CRANAD-28: A Robust Fluorescent Compound for Visualization of Amyloid Beta Plaques
Abstract
1. Introduction
2. Results
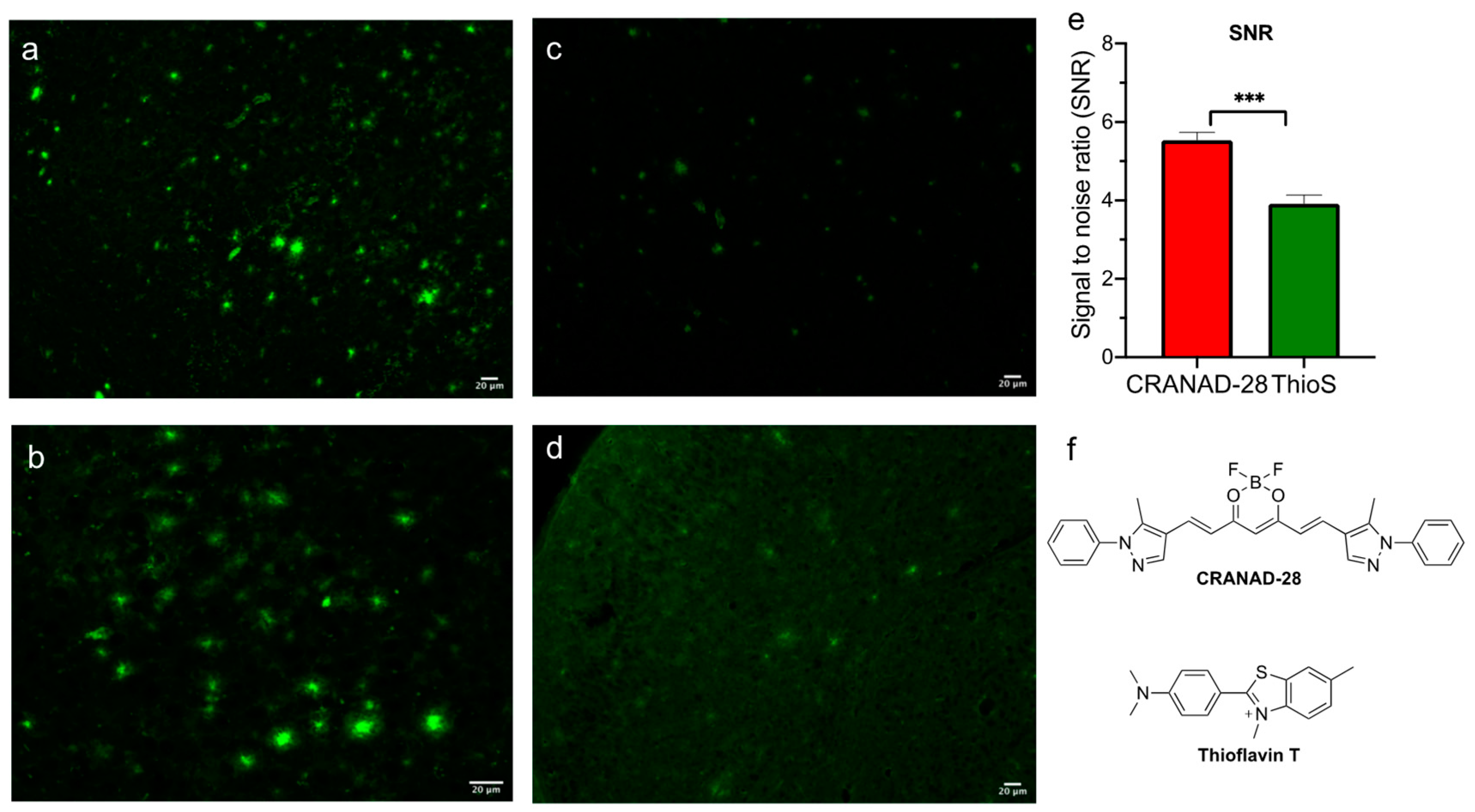
2.1. Brightness Comparison between CRANAD-28 and Thioflavin S
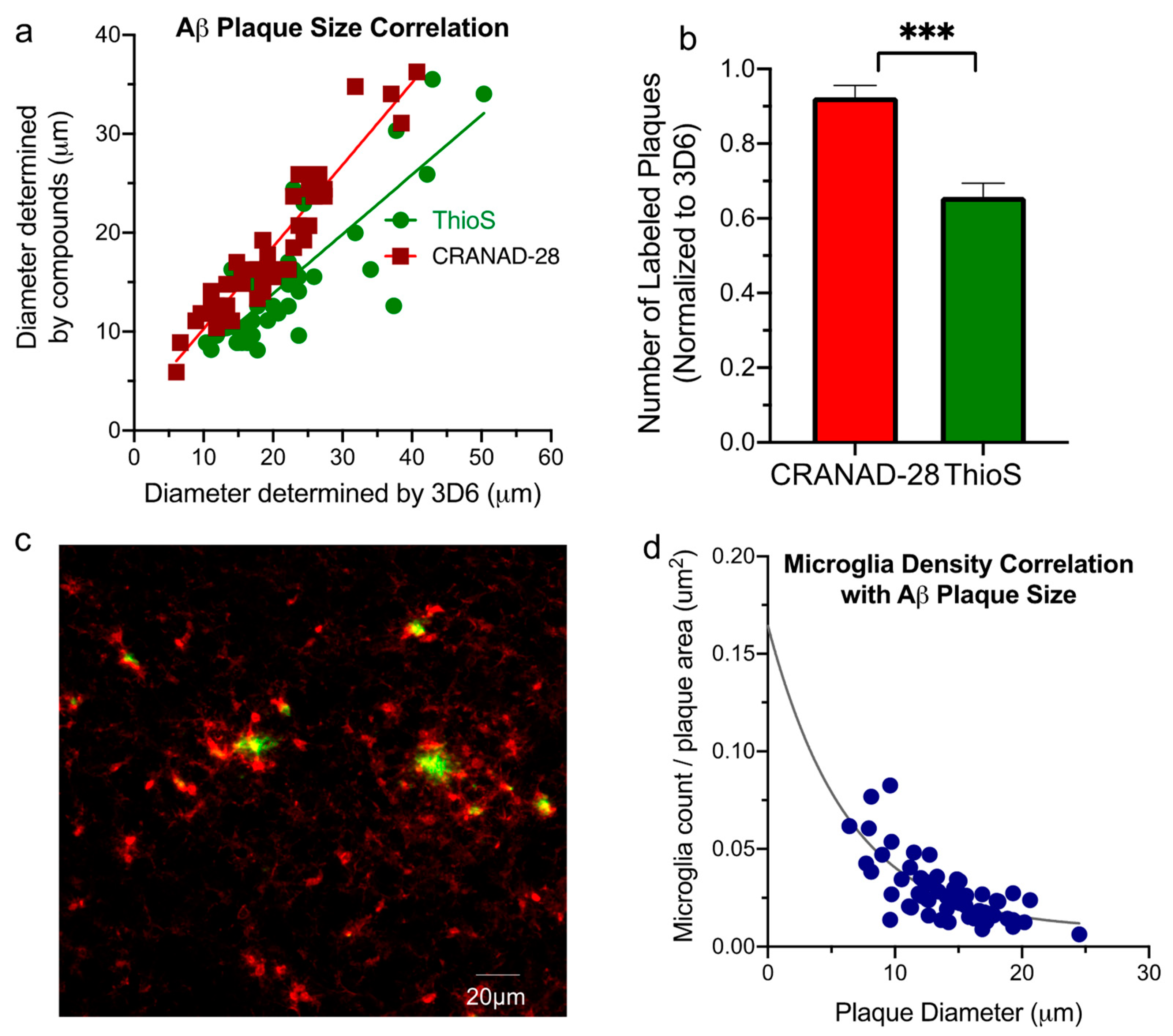
2.2. Aβ Plaque Morphology and Quantification Assay
2.3. Aβ Plaque and Microglia Staining
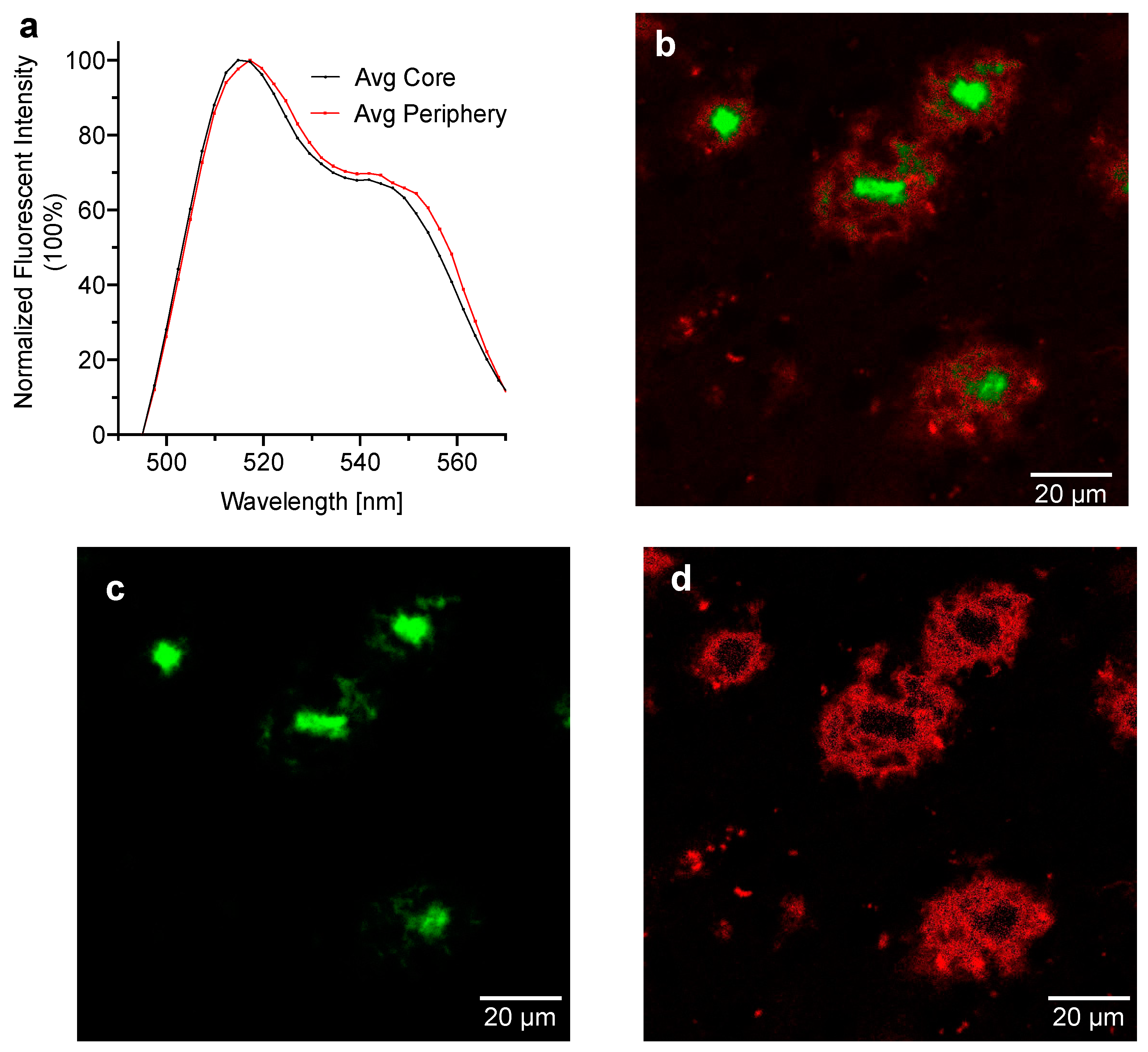
2.4. Spectral Signatures of CRANAD-28 for Core and Periphery of the Aβ Plaques
3. Discussion
4. Materials and Methods
4.1. Transgenic Mouse Brain Slide Preparation
4.2. Slide Staining and Fluorescence Imaging with CRANAD-28 and Thioflavin S
4.3. Slide Staining with 3D6 Antibody and IBA-1 Antibody
4.4. Confocal Microscope and Spectrum Extraction
Author Contributions
Funding
Conflicts of Interest
References
- Selkoe, D.; Hardy, J. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol. Med. 2016, 8, 595–608. [Google Scholar] [CrossRef]
- Karran, E.; Mercken, M.; De Strooper, B. The amyloid cascade hypothesis for Alzheimer’s disease: An appraisal for the development of therapeutics. Nat. Rev. Drug Discov. 2011, 10, 698–712. [Google Scholar] [CrossRef] [PubMed]
- Herrup, K. The case for rejecting the amyloid cascade hypothesis. Nat. Neurosci. 2015, 18, 794–799. [Google Scholar] [CrossRef] [PubMed]
