Playing with Structural Parameters: Synthesis and Characterization of Two New Maltol-Based Ligands with Binding and Antineoplastic Properties
Abstract
1. Introduction
2. Results and Discussion
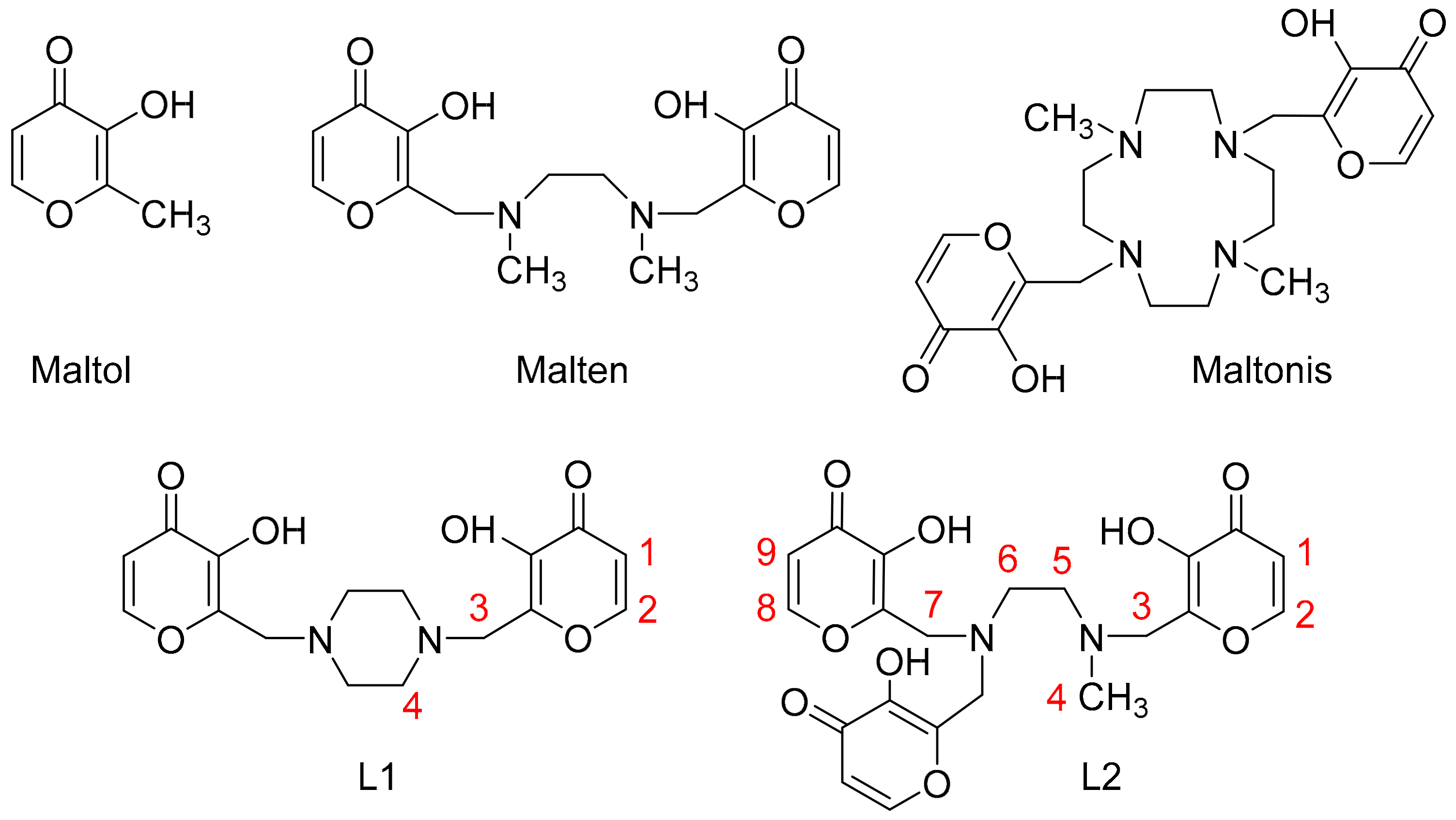
2.1. Synthesis
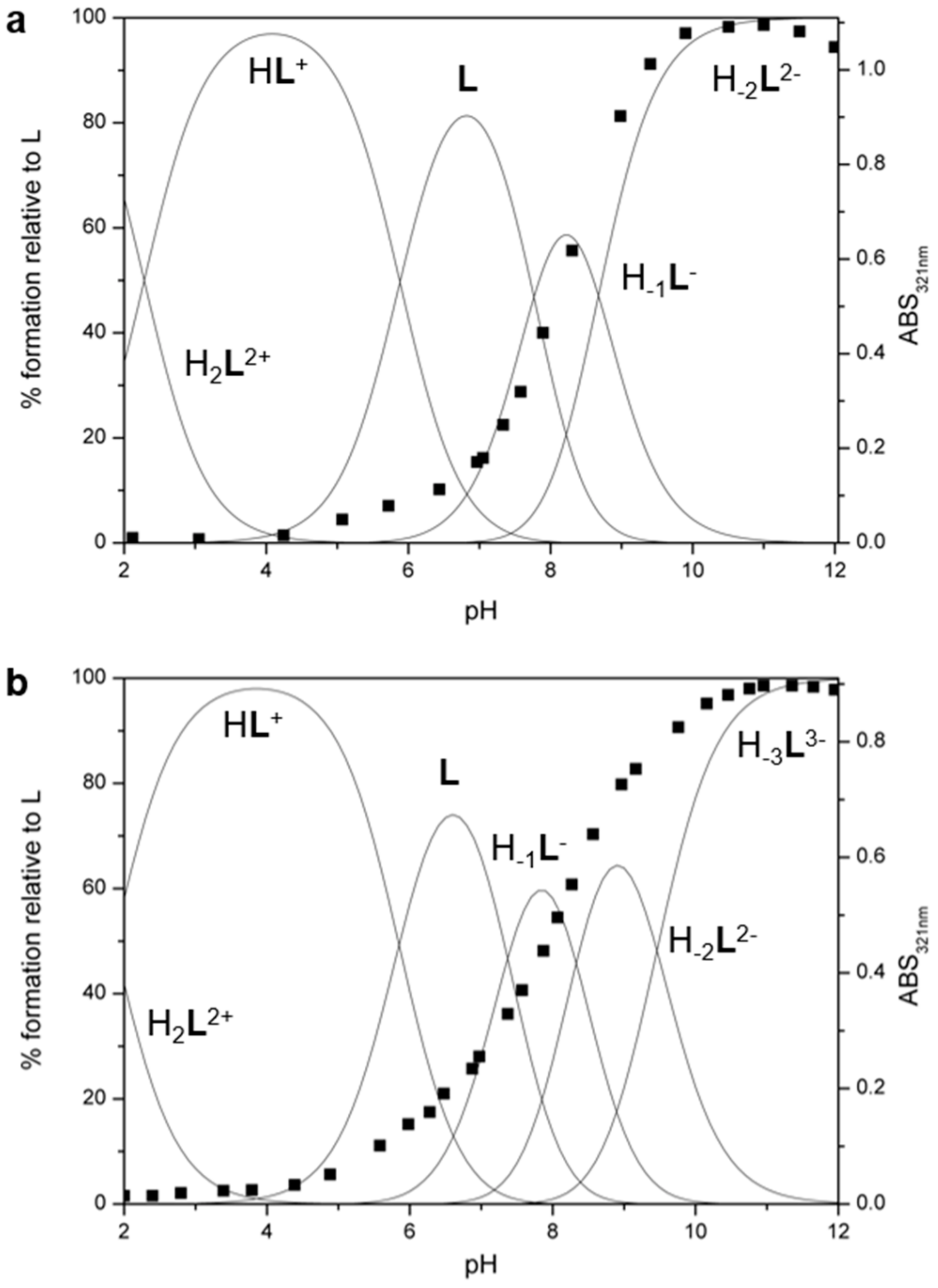
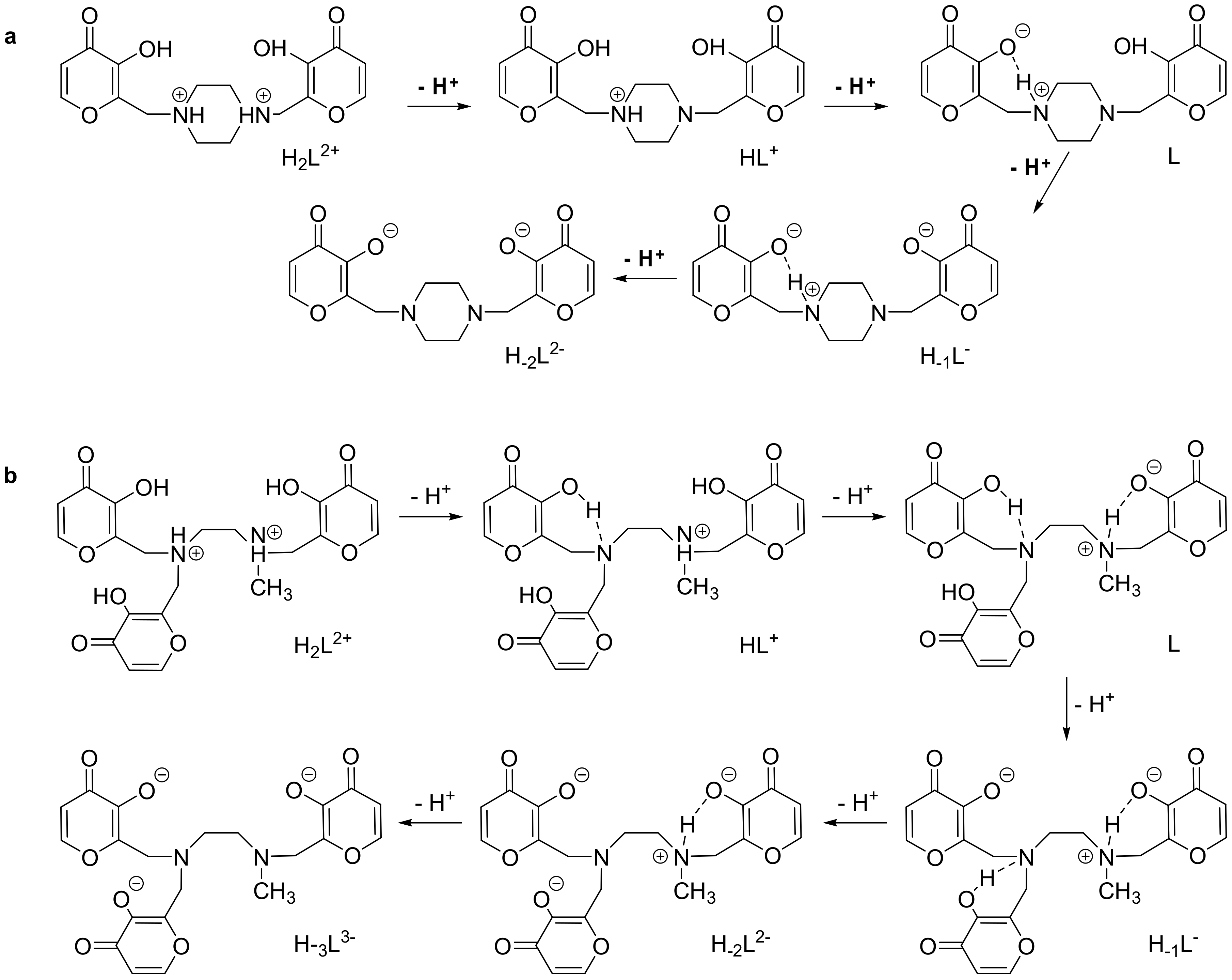
2.2. Acid-base Behavior
UV-VIS and 1H–NMR Studies
2.3. Stability
2.4. Coordination of Metal Ions
2.4.1. Mononuclear Complexes
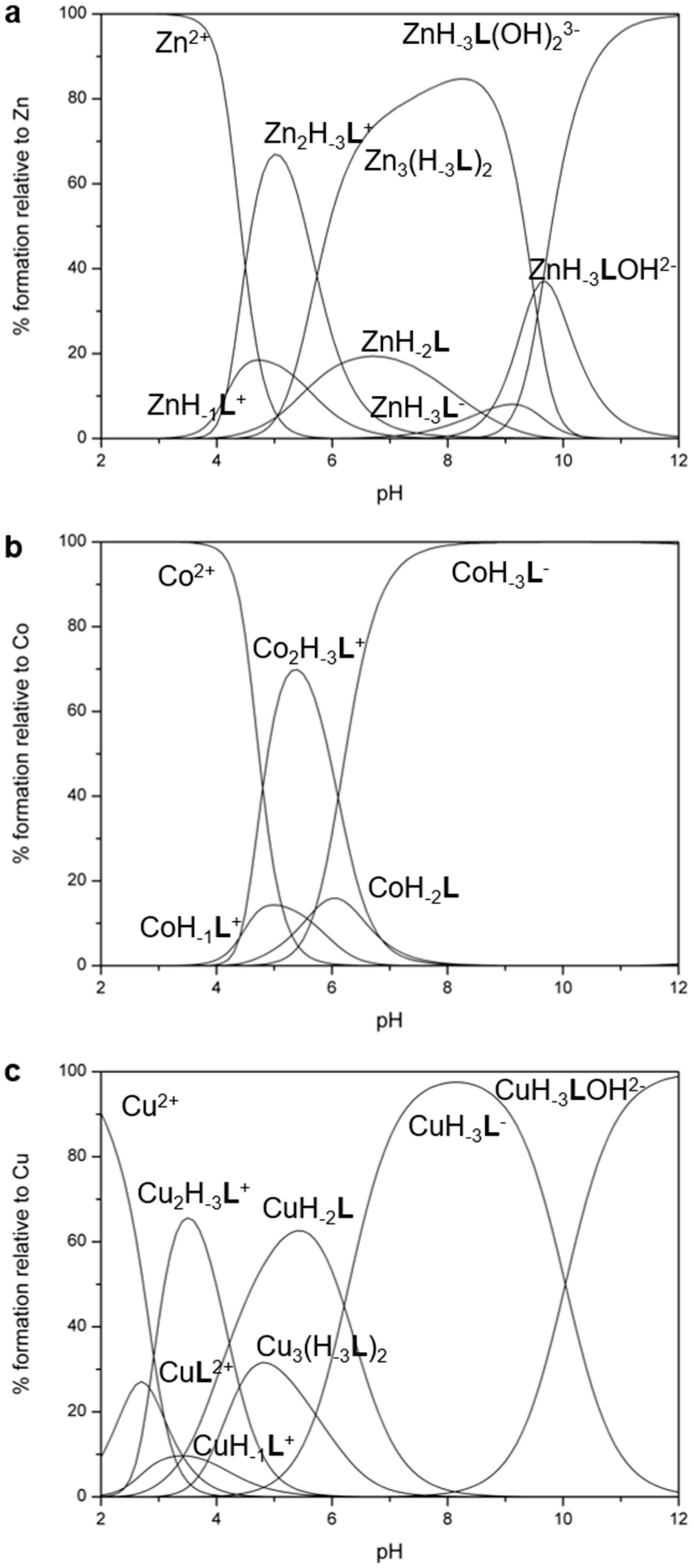
2.4.2. Polynuclear Complexes
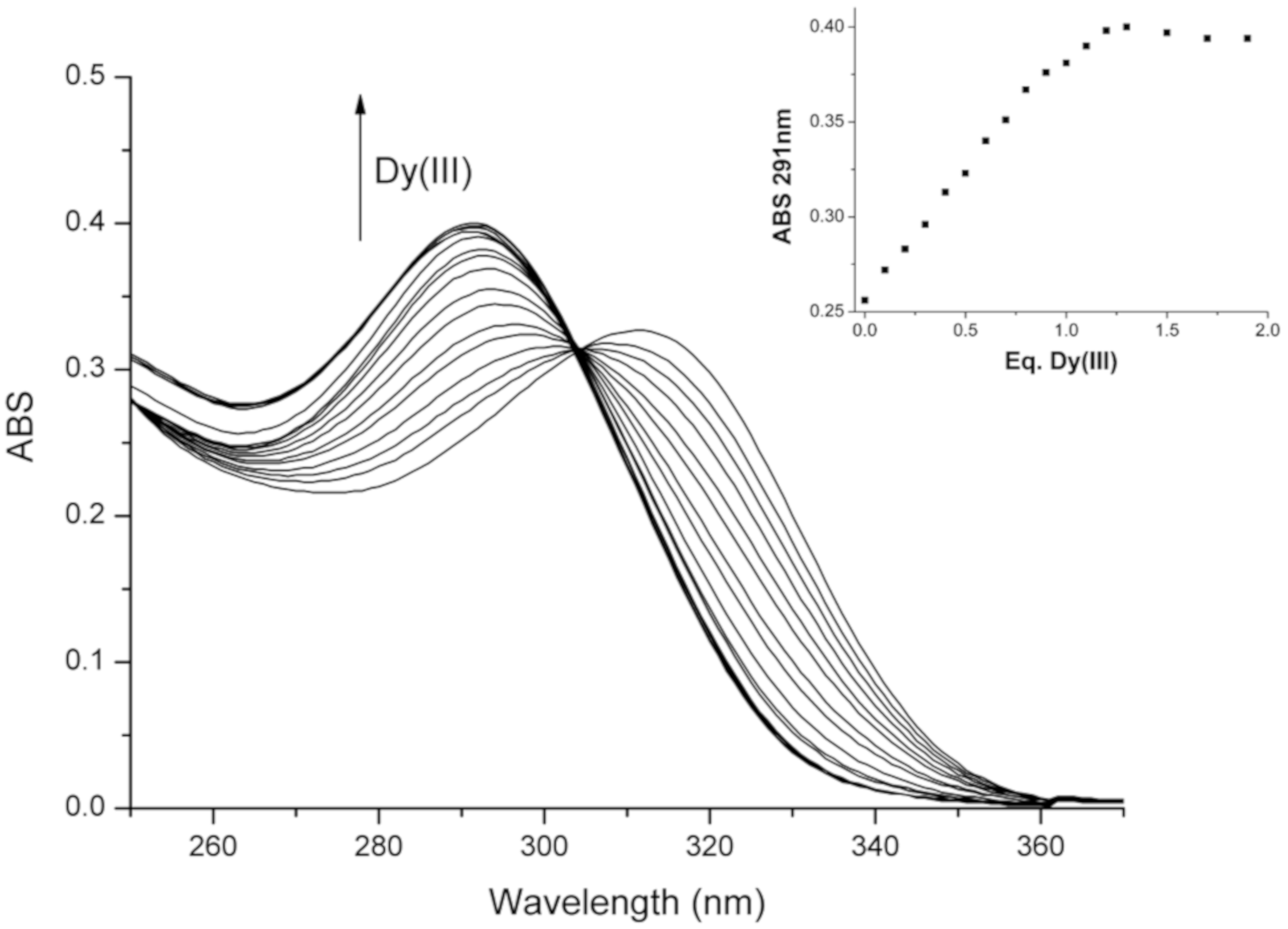
2.4.3. Interaction with Alkaline-Earth (AE) and Rare Earth (RE) Ions
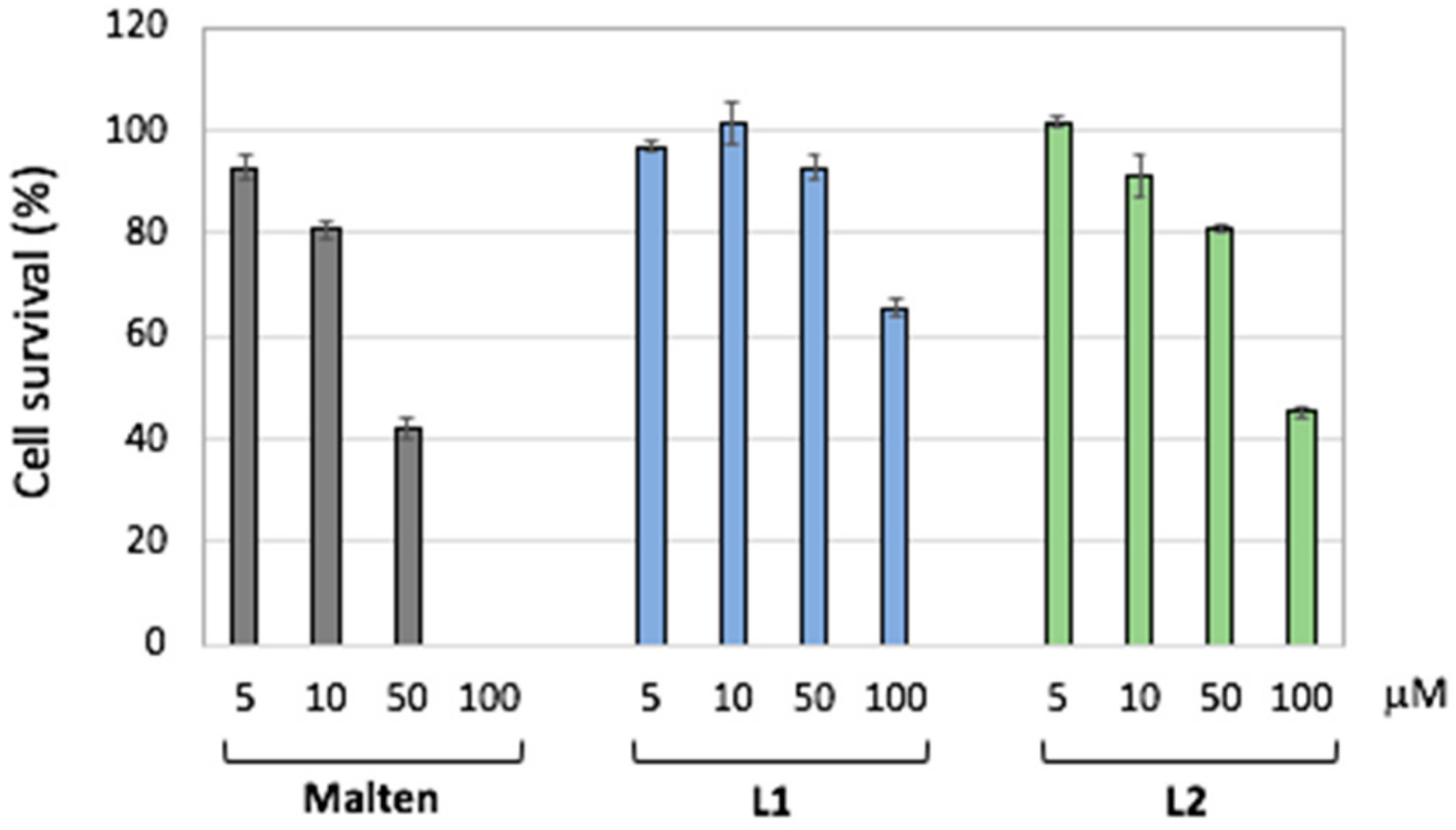
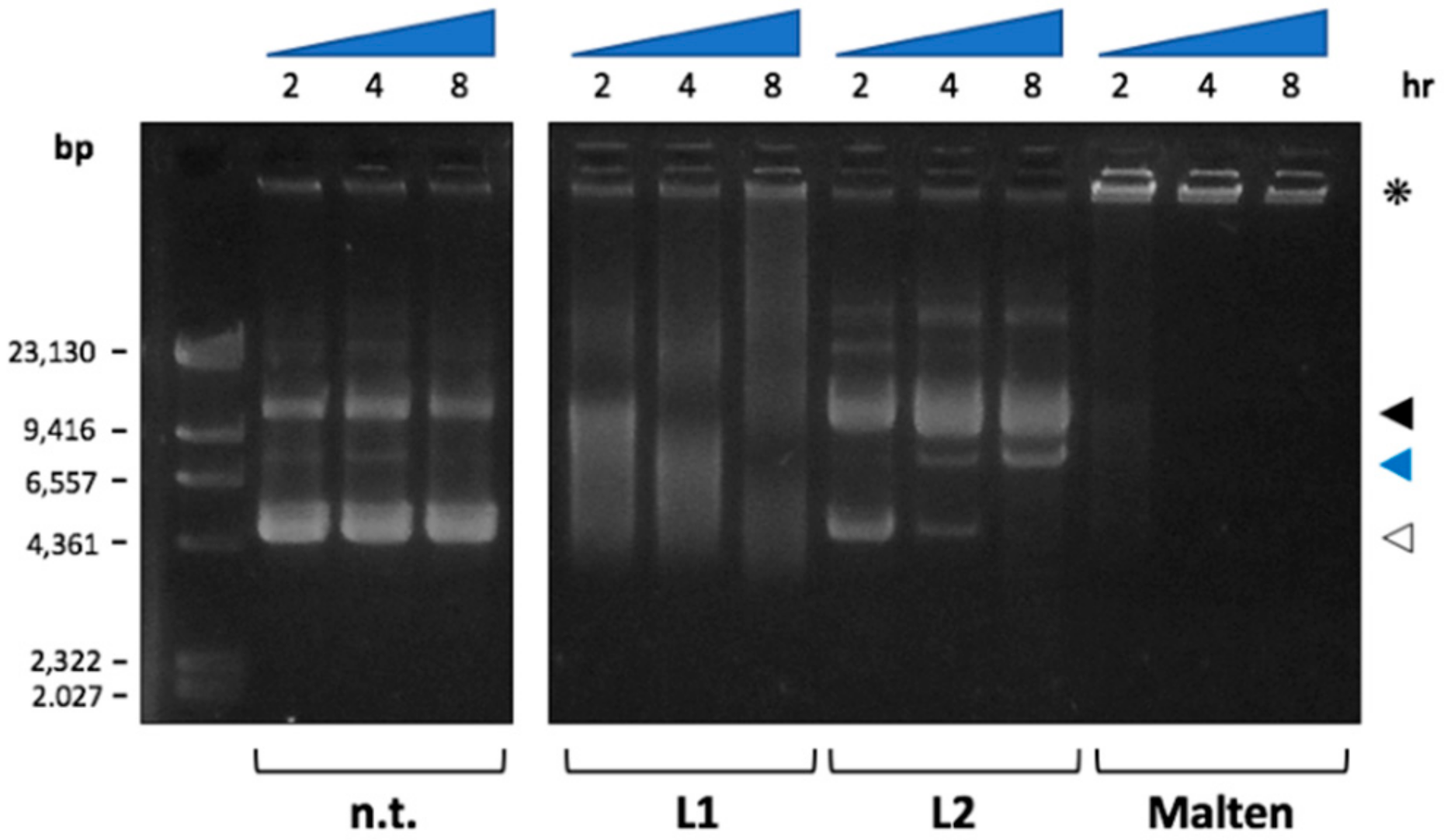
2.5. Biological Data
3. Experimental
3.1. UV-Vis and NMR Experiments
3.2. EMF Measurements
3.3. Synthesis
3.4. Biological Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cancer Research UK. Available online: https://www.cancerresearchuk.org/health-professional/cancer-statistics/worldwide-cancer/incidence#headng-One (accessed on 16 August 2019).
- National Cancer Institute (USA). Available online: https://www.cancer.gov/about-cancer/understanding/statistics (accessed on 16 August 2019).
- Joint FAO/WHO Expert Committee on Food Additives. Evaluation of certain veterinary drug residues in food: Sixty-sixth report of the Joint FAO/WHO Expert Committee on Food Additives. World Health Organ. Tech. Rep. Ser. 2006, 939, 1.
- Harvey, R.S.; Reffitt, D.M.; Doig, L.A.; Meenan, J.; Ellis, R.D.; Thompson, R.P.; Powell, J.J. Ferric trimaltol corrects iron deficiency anaemia in patients intolerant of iron. Aliment. Pharmacol. Ther. 1998, 12, 845–848. [Google Scholar] [CrossRef] [PubMed]
- Thompson, K.H.; Liboiron, B.D.; Sun, Y.; Bellman, K.D.D.; Setyawati, I.A.; Patrick, B.O.; Karunaratne, V.; Rawji, G.; Wheeler, J.; Sutton, K.; et al. Preparation and characterization of vanadyl complexes with bidentate maltol-type ligands; in vivo comparisons of anti-diabetic therapeutic potential. Biol. Inorg. Chem. 2003, 8, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Krishnakumar, V.; Barathi, D.; Mathammal, R.; Balamani, J.; Jayamani, N. Spectroscopic properties, NLO, HOMO-LUMO and NBO of maltol. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2014, 121, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Anwar-Mohamed, A.; El-Kadi, A.O. Induction of cytochrome P450 1a1 by the food flavoring agent, maltol. Toxicol In Vitro 2007, 21, 685–690. [Google Scholar] [CrossRef]
