.K.; Grimwood, D.J.; Spackman, P.R.; Jayatilaka, D.; Spackman, M.A. CrystalExplorer17.5; University of Western Australia: Perth, Australia, 2017. [Google Scholar]
- Hao, H.; Ma, G. Research at the University of Western Australia on structure protections against blast and impact loads. Aust. J. Struct. Eng. 2012, 13. [Google Scholar] [CrossRef]
- Jayatilaka, D.; Wolff, S.K.; Grimwood, D.J.; McKinnon, J.J.; Spackman, M.A. CrystalExplorer: A tool for displaying Hirshfeld surfaces and visualizing intermolecular interactions in molecular crystals. Acta Cryst. A. 2006, 62, 90. [Google Scholar] [CrossRef]
- McKinnon, J.; Spackman, M.; Mitchell, A.S. Novel tools for visualizing and exploring intermolecular interactions in molecular crystals. Acta Crystallogr. Sect. B Struct. Sci. 2004, 60, 627–668. [Google Scholar] [CrossRef]
- Allen, F.H.; Baalham, C.A.; Lommerse, J.P.M.; Raithby, P.R. Carbonyl–Carbonyl Interactions can be Competitive with Hydrogen Bonds. Acta Crystallogr. Sect. B Struct. Sci. 1998, 54, 320–329. [Google Scholar] [CrossRef]
- Ravikumar, B.; Athimoolam, S.; Rajaram, R.K. L-Ornithinium sulfate monohydrate. Acta Crystallogr. Sect. E Struct. Rep. Online 2004, 60, 2093–2095. [Google Scholar] [CrossRef]
- Ramaswamy, S.; Sridhar, B.; Ramakrishnan, V.; Rajaram, R.K. L-Ornithine nitrate. Acta Crystallogr. Sect. E Struct. Rep. Online 2002, 58, 646–648. [Google Scholar] [CrossRef]
- Fleck, M.; Ghazaryan, V.V.; Petrosyan, A.M. L-Ornithinium(2+) Sulfate Hydrogen Fluoride and Tri-L-Ornithinium(2+) Dinitrate Disulfate. JUET Res. J. Sci. Tech. 2014, 1, 11–25. [Google Scholar]
- Salunke, D.M.; Vijayan, M. X-ray studies on crystalline complexes involving amino acids and peptides. IX. Crystal structure of L-ornithine L-aspartate hemihydrate. Int. J. Pept. Protein Res. 1983, 22, 154–160. [Google Scholar] [CrossRef]
- Salunke, D.M.; Vijayan, M. preliminary X-ray studies on n-acetylglycyl-L-lysine methylester acetate, L-arginite-L-aspartate and L-ornithine L-aspartate. Curr. Sci. 1979, 48, 1071–1079. [Google Scholar]
- Soman, J.; Vijayan, M. X-ray studies on crystalline complexes involving amino acids and peptides. XVI. Structure of L-ornithine D-aspartate monohydrate. Acta Crystallogr. Sect. C Cryst. Struct. Commun. 1988, 44, 1794–1797. [Google Scholar] [CrossRef]
- Allouchi, H.; Ceolin, R.; Berthon, L.; Tombret, F.; Rietveld, I. Characterization of molecular associations involving l-ornithine and α-ketoglutaric acid: Crystal structure of L-ornithinium α-ketoglutarate. Ann. Pharm. Fr. 2014, 72, 238–243. [Google Scholar] [CrossRef]
