Untargeted HILIC-MS-Based Metabolomics Approach to Evaluate Coffee Roasting Process: Contributing to an Integrated Metabolomics Multiplatform
Abstract
1. Introduction
2. Results and Discussion
2.1. Metabolomics Analysis of Coffee Samples by HILIC-MS
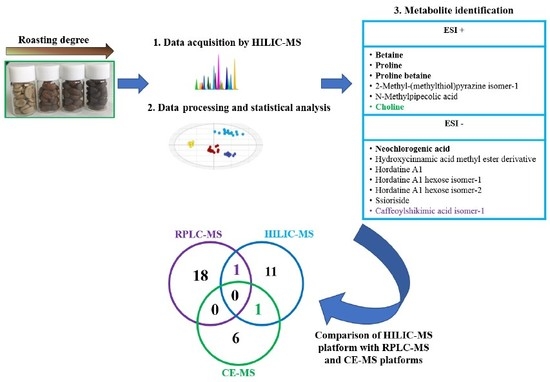
2.2. Metabolite Identification of Coffee Roasting Process
2.3. Integration of the Untargeted HILIC-MS Strategy Developed in a Metabolomics Multiplatform for the Search of Markers of the Coffee Roasting Process
3. Materials and Methods
3.1. Chemicals and samples
3.2. Sample Preparation
3.3. HILIC-MS Analysis
3.4. Metabolomics Sequence
3.5. Data Processing and Multivariate Analysis
3.6. Metabolite Identification
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Valduga, A.T.; Gonçalves, I.L.; Magri, E.; Delalibera Finzer, J.R. Chemistry, pharmacology and new trends in traditional functional and medicinal beverages. Food Res. Int. 2019, 120, 478–503. [Google Scholar] [CrossRef]
- Ludwig, I.A.; Clifford, M.N.; Lean, M.E.J.; Ashihara, H.; Crozier, A. Coffee: Biochemistry and potential impact on health. Food Funct. 2014, 5, 1695. [Google Scholar] [CrossRef] [PubMed]
- Romualdo, G.R.; Rocha, A.B.; Vinken, M.; Cogliati, B.; Moreno, F.S.; Chaves, M.A.G.; Barbisan, L.F. Drinking for protection? Epidemiological and experimental evidence on the beneficial effects of coffee or major coffee compounds against gastrointestinal and liver carcinogénesis. Food Res. Int. 2019, 123, 567–589. [Google Scholar] [CrossRef] [PubMed]
- Putri, S.P.; Irifune, T.; Fukusaki, E. GC/MS based metabolite profiling of Indonesian specialty coffee from different species and geographical origin. Metabolomics 2019, 15, 126. [Google Scholar] [CrossRef] [PubMed]
- Ciaramelli, C.; Palmioli, A.; Airoldi, C. Coffee variety, origin and extraction procedure: Implications for coffee beneficial effects on human health. Food Chem. 2019, 278, 47–55. [Google Scholar] [CrossRef]
- Hoyos Ossa, D.E.; Gil-Solsona, R.; Peñuela, G.A.; Sancho, J.V.; Hernández, F.J. Assessment of protected designation of origin for Colombian coffees based on HRMS-based metabolomics. Food Chem. 2018, 250, 89–97. [Google Scholar] [CrossRef]
- Wei, F.; Furihata, K.; Koda, M.; Hu, F.; Kato, R.; Miyakawa, T.; Tanokura, M. 13C NMR-based metabolomics for the classification of green coffee beans according to variety and origin. J. Agric. Food Chem. 2012, 60, 10118–10125. [Google Scholar] [CrossRef]
- Choi, M.-Y.; Choi, W.; Park, J.H.; Lim, J.; Kwon, S.W. Determination of coffee origins by integrated metabolomic approach of combining multiple analytical data. Food Chem. 2010, 121, 1260–1268. [Google Scholar] [CrossRef]
- Arana, V.A.; Medina, J.; Alarcon, R.; Moreno, E.; Heintz, L.; Schafer, H.; Wist, J. Coffee’s country of origin determined by NMR: The Colombian case. Food Chem. 2015, 175, 500–506. [Google Scholar] [CrossRef]
- Garrett, R.; Schmidt, E.M.; Pereira, L.F.P.; Kitzberger, C.S.G.; Scholz, M.B.S.; Eberlin, M.N.; Rezende, C.M. Discrimination of arabica coffee cultivars by electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry and chemometrics. LWT-Food Sci. Technol. 2013, 50, 496–502. [Google Scholar] [CrossRef]