- Hong, S.; Iizuka, Y.; Lee, T.; Kim, C.Y.; Seong, G.J. Neuroprotective and neurite outgrowth effects of maltol on retinal ganglion cells under oxidative stress. Mol. Vis. 2014, 20, 1456–1462, PMID: 25352751. [Google Scholar]
- Kandioller, W.; Hartinger, C.G.; Nazarov, A.A.; Bartel, C.; Skocic, M.; Jakupec, M.A.; Arion, V.B.; Keppler, B.K. Maltol-Derived Ruthenium–Cymene Complexes with Tumor Inhibiting Properties: The Impact of Ligand–Metal Bond Stability on Anticancer Activity In Vitro. Chemistry 2009, 15, 12283–12291. [Google Scholar] [CrossRef]
- Song, Y.; Hong, S.; Iizuka, Y.; Kim, C.Y.; Seong, G.J. The Neuroprotective Effect of Maltol against Oxidative Stress on Rat Retinal Neuronal Cells. Korean J. Ophthalmol. 2015, 29, 58–65. [Google Scholar] [CrossRef]
- Kang, K.S.; Yamabe, N.; Kim, H.Y.; Yokozawa, T. Role of maltol in advanced glycation end products and free radicals: In-vitro and in-vivo studies. J. Pharm. Pharmacol. 2008, 60, 445–452. [Google Scholar] [CrossRef]
- Patton, S. The formation of maltol in certain carbohydrate-glycine systems. J. Biol. Chem. 1950, 184, 131–134. [Google Scholar]
- Hironishi, M.; Kordek, R.; Yanagihara, R.; Garruto, R.M. Maltol (3-hydroxy-2-methyl-4-pyrone) toxicity in neuroblastoma cell lines and primary murine fetal hippocampal neuronal cultures. Neurodegeneration 1996, 5, 325–329. [Google Scholar] [CrossRef] [PubMed]
- Yasumoto, E.; Nakano, K.; Nakayachi, T.; Morshed, S.R.; Hashimoto, K.; Kikuchi, H.; Nishikawa, H.; Kawase, M.; Sakagami, H. Cytotoxic activity of deferiprone, maltol and related hydroxyketones against human tumor cell lines. Anticancer Res. 2004, 24, 755–762, PMID: 15161023. [Google Scholar]
- Murakami, K.; Ishida, K.; Watakabe, K.; Tsubouchi, R.; Haneda, M.; Yoshino, M. Prooxidant Action of Maltol: Role of Transition Metals in the Generation of Reactive Oxygen Species and Enhanced Formation of 8-hydroxy-2′-deoxyguanosine Formation in DNA. Biometals 2006, 19, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Murakami, K.; Ishida, K.; Watakabe, K.; Tsubouchi, R.; Naruse, M.; Yoshino, M. Maltol/iron-mediated