- Schaffrin, R.; Trotter, J. Crystal structure of D,L-ornithine hydrobromide. J. Chem. Soc. 1970, 1561–1565. [Google Scholar] [CrossRef]
- Chiba, A.; Ueki, T.; Ashida, T.; Sasada, Y.; Kakudo, M. The crystal structure of L-ornithine hydrochloride. Acta Crystallogr. 1967, 22, 863–870. [Google Scholar] [CrossRef]
- Guha, S.; Mazumdar, S.; SahaΝ. The crystal and molecular structure of L-ornithine hydrochloride. Z. Kristallogr. 1969, 129, 84–100. [Google Scholar] [CrossRef]
- Dittrich, B.; Munshi, P.; Spackman, M. Redetermination, invariom-model and multipole refinement of L -ornithine hydrochloride. Acta Crystallogr. Sect. B Struct. Sci. 2007, 63, 505–509. [Google Scholar] [CrossRef]
- Woińska, M.; Grabowsky, S.; Dominiak, P.M.; Woźniak, K.; Jayatilaka, D. Hydrogen atoms can be located accurately andprecisely by x-ray crystallography. Sci. Adv. 2016, 1–8. [Google Scholar]
- Hempel, A.; Dauter, Z.; Szwabski, S. Preliminary crystal data on hexabromoselenates(IV) of some amino acids. Z. Kristallogr. 1977, 146, 318–319. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds 1 and 2 are available from the authors. |


| Chemical Formula | C20 H22 N2 O5 |
|---|---|
| molecular weight (g/mol) | 370.40 |
| crystal system, space group | orthorhombic, P212121 |
| T (K) | 293 |
| a, b, c (Ǻ) | 8.9040(10) 9.9480(10) 22.489(3) |
| α, β, γ (o) | 90, 90, 90 |
| Z | 4 |
| Volume (Ǻ3) | 1992.0(4) |
| radiation type | Mo K α |
| µ (mm−1) | 0.089 |
| diffractometer | Siemens P3 |
| no. of total and independent reflections | 3647, 2396 |
| R [F2 > 4 σ(F2)], wR2, S | 0.0432, 0.1307, 0.974 |
| no. of reflections and parameters | 3174, 301 |
| ∆ρmax, ∆ρmin (e−3) | 0.196; −0.152 |
| D-H (Å) | H…A (Å) | D…A (Å) | D-H…A (o) | |
|---|---|---|---|---|
| 1 | ||||
| N1-H1…O3* | 0.86 | 2.17 | 2.608(2) | 111 |
| N2-H20…O5i | 0.86 | 1.96 | 2.801(4) | 166 |
| O2-H21…O1ii | 0.82 | 1.84 | 2.661(2) | 174 |
| O4-H41…O2ii | 0.82 | 1.79 | 2.600(2) | 172 |
| C3-H3…O3iii | 0.93 | 2.56 | 3.246(4) | 131 |
| C9-H91…O1* | 0.97 | 2.57 | 3.093(3) | 114 |
| C9-H92…O4* | 0.97 | 2.57 | 2.896(3) | 100 |
| C11-H112…O1* | 0.97 | 2.52 | 3.087(3) | 117 |
| C12-H121…N1* | 0.97 | 2.61 | 2.992(3) | 104 |
| Physicochemical Properties | Pharmacokinetics | Druglikeness | Medicinal Chemistry | ||||
|---|---|---|---|---|---|---|---|
| Molecular weight [g/mol] | 370.40 | GI absorption | high | Lipinski violation | yes, 0 | PAINS | 0 |
| Heavy atoms | 27 | BBB permeant | no | Ghose | yes | leadlikeness | no, 2 |
| Aromatic heavy atoms | 12 | Pgp substrate | no | Veber | no, 1 | synth. accessibility | 2.61 |
| Fraction Csp3 | 0.25 | CYP1A2 inhibitor | no | Egan | yes | ||