- Hardy, J.A.; Higgins, G.A. Alzheimer’s disease: The amyloid cascade hypothesis. Science 1992, 256, 184–185. [Google Scholar] [CrossRef] [PubMed]
- Hardy, J.; Selkoe, D.J. The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science 2002, 297, 353–356. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, U.; Nilson, A.N.; Kayed, R. The Role of Amyloid-beta Oligomers in Toxicity, Propagation, and Immunotherapy. EBioMedicine 2016, 6, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Telling, N.D.; Everett, J.; Collingwood, J.F.; Dobson, J.; van der Laan, G.; Gallagher, J.J.; Wang, J.; Hitchcock, A.P. Iron Biochemistry is Correlated with Amyloid Plaque Morphology in an Established Mouse Model of Alzheimer’s Disease. Cell Chem. Biol. 2017, 24, 1205–1215 e3. [Google Scholar] [CrossRef]
- Shea, D.; Hsu, C.C.; Bi, T.M.; Paranjapye, N.; Childers, M.C.; Cochran, J.; Tomberlin, C.P.; Wang, L.; Paris, D.; Zonderman, J.; et al. Alpha-Sheet secondary structure in amyloid beta-peptide drives aggregation and toxicity in Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2019, 116, 8895–8900. [Google Scholar] [CrossRef]