apoptosis in HL60 cells: Participation of reactive oxygen species. Toxicol. Lett. 2006, 161, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.L.; Pan, H.Z.; Scott, M.D.; Meshnick, S.R. Activated oxygen generation by a primaquine metabolite: Inhibition by antioxidants derived from Chinese herbal remedies. Free Radic. Biol. Med. 1992, 12, 213–218. [Google Scholar] [CrossRef]
- Kim, Y.B.; Oh, S.H.; Sok, D.-I.; Kim, M.R. Neuroprotective effect of maltol against oxidative stress in brain of mice challenged with kainic acid. Nutr. Neurosci. 2004, 7, 33–39. [Google Scholar]
- Yang, Y.; Wang, J.; Xu, C.; Pan, H.; Zhang, Z.J. Maltol inhibits apoptosis of human neuroblastoma cells induced by hydrogen peroxide. Biochem. Mol. Biol. 2006, 39, 145–149. [Google Scholar] [CrossRef]
- Li, M.; Lu, Y.; Hu, Y.; Zhai, X.; Xu, W.; Jing, H.; Tian, X.; Lin, Y.; Gao, D.; Yao, J. Salvianolic acid B protects against acute ethanol-induced liver injury through SIRT1-mediated deacetylation of p53 in rats. Toxicol. Lett. 2014, 228, 67–74. [Google Scholar] [CrossRef]
- Liu, H.; Qi, X.; Cao, S.; Li, P. Protective effect of flavonoid extract from Chinese bayberry (Myrica rubra Sieb. et Zucc.) fruit on alcoholic liver oxidative injury in mice. J. Nat. Med. 2014, 68, 521–529. [Google Scholar] [CrossRef]
- Han, Y.; Xu, Q.; Hu, J.N.; Han, X.Y.; Li, W.; Zhao, L.C. Maltol, a food favoring agent, attenuates acute alcohol-induced oxidative damage in mice. Nutrients 2015, 7, 682–696. [Google Scholar] [CrossRef]
- Dömötör, O.; Aicher, S.; Schmidlehner, M.; Novak, M.S.; Roller, A.; Jakupec, M.A.; Kandioller, W.; Hartinger, C.G.; Keppler, B.K.; Enyedy, E.A. Antitumor pentamethylcyclopentadienyl rhodium complexes of maltol and allomaltol: Synthesis, solution speciation and bioactivity. J. Inorg. Biochem. 2014, 134, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Bransovà, J.; Brtko, J.; Uher, M.; Novotny, L. Antileukemic activity of 4-pyranone derivatives. Int. J. Biochem. Cell. Biol. 1995, 7, 701–706. [Google Scholar] [CrossRef]
- Jakupec, M.A.; Keppler, B.K. Gallium in Cancer Treatment. Curr. Top. Med. Chem. 2004, 4, 1575–1583. [Google Scholar] [CrossRef] [PubMed]
- Barve, A.; Kumbhar, A.; Bhat, M.; Joshi, B.; Butcher, R.; Sonawane, U.; Joshi, R. Mixed-ligand copper(II) maltolate complexes: Synthesis, characterization, DNA binding and cleavage, and cytotoxicity. Inorg. Chem. 2009, 48, 9120–9132. [Google Scholar] [CrossRef] [PubMed]