| Rotatable bonds | 11 | CYP2C19 inhibitor | no | Muegge | yes | ||
| H-bond acceptors | 5 | CYP2C9 inhibitor | no | Bioavailability score | 0.56 | ||
| H-bond donors | 4 | CYP2D6 inhibitor | no | ||||
| Molar refractivity | 98.96 | CYP3A4 inhibitor | no | ||||
| TPSA [Ǻ2] | 115.73 | Log Kp [cm/s] | −7.39 | ||||
| Consensus log P 0/w | 1.81 | ||||||
| Comp. Name/CSD Ref. Code | iLog P | Molar Refractivity | Log S | WLOGP | TPSA [Ǻ2] | In Silico% Absorption | GI Absorption | P-gp Substrate | Log Kp |
|---|---|---|---|---|---|---|---|---|---|
| DFMO | 0.53 | 38.28 | S | 0.61 | 89.34 | 78.17 | high | no | −9.48 |
| 1 | 2.30 | 98.96 | MS | 1.44 | 115.73 | 69.08 | high | no | −7.39 |
| EXOFAY | 0.00 | 128.51 | S | −0.42 | 112.50 | 70.19 | high | yes | −7.90 |
| GOTFAY | −2.81 | 88.26 | S | −1.19 | 247.11 | 23.75 | low | no | −10.55 |
| TEFMIA | 0.64 | 88.89 | S | 0.83 | 253.10 | 21.69 | low | no | −10.11 |
| 2 | 0.00 | 39.76 | S | −6.63 | 95.41 | 76.09 | low | no | −9.89 |
| ORNBDL10 | 0.00 | 42.84 | S | −6.63 | 95.41 | 76.09 | low | no | −10.04 |
| BAPKUB | 0.00 | 55.30 | S | −5.55 | 181.22 | 46.48 | low | yes | −13.23 |
| BAPKOV | 0.00 | 52.22 | S | −5.55 | 181.22 | 46.48 | low | yes | −13.08 |
| EVIJAU | 0.00 | 64.75 | S | −5.27 | 250.10 | 22.72 | low | yes | −13.34 |
| BEZQOO | 0.00 | 51.30 | S | 2.30 | 101.81 | 73.88 | high | no | −9.20 |
| YIGMAE | 0.00 | 46.31 | S | 1.03 | 95.41 | 76.09 | high | no | −8.75 |
| IHEPES | 0.00 | 58.60 | S | −1.66 | 89.19 | 78.23 | high | no | −7.81 |
| VUYHII | 0.42 | 61.92 | S | −8.21 | 212.54 | 35.68 | low | yes | −15.30 |
| CAPRAM | 0.42 | 61.92 | S | −8.21 | 212.54 | 35.68 | low | yes | −15.30 |
| BIHYEX | −0.97 | 49.42 | S | −2.62 | 190.45 | 43.30 | low | yes | −13.88 |
| BAPKIP | −1.31 | 58.90 | S | −2.27 | 250.10 | 22.72 | low | yes | −13.69 |
| JADGED | 0.06 | 61.86 | S | −5.46 | 189.91 | 43.49 | low | yes | −12.80 |
| PUYVUA | 0.69 | 46.44 | S | −3.35 | 164.29 | 52.32 | low | no | −10.50 |
| Comp. Name/CSD Rref. Rcode | Mol. Weight [g/mol] | Liphophilicity (MLogP) | H-Bond Donors | H-Bond Acceptors | Lipinski Violations | Ghose Viol. | Veber Viol. | Egan Viol. | Muegge Viol. | Bioavailability Score |
|---|---|---|---|---|---|---|---|---|---|---|
| Lipinski`s Rule of Five | ||||||||||
| DFMO | 182.17 | −2.32 | 3 | 6 | yes, 0 | no, 1 | yes | yes | no, 2 | 0.55 |
| 1 | 370.40 | 1.61 | 4 | 5 | yes, 0 | yes | no, 1 | yes | yes | 0.56 |
| EXOFAY | 469.01 | −1.28 | 3 | 5 | yes, 0 | no, 1 | no, 1 | yes | yes | 0.56 |
| GOTFAY | 363.28 | −5.79 | 3 | 9 | yes, 1 | no, 1 | no, 1 | no, 1 | no, 2 | 0.55 |
| TEFMIA | 362.27 | −4.33 | 3 | 9 | yes, 1 | yes | no, 1 | no, 1 | no, 2 | 0.55 |