- Consonni, R.; Ruth, L.; Cogliati, C.C. NMR based geographical characterization of roasted coffee. Talanta 2012, 88, 420–426. [Google Scholar] [CrossRef] [PubMed]
- Setoyama, D.; Iwasa, K.; Seta, H.; Shimizu, H.; Fujimura, Y.; Miura, D.; Wariishi, H.; Nagai, C.; Nakahara, K. High-throughput metabolic profiling of diverse green Coffea arabica beans identified tryptophan as a universal discrimination factor for immature beans. PLoS ONE 2013, 9, e70098. [Google Scholar] [CrossRef] [PubMed]
- Da Silva Taveira, J.H.; Meira Borém, F.; Pereira Figueiredo, L.; Reis, N.; Franca, A.S.; Harding, S.A.; Tsai, C.J. Potential markers of coffee genotypes grown in different Brazilian regions: A metabolomics approach. Food Res. Int. 2014, 61, 75–82. [Google Scholar] [CrossRef]
- Chang, K.L.; Ho, P.C. Gas chromatography time-of-flight mass spectrometry (GC-TOF-MS)-based metabolomics for comparison of caffeinated and decaffeinated coffee and its implications for Alzheimer’s disease. PLoS ONE 2014, 9, e104621. [Google Scholar] [CrossRef]
- Villalón-López, N.; Serrano-Contreras, J.I.; Téllez-Medina, D.I.; Gerardo Zepeda, L. An 1H NMR-based metabolomic approach to compare the chemical profiling of retail samples of ground roasted and instant coffees. Food Res. Int. 2018, 106, 263–270. [Google Scholar]
- Xu, L.; Lao, F.; Xu, Z.; Wang, X.; Chen, F.; Liao, X.; Chen, A.; Yang, S. Use of liquid chromatography quadrupole time-of-flight mass spectrometry and metabolomic approach to discriminate coffee brewed by different methods. Food Chem. 2019, 286, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Rothwell, J.A.; Loftfield, E.; Wedekind, R.; Freedman, N.; Kambanis, C.; Scalbert, A.; Sinha, R. A metabolomic study of the variability of the chemical composition of commonly consumed coffee brews. Metabolites 2019, 9, 17. [Google Scholar] [CrossRef]
- Wei, F.; Furihata, K.; Koda, M.; Hu, F.; Miyakawa, T.; Tanokura, M. Roasting process of coffee beans as studied by nuclear magnetic resonance: Time course of changes in composition. J. Agric. Food Chem. 2012, 60, 1005–1012. [Google Scholar] [CrossRef]
- Wei, F.; Furihata, K.; Miyakawa, T.; Tanokura, M. A pilot study of NMR-based sensory prediction of roasted coffee bean extracts. Food Chem. 2014, 152, 363–369. [Google Scholar] [CrossRef]
- Gloess, A.N.; Yeretzian, C.; Knochenmuss, R.; Groessl, M. On-line analysis of coffee roasting with ion mobility spectrometry–mass spectrometry (IMS–MS). Int. J. Mass. Spectrom. 2018, 424, 49–57. [Google Scholar] [CrossRef]
- Jaiswal, R.; Matei, M.F.; Golon, A.; Witt, M.; Kuhnert, N. Understanding the fate of chlorogenic acids in coffee roasting using mass spectrometry based targeted and non-targeted analytical strategies. Food Funct. 2012, 3, 976–984. [Google Scholar] [CrossRef] [PubMed]