- Kandioller, W.; Hartinger, C.G.; Nazarov, A.A.; Kasser, J.; John, R.; Jakupec, M.A.; Arion, V.B.; Dyson, P.J.; Keppler, B.K. Tuning the anticancer activity of maltol-derived ruthenium complexes by derivatization of the 3-hydroxy-4-pyrone moiety. J. Organomet. Chem. 2009, 694, 922–929. [Google Scholar] [CrossRef]
- Song, B.; Saatchi, K.; Rawji, G.H.; Orvig, C. Synthesis and solution studies of the complexes of pyrone analogue ligands with vanadium(IV) and vanadium(V). Inorg. Chim. Acta 2002, 339, 393–399. [Google Scholar] [CrossRef]
- Saatchi, K.; Thompson, K.H.; Patrick, B.O.; Pink, M.; Yuen, V.G.; McNeill, J.H.; Orvig, C. Coordination Chemistry and Insulin-Enhancing Behavior of Vanadium Complexes with Maltol C6H6O3 Structural Isomers. Inorg. Chem. 2005, 44, 2689–2697. [Google Scholar] [CrossRef]
- Liang, F.; Wan, S.; Li, Z.; Xiong, X.; Yang, L.; Zhou, X.; Wu, C. Medical Applications of Macrocyclic Polyamines. Curr. Med. Chem. 2006, 13, 711–727. [Google Scholar] [CrossRef]
- Casero, R.A.J.; Woster, P.M. Terminally alkylated polyamine analogues as chemotherapeutic agents. J. Med. Chem. 2001, 44, 1–26. [Google Scholar] [CrossRef]
- Parker, D. Preparation of azamacrocycles for imaging and treatment of tumors. Eur. Pat. Appl. EP 382583 A1 19900816 1990. [Google Scholar]
- Ambrosi, G.; Formica, M.; Fusi, V.; Giorgi, L.; Macedi, E.; Micheloni, M.; Paoli, P.; Rossi, P. A biphenol-based chemosensor for ZnII and CdII metal ions: Synthesis, potentiometric studies, and crystal structures. Inorg. Chem. 2016, 55, 7676–7687. [Google Scholar] [CrossRef] [PubMed]
- Amatori, S.; Ambrosi, G.; Borgogelli, E.; Fanelli, M.; Formica, M.; Fusi, V.; Giorgi, L.; Macedi, E.; Micheloni, M.; Paoli, P.; et al. Modulating the sensor response to halide using NBD-based azamacrocycles. Inorg. Chem. 2014, 53, 4560–4569. [Google Scholar] [CrossRef] [PubMed]
- Amatori, S.; Ambrosi, G.; Fanelli, M.; Formica, M.; Fusi, V.; Giorgi, L.; Macedi, E.; Micheloni, M.; Paoli, P.; Pontellini, R.; et al. Multi-use NBD-based tetra-amino macrocycle: Fluorescent probe for metals and anions and live cell marker. Chem. Eur. J. 2012, 18, 4274–4284. [Google Scholar] [CrossRef] [PubMed]