- Zhang, X.; Tian, Y.; Yuan, P.; Li, Y.; Yaseen, M.A.; Grutzendler, J.; Moore, A.; Ran, C. A bifunctional curcumin analogue for two-photon imaging and inhibiting crosslinking of amyloid beta in Alzheimer’s disease. Chem. Comm. 2014, 50, 11550–11553. [Google Scholar] [CrossRef]
- Bussiere, T.; Bard, F.; Barbour, R.; Grajeda, H.; Guido, T.; Khan, K.; Schenk, D.; Games, D.; Seubert, P.; Buttini, M. Morphological characterization of Thioflavin-S-positive amyloid plaques in transgenic Alzheimer mice and effect of passive Abeta immunotherapy on their clearance. Am. J. Pathol. 2004, 165, 987–995. [Google Scholar] [CrossRef]
- Saeed, S.M.; Fine, G. Thioflavin-T for amyloid detection. Am. J. Clin. Pathol. 1967, 47, 588–593. [Google Scholar] [CrossRef] [PubMed]
- Khurana, R.; Coleman, C.; Ionescu-Zanetti, C.; Carter, S.A.; Krishna, V.; Grover, R.K.; Roy, R.; Singh, S. Mechanism of thioflavin T binding to amyloid fibrils. J. Struct. Biol. 2005, 151, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Johnson-Wood, K.; Lee, M.; Motter, R.; Hu, K.; Gordon, G.; Barbour, R.; Khan, K.; Gordon, M.; Tan, H.; Games, D.; et al. Amyloid precursor protein processing and A beta42 deposition in a transgenic mouse model of Alzheimer disease. Proc. Natl. Acad. Sci. USA 1997, 94, 1550–1555. [Google Scholar] [CrossRef] [PubMed]