- Cordero, C.; Liberto, E.; Bicchi, C.; Rubiolo, P.; Reichenbach, S.E.; Tian, X.; Tao, Q. Targeted and non-targeted approaches for complex natural sample profiling by GC×GC-qMS. J. Chromatogr. Sci. 2010, 48, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Ruosi, M.R.; Cordero, C.; Cagliero, C.; Rubiolo, P.; Bicchi, C.; Sgorbini, B.; Liberto, E. A further tool to monitor the coffee roasting process: Aroma composition and chemical indices. J. Agric. Chem. 2012, 60, 11283–11291. [Google Scholar] [CrossRef] [PubMed]
- Santos da Rosa, J.; Freitas-Silva, O.; Costa Rouws, J.R.; Gonçalves da Silva Moreira, I.; Moreira Novaes, F.J.; De Almeida Azevedo, D.; Schwab, N.; Oliveira Godoy, R.L.; Nogueira Eberlin, M.; Moraes de Rezende, C. Mass spectrometry screening of Arabica coffee roasting: A non-target and non-volatile approach by EASI-MS and ESI-MS. Food Res. Int. 2016, 89, 967–975. [Google Scholar] [CrossRef]
- Pérez-Míguez, R.; Sánchez-López, E.; Plaza, M.; Castro-Puyana, M.; Marina, M.L. A non-targeted metabolomics approach based on reverse-phase liquid chromatography-mass spectrometry to evaluate coffee roasting process. Anal. Bioanal. Chem. 2018, 410, 7859–7890. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Míguez, R.; Sánchez-López, E.; Plaza, M.; Marina, M.L.; Castro-Puyana, M. Capillary electrophoresis-mass spectrometry metabolic fingerprinting of green and roasted coffee. J. Chromatogr. A 2019, 360353. [Google Scholar] [CrossRef]
- Bernardo-Bermejo, S.; Sánchez-López, E.; Castro-Puyana, M.; Benito, S.; Lucio-Cazaña, F.J.; Marina, M.L. An untargeted metabolomics strategy based on liquid chromatography-mass spectrometry to study high glucose-induced changes in HK-2 cells. J. Chromatogr. A 2019, 1596, 124–133. [Google Scholar] [CrossRef]
- Guijarro-Díez, M.; Nozal, L.; Marina, M.L.; Crego, A.L. Metabolomic fingerprinting of saffron by LC/MS: Novel authenticity markers. Anal. Bioanal. Chem. 2015, 407, 7197–7213. [Google Scholar]
- Król, K.; Gantner, M.; Tatarak, A.; Hallmann, E. The content of polyphenols in coffee beans as roasting, origin and storage effect. Eur. Food Res. Tchnol. 2019. [Google Scholar] [CrossRef]
- Ginz, M.; Engelhardt, U.H. Identification of proline-based diketopiperazines in roasted coffee. J. Agric. Food Chem. 2000, 48, 3528–3532. [Google Scholar] [CrossRef]
- Lang, R.; Lang, T.; Bader, M.; Beusch, A.; Schlagbauer, V.; Hofmann, T. High-throughput quantification of proline betaine in foods and suitability as a valid biomarker for citrus consumption. J. Agric. Food Chem. 2017, 65, 1613–1619. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Lee, S.; Bang, E.; Lee, S.; Rhee, J.K.; Na, Y.C. Comparative evaluation of flavor compounds in fermented green and roasted coffee beans by solid phase microextraction-gas chromatography/mass spectrometry. Flavour Frag. J. 2019, 34, 365–376. [Google Scholar] [CrossRef]
- De Carvalho Martins, V.; de Oliveira Godoy, R.L.; Miranda Senna Gouvêa, A.C.; Pessanha de Araujo Santiago, M.C.; Galhardo Borguini, R.; de Oliveira Braga, E.C.; Pacheco, S.; da Silva de Mattos do Nascimento, L. Fraud investigation in commercial coffee by chromatography. Food Qual. Saf. 2018, 3, 121–133. [Google Scholar] [CrossRef]