- Fanelli, M.; Fusi, V. Derivatives of [(3-hydroxy-4 pyron-2-yl)methyl]-amine and use thereof as anti-neoplastic drugs. PCT Int. Appl. WO 2010061282A1 20100603 2010. [Google Scholar]
- Fanelli, M.; Fusi, V. Derivatives of [(3-hydroxy-4-piron-2-yl) methyl] -amine and their use as anti-cancer drugs. Ital. IT 1392249 B1 20120222 2012. [Google Scholar]
- Amatori, S.; Bagaloni, I.; Macedi, E.; Formica, M.; Giorgi, L.; Fusi, V.; Fanelli, M. Malten, a new synthetic molecule showing in vitro antiproliferative activity against tumour cells and induction of complex DNA structural alterations. Br. J. Cancer 2010, 103, 239–248. [Google Scholar] [CrossRef][Green Version]
- Amatori, S.; Ambrosi, G.; Fanelli, M.; Formica, M.; Fusi, V.; Giorgi, L.; Macedi, E.; Micheloni, M.; Paoli, P.; Pontellini, R.; et al. Synthesis, basicity, structural characterization, and biochemical properties of two [(3-Hydroxy-4-pyron-2-yl)methyl]amine derivatives showing antineoplastic features. J. Org. Chem. 2012, 77, 2207–2218. [Google Scholar] [CrossRef]
- Guerzoni, C.; Amatori, S.; Giorgi, L.; Manara, M.C.; Landuzzi, L.; Lollini, P.-L.; Tassoni, A.; Balducci, M.; Manfrini, M.; Pratelli, L.; et al. An aza-macrocycle containing maltolic side-arms (maltonis) as potential drug against human pediatric sarcomas. BMC Cancer 2014, 14, 137. [Google Scholar] [CrossRef][Green Version]
- Benelli, C.; Borgogelli, E.; Formica, M.; Fusi, V.; Giorgi, L.; Macedi, E.; Micheloni, M.; Paoli, P.; Rossi, P. Di-maltol-polyamine ligands to form heterotrinuclear metal complexes: Solid state, aqueous solution and magnetic characterization. Dalton Trans. 2013, 42, 5848–5859. [Google Scholar] [CrossRef]
- Amatori, S.; Ambrosi, G.; Fanelli, M.; Formica, M.; Fusi, V.; Giorgi, L.; Macedi, E.; Micheloni, M.; Paoli, P.; Rossi, P. A Preorganized metalloreceptor for alkaline earth ions showing calcium versus magnesium selectivity in water: Biological activity of selected metal complexes. Chem. Eur. J. 2014, 20, 11048–11057. [Google Scholar] [CrossRef]
- Borgogelli, E.; Formica, M.; Fusi, V.; Giorgi, L.; Macedi, E.; Micheloni, M.; Paoli, P.; Rossi, P. Preorganizing binding side-arms on a cyclen scaffold: The choice of a suitable metal ion. Dalton Trans. 2013, 42, 2902–2912. [Google Scholar] [CrossRef] [PubMed]