- DeMattos, R.B.; Bales, K.R.; Cummins, D.J.; Dodart, J.C.; Paul, S.M.; Holtzman, D.M. Peripheral anti-A beta antibody alters CNS and plasma A beta clearance and decreases brain A beta burden in a mouse model of Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2001, 98, 8850–8855. [Google Scholar] [CrossRef] [PubMed]
- Demattos, R.B.; Lu, J.; Tang, Y.; Racke, M.M.; Delong, C.A.; Tzaferis, J.A.; Hole, J.T.; Forster, B.M.; McDonnell, P.C.; Liu, F.; et al. A plaque-specific antibody clears existing beta-amyloid plaques in Alzheimer’s disease mice. Neuron 2012, 76, 908–920. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, X.; Yuan, P.; Yang, J.; Xu, Y.; Grutzendler, J.; Shao, Y.; Moore, A.; Ran, C. Oxalate-curcumin-based probe for micro- and macroimaging of reactive oxygen species in Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2017, 114, 12384–12389. [Google Scholar] [CrossRef]
- Rasmussen, J.; Mahler, J.; Beschorner, N.; Kaeser, S.A.; Hasler, L.M.; Baumann, F.; Nystrom, S.; Portelius, E.; Blennow, K.; Lashley, T.; et al. Amyloid polymorphisms constitute distinct clouds of conformational variants in different etiological subtypes of Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2017, 114, 13018–13023. [Google Scholar] [CrossRef]