- Gil de la Fuente, A.; Godzien, J.; Fernández López, M.; Rupérez, F.J.; Barbas, C.; Otero, A. Knowledge-based metabolite annotation tool: CEU Mass Mediator. J. Pharm. Biomed. Anal. 2018, 154, 138–149. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |



| ESI+ | ESI− | |||
|---|---|---|---|---|
| Quality Parameters | F (and p-values) of Cross-Validated ANOVA | Quality Parameters | F (and p-values) of Cross-Validated ANOVA | |
| GCB vs LRC | R2X = 0.485 R2Y = 0.989 Q2 = 0.977 | 561.6 (1.0 × 10−22) | R2X = 0.747 R2Y = 0.995 Q2 = 0.987 | 466.0 (2.3 × 10−22) |
| GCB vs MRC | R2X = 0.347 R2Y = 0.972 Q2 = 0.903 | 125.7 (2.1 × 10−14) | R2X = 0.676 R2Y = 0.993 Q2 = 0.980 | 220.2 (1.5 × 10−18) |
| GCB vs DRC | R2X = 0.637 R2Y = 0.990 Q2 = 0.980 | 197.1 (1.6 × 10−18) | R2X = 0.625 R2Y = 0.988 Q2 = 0.962 | 120.8 (1.6 × 10−15) |
| VIP Values | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| # | RT (min) | Molecular Formula | Tentative Identification | [M−H]− | Mass Error (ppm) | Main MS/MS Fragments | GCB vs. LRC | GCB vs. MRC | GCB vs. DRC | Roasting Trend |
| 1 | 2.2 | C10H10O4 | Hydroxycinnamic acid methyl ester derivative | 193.0483 | 11 | 65.0388 121.0280 93.0334 133.0277 | 0.78670 | 1.12365 | 1.49312 | ↑ |
| 2 | 2.5 | C16H16O8 | Caffeoylshikimic acid isomer | 335.0708 | 11 | 161.0238 135.0440 179.0333 | 1.81873 | 2.19334 | 2.41682 | ↓ |
| 3 | 2.7 | C28H38N8O5 | Hordatine A1 | 565.2902 | 11 | 59.0136 101.0589 113.0234 | 1.82774 | 2.19590 | 1.86040 | ↓ |
| 4 | 13.2 | C34H48N8O10 | Hordatine A1 hexose isomer | 727.3413 | 16 | 643.2944 113.0236 | 0.81788 | 0.95373 | 0.94533 | ↓ |
| 5 | 14.9 | Unknown | 481.2371 | 59.0133 89.0235 | 2.41084 | 2.85921 | 3.12239 | ↓ | ||
| 6 | 14.9 | C34H48N8O10 | Hordatine A1 hexose isomer | 727.3413 | 16 | 643.2942 113.0235 89.0235 | 2.32700 | 2.67836 | 2.78861 | ↓ |
| 7 | 16.4 | C27H38O12 | Ssioriside | 553.2225 | 3 | 44.9983 89.0239 59.0143 391.0400 119.0358 | 1.54553 | 2.49650 | 2.52013 | - |
| 8 | 17.7 | Unknown | 135.0436 | 67.0185 | 1.38856 | 1.31171 | 0.51246 | ↓ | ||
| 9 | 18.5 | Unknown | 135.0448 | 134.0358 89.0381 | 1.75153 | 1.26031 | 1.05637 | ↓ | ||
| 10 | 18.6 | C16H18O9 | Neochlorogenic acid * | 353.0803 | 7 | 191.0553 179.0339 135.0439 | 2.04079 | 1.59240 | 0.56977 | ↓ |
| 11 | 21.6 | Unknown | 705.3292 | 659.3259 335.2215 323.0965 | 1.80217 | 9.9 × 10−9 | 9.7 × 10−9 | ↑ | ||
| 12 | 25.1 | Unknown | 341.0528 | 89.0242 59.0138 71.0137 | 1.98371 | 2.6 × 10−8 | 2.2 × 10−8 | ↑ | ||
| 13 | 29.0 | Unknown | 242.0787 | 78.9588 168.0424 | 0.82795 | 0.43160 | 1.35421 | ↓ | ||
| VIP Values | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| # | RT (min) | Molecular Feature | Tentative Identification | [M + H]+ | Mass Error (ppm) | Main MS/MS Fragments | GCB vs. LRC | GCB vs. MRC | GCB vs. DRC | Roasting Trend |
| 1 | 12.3 | Unknown | 177.0561 | 117.0327 89.0377 145.0268 | 1.719010 | 2.00514 | 1.60664 | ↓ | ||
| 2 | 14.9 | Unknown | 679.5175 | - | 1.69434 | 1.90782 | 1.79200 | ↓ | ||