| 2 | 168.62 | −7.83 | 2 | 2 | yes, 0 | no, 2 | yes | yes | no, 2 | 0.55 |
| ORNBDL10 | 213.07 | −7.65 | 2 | 2 | yes, 0 | no, 1 | yes | yes | no, 1 | 0.55 |
| BAPKUB | 310.14 | −9.05 | 3 | 6 | yes, 0 | no, 1 | no, 1 | no, 1 | no, 2 | 0.11 |
| BAPKOV | 265.69 | −9.23 | 3 | 6 | yes, 0 | no, 1 | no, 1 | no, 1 | no, 2 | 0.11 |
| EVIJAU | 327.70 | −9.01 | 3 | 9 | yes, 1 | no, 1 | no, 1 | no, 1 | no, 2 | 0.11 |
| BEZQOO | 294.27 | −7.97 | 4 | 9 | yes, 0 | yes | yes | yes | yes | 0.56 |
| YIGMAE | 275.24 | −7.14 | 2 | 8 | yes, 0 | yes | yes | yes | yes | 0.56 |
| IHEPES | 433.63 | −3.66 | 3 | 6 | yes, 0 | no, 1 | yes | yes | yes | 0.55 |
| VUYHII | 283.28 | −13.86 | 4 | 7 | yes, 0 | no, 1 | no, 1 | no, 1 | no, 2 | 0.55 |
| CAPRAM | 283.28 | −13.86 | 4 | 7 | yes, 0 | no, 1 | no, 1 | no, 1 | no, 2 | 0.55 |
| BIHYEX | 248.25 | −10.39 | 4 | 4 | yes, 0 | no, 1 | no, 1 | no, 1 | no, 2 | 0.11 |
| BAPKIP | 292.24 | −9.38 | 3 | 9 | yes, 1 | no, 1 | no, 1 | no, 1 | no, 2 | 0.11 |
| JADGED | 278.26 | −9.20 | 3 | 7 | yes, 0 | no, 1 | no, 1 | no, 1 | no, 2 | 0.11 |
| PUYVUA | 195.17 | −8.17 | 2 | 5 | yes, 0 | no, 2 | no, 1 | no, 1 | no, 3 | 0.55 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bojarska, J.; Remko, M.; Breza, M.; Madura, I.D.; Kaczmarek, K.; Zabrocki, J.; Wolf, W.M. A Supramolecular Approach to Structure-Based Design with A Focus on Synthons Hierarchy in Ornithine-Derived Ligands: Review, Synthesis, Experimental and in Silico Studies. Molecules 2020, 25, 1135. https://doi.org/10.3390/molecules25051135
Bojarska J, Remko M, Breza M, Madura ID, Kaczmarek K, Zabrocki J, Wolf WM. A Supramolecular Approach to Structure-Based Design with A Focus on Synthons Hierarchy in Ornithine-Derived Ligands: Review, Synthesis, Experimental and in Silico Studies. Molecules. 2020; 25(5):1135. https://doi.org/10.3390/molecules25051135
Chicago/Turabian StyleBojarska, Joanna, Milan Remko, Martin Breza, Izabela D. Madura, Krzysztof Kaczmarek, Janusz Zabrocki, and Wojciech M. Wolf. 2020. "A Supramolecular Approach to Structure-Based Design with A Focus on Synthons Hierarchy in Ornithine-Derived Ligands: Review, Synthesis, Experimental and in Silico Studies" Molecules 25, no. 5: 1135. https://doi.org/10.3390/molecules25051135
APA StyleBojarska, J., Remko, M., Breza, M., Madura, I. D., Kaczmarek, K., Zabrocki, J., & Wolf, W. M. (2020). A Supramolecular Approach to Structure-Based Design with A Focus on Synthons Hierarchy in Ornithine-Derived Ligands: Review, Synthesis, Experimental and in Silico Studies. Molecules, 25(5), 1135. https://doi.org/10.3390/molecules25051135