- Condello, C.; Lemmin, T.; Stohr, J.; Nick, M.; Wu, Y.; Maxwell, A.M.; Watts, J.C.; Caro, C.D.; Oehler, A.; Keene, C.D.; et al. Structural heterogeneity and intersubject variability of Abeta in familial and sporadic Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2018, 115, E782–E791. [Google Scholar] [CrossRef]
- McLean, C.A.; Cherny, R.A.; Fraser, F.W.; Fuller, S.J.; Smith, M.J.; Beyreuther, K.; Bush, A.I.; Masters, C.L. Soluble pool of Abeta amyloid as a determinant of severity of neurodegeneration in Alzheimer’s disease. Ann. Neurol. 1999, 46, 860–866. [Google Scholar] [CrossRef]
- Lue, L.F.; Kuo, Y.M.; Roher, A.E.; Brachova, L.; Shen, Y.; Sue, L.; Beach, T.; Kurth, J.H.; Rydel, R.E.; Rogers, J. Soluble amyloid beta peptide concentration as a predictor of synaptic change in Alzheimer’s disease. Am. J. Pathol. 1999, 155, 853–862. [Google Scholar] [CrossRef]
- Terry, R.; Masliah, E.; Salmon, D.P.; Butters, N.; DeTeresa, R.; Hill, R.; Hansen, L.A.; Katzman, R. Physical basis of cognitive alterations in Alzheimer’s disease: Synapse loss is the major correlate of cognitive impairment. Ann. Neurol. 1991, 30, 572–580. [Google Scholar] [CrossRef] [PubMed]
- Guillozet, A.L.; Weintraub, S.; Mash, D.C.; Mesulam, M.M. Neurofibrillary tangles, amyloid, and