| 3 | 18.3 | Unknown | 393.0609 | 38.9629 | 1.70296 | 1.80418 | 0.60806 | - | ||
| 4 | 18.3 | Unknown | 163.0394 | 89.0380 117.0327 135.0431 | 1.71644 | 1.86415 | 0.51289 | - | ||
| 5 | 19.1 | C7H13NO2 | N-methylpipecolic acid | 144.1012 | 6 | 58.0649 84.0797 98.0973 | 1.71129 | 1.56815 | 1.76873 | ↓ |
| 6 | 19.4 | C5H11NO2 | Betaine * | 118.0858 | 10 | 58.0646 | 1.76821 | 2.079 | 1.8509 | ↓ |
| 7 | 19.9 | C7H13NO2 | Proline betaine * | 144.1019 | 6 | 98.0958 58.0637 84.0814 | 1.56002 | 1.30522 | 1.66228 | ↓ |
| 8 | 21.6 | Unknown | 295.1666 | 121.0275 175.1433 84.0804 | 1.53231 | 1.95759 | 1.7594 | - | ||
| 9 | 22.4 | C5H9NO2 | Proline * | 116.0703 | 2 | 70.0641 | 1.53958 | 3.55 × 10−8 | 1.49 × 10−8 | ↓ |
| 10 | 23.5 | Unknown | 200.1220 | - | 0.40495 | 1.77901 | 1.77808 | ↓ | ||
| 11 | 23.5 | Unknown | 244.1122 | 141.0470 126.0236 | 0.31289 | 1.88784 | 1.80463 | ↓ | ||
| 12 | 23.5 | C6H8N2S | 2-Methyl-(methylthiol)pyrazine isomer | 141.0483 | 10 | 126.0239 99.0134 82.0522 | 1.29913 | 1.81396 | 1.83413 | ↓ |
| 13 | 23.8 | Unknown | 236.1503 | 58.0647 57.0329 59.0723 | 1.7175 | 2.04337 | 1.86042 | ↓ | ||
| 14 | 24.2 | Unknown | 520.1954 | 123.0905 296.1594 | 0.986242 | 0 | 5.79 × 10−9 | - | ||
| 15 | 24.3 | Unknown | 266.1605 | 104.1064 60.0800 | 1.4989 | 1.60253 | 1.29524 | ↓ | ||
| 16 | 24.7 | Unknown | 226.1186 | 123.0915 110.0830 | 1.64839 | 1.91133 | 1.67194 | ↑ | ||
| 17 | 25.6 | Unknown | 129.0654 | - | 1.76951 | 9.33 × 10−9 | 3.66 × 10−8 | ↓ | ||
| 18 | 27.8 | Unknown | 198.1236 | - | 1.7959 | 2.11365 | 1.87729 | ↓ | ||
| 19 | 28.8 | C5H14NO | Choline *,a | 104.1065 | 7 | 58.0647 60.0804 45.0328 | 1.63786 | 0.18465 | 1.78252 | ↓ |
| 20 | 28.8 | Unknown | 258.1106 | 104.1070 124.9995 86.0961 184.0730 | 1.53731 | 0.15982 | 1.77857 | ↓ | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Míguez, R.; Castro-Puyana, M.; Sánchez-López, E.; Plaza, M.; Marina, M.L. Untargeted HILIC-MS-Based Metabolomics Approach to Evaluate Coffee Roasting Process: Contributing to an Integrated Metabolomics Multiplatform. Molecules 2020, 25, 887. https://doi.org/10.3390/molecules25040887
Pérez-Míguez R, Castro-Puyana M, Sánchez-López E, Plaza M, Marina ML. Untargeted HILIC-MS-Based Metabolomics Approach to Evaluate Coffee Roasting Process: Contributing to an Integrated Metabolomics Multiplatform. Molecules. 2020; 25(4):887. https://doi.org/10.3390/molecules25040887
Chicago/Turabian StylePérez-Míguez, Raquel, María Castro-Puyana, Elena Sánchez-López, Merichel Plaza, and María Luisa Marina. 2020. "Untargeted HILIC-MS-Based Metabolomics Approach to Evaluate Coffee Roasting Process: Contributing to an Integrated Metabolomics Multiplatform" Molecules 25, no. 4: 887. https://doi.org/10.3390/molecules25040887
APA StylePérez-Míguez, R., Castro-Puyana, M., Sánchez-López, E., Plaza, M., & Marina, M. L. (2020). Untargeted HILIC-MS-Based Metabolomics Approach to Evaluate Coffee Roasting Process: Contributing to an Integrated Metabolomics Multiplatform. Molecules, 25(4), 887. https://doi.org/10.3390/molecules25040887