- Kendall, P.M.; Johnson, J.V.; Cook, C.E. Synthetic route to an aromatic analog of strigol. J. Org. Chem. 1979, 44, 1421–1424. [Google Scholar] [CrossRef]
- Rodríguez, J.R.; Rumbo, A.; Castedo, L.; Mascarenas, J.L. Straightforward construction of fused 6,7,5-tricarbocyclic systems by tandem [5 + 2]/[4 + 2] cycloadditions. J. Org. Chem. 1999, 64, 966–970. [Google Scholar] [CrossRef] [PubMed]
- Khalili, F.; Henni, A.; East, A.L.L. pKa values of some piperazines at (298, 303, 313, and 323) K. J. Chem. Eng. Data 2009, 54, 2914–2917. [Google Scholar] [CrossRef]
- Paoletti, P.; Barbucci, R.; Vacca, A.; Dei, A. Thermodynamics of protonation of amines. Values of log K, ΔH, and ΔS for the protonation of NN′- and NN-dimethylethylenediamine and NNN′N′-tetramethylethylenediamine. J. Chem. Soc. A. 1971, 310–313. [Google Scholar] [CrossRef]
- Elvingson, E.; Baro, L.; Petterson, L. Speciation in vanadium bioinorganic systems. 2. An NMR, ESR, and potentiometric study of the aqueous H+−vanadate−maltol system. Inorg. Chem. 1996, 35, 3388–3393. [Google Scholar] [CrossRef]
- Covington, A.K.; Paabo, M.; Robinson, R.A.; Bates, R.G. Use of the glass electrode in deuterium oxide and the relation between the standardized pD (paD) scale and the operational pH in heavy water. Anal. Chem. 1968, 40, 700–706. [Google Scholar] [CrossRef]
- Becatti, M.; Bencini, A.; Nistri, S.; Conti, L.; Fabbrini, M.G.; Lucarini, L.; Ghini, V.; Severi, M.; Fiorillo, C.; Giorgi, C.; et al. Different antioxidant efficacy of two MnII-containing superoxide anion scavengers on hypoxia/reoxygenation-exposed cardiac muscle cells. Sci. Rep. 2019, 9, 10320. [Google Scholar] [CrossRef]
- Montis, R.; Bencini, A.; Coles, S.J.; Conti, L.; Fusaro, L.; Gale, P.A.; Giorgi, C.; Horton, P.N.; Lippolis, V.; Mapp, L.K.; et al. Fluoride binding by an anionic receptor: Tuning the acidity of amide NH groups for basic anion hydrogen bonding and recognition. Chem. Commun. 2019, 55, 2745–2748. [Google Scholar] [CrossRef]
- Bettazzi, F.; Voccia, D.; Bencini, A.; Giorgi, C.; Palchetti, I.; Valtancoli, B.; Conti, L. Optical and electrochemical study of acridine-based polyaza ligands for anion sensing. Eur. J. Inorg. Chem. 2018, 2675–2679. [Google Scholar] [CrossRef]
- Bettoschi, A.; Bencini, A.; Berti, D.; Caltagirone, C.; Conti, L.; Demurtas, D.; Giorgi, C.; Isaia, F.; Lippolis, V.; Mamusa, M.; et al. Highly stable ionic liquid-in-water emulsions as a new class of fluorescent sensors for metal ions: The case study of Fe3+ sensing. RSC Adv. 2015, 5, 37385–37391. [Google Scholar] [CrossRef][Green Version]
- Dapporto, P.; Fusi, V.; Micheloni, M.; Palma, P.; Paoli, P.; Pontellini, R. Binding properties and crystal structures of azamacrocycles containing nitrophenol side arms. Inorg. Chim. Acta 1998, 275–276, 168–174. [Google Scholar] [CrossRef]
- Rossotti, F.J.C.; Rossotti, H. Potentiometric titrations using Gran plots: A textbook omission. J. Chem. Educ. 1965, 42, 375–378. [Google Scholar] [CrossRef]
- Gans, P.; Sabatini, A.; Vacca, A. Investigation of equilibria in solution. Determination of equilibrium constants with the HYPERQUAD suite of programs. Talanta 1996, 43, 1739–1753. [Google Scholar] [CrossRef]
- Alderighi, L.; Gans, P.; Ienco, A.; Peters, D.; Sabatini, A.; Vacca, A. Hyperquad simulation and speciation (HySS): A utility program for the investigation of equilibria involving soluble and partially soluble species. Coord. Chem. Rev. 1999, 184, 311–318. [Google Scholar] [CrossRef]
- Amatori, S.; Ambrosi, G.; Errico Provenzano, A.; Fanelli, M.; Formica, M.; Fusi, V.; Giorgi, L.; Macedi, E.; Micheloni, M.; Paoli, P.; et al. PdII and PtII complexes with a thio-aza macrocycle ligand containing an intercalating fragment: Structural and antitumor activity studies. Inorg. Biochem. 2016, 162, 154–161. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are available from the authors. |



| Reaction | log K | |
|---|---|---|
| L = L1 | L = L2 | |
| H−3L3− + H+ = H−2L2− | - | 9.46(1)1 |
| H−2L2− + H+ = H−1L− | 8.67(9) | 8.33(1) |
| H−1L− + H+ = L | 7.77(7) | 7.37(1) |
| L + H+ = HL+ | 5.88(8) | 5.85(1) |
| HL+ + H+ = H2L2+ | 2.28(8) | 1.86(2) |
| Reaction | log K | |||||
|---|---|---|---|---|---|---|
| Co(II) | Cu(II) | Zn(II) | ||||
| L = L1 | L = Malten | L = L1 | L = Malten [41] | L = L1 | L = Malten | |
| M2+ + H−2L2− = [M(H−2L)] | 7.46(9)1 | 10.35(1) | 13.22(9) | 17.53(1) | 9.22(8) | 10.29(1) |
| [M(H−2L)] + H+ = [M(H−1L)]+ | - | 5.95(2) | 5.1(1) | 3.84(1) | - | 6.27(2) |
| [M(H-1L)]+ + H+ = [ML]2+ | - | - | - | 2.89(2) | - | - |
| [M(H−2L)] + OH− = [M(H−2L)OH]− | - | 2.92(4) | - | 2.95(1) | - | 2.95(4) |
| Reaction | log K | ||
|---|---|---|---|
| Co(II) | Cu(II) | Zn(II) | |
| M2+ + H−3L3− = [M(H−3L)]− | 12.88(1)1 | 18.25(2) | 10.74(1) |
| [M(H−3L)]− + H+ = [M(H−2L)] | 5.71(1) | 6.21(1) | 8.58(2) |
| [M(H−2L)] + H+ = [M(H−1L)]+ | 5.57(2) | 3.43(2) | 5.57(2) |
| [M(H−1L)]+ + H+ = [ML]2+ | - | 3.41(2) | - |
| [M(H−3L)]− + OH− = [M(H−3L)OH]2− | - | 3.79(3) | 5.07(3) |
| [M(H−3L)OH]2− + OH− = [M(H−3L)(OH)2]3− | - | - | 4.18(2) |
| M2+ + [M(H−3L)]− = [M2(H−3L)]+ | 5.24(4) | 7.63(3) | 8.62(3) |
| [M2(H−3L)]+ + OH− = [M2(H3L)OH] | 3.57(3) | - | - |
| [M2(H−3L)OH] + OH− = [M2(H−3L)(OH)2]− | 2.47(4) | - | - |
| [M(H−3L)]− + [M2(H−3L)]+ = [M3(H−3L)2] | - | 5.02(5) | 6.53(4) |
| [M3(H−3L)2] + OH− = [M3(H−3L)2OH]− | - | 4.59(6) | - |
| [M3(H−3L)2OH]− + OH− = [M3(H−3L)2(OH)2]2− | - | 3.39(4) | - |
| [M3(H−3L)2(OH)2]2− + OH− = [M3(H−3L)2(OH)3]3− | - | 3.22(5) | - |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Macedi, E.; Paderni, D.; Formica, M.; Conti, L.; Fanelli, M.; Giorgi, L.; Amatori, S.; Ambrosi, G.; Valtancoli, B.; Fusi, V. Playing with Structural Parameters: Synthesis and Characterization of Two New Maltol-Based Ligands with Binding and Antineoplastic Properties. Molecules 2020, 25, 943. https://doi.org/10.3390/molecules25040943
Macedi E, Paderni D, Formica M, Conti L, Fanelli M, Giorgi L, Amatori S, Ambrosi G, Valtancoli B, Fusi V. Playing with Structural Parameters: Synthesis and Characterization of Two New Maltol-Based Ligands with Binding and Antineoplastic Properties. Molecules. 2020; 25(4):943. https://doi.org/10.3390/molecules25040943
Chicago/Turabian StyleMacedi, Eleonora, Daniele Paderni, Mauro Formica, Luca Conti, Mirco Fanelli, Luca Giorgi, Stefano Amatori, Gianluca Ambrosi, Barbara Valtancoli, and Vieri Fusi. 2020. "Playing with Structural Parameters: Synthesis and Characterization of Two New Maltol-Based Ligands with Binding and Antineoplastic Properties" Molecules 25, no. 4: 943. https://doi.org/10.3390/molecules25040943
APA StyleMacedi, E., Paderni, D., Formica, M., Conti, L., Fanelli, M., Giorgi, L., Amatori, S., Ambrosi, G., Valtancoli, B., & Fusi, V. (2020). Playing with Structural Parameters: Synthesis and Characterization of Two New Maltol-Based Ligands with Binding and Antineoplastic Properties. Molecules, 25(4), 943. https://doi.org/10.3390/molecules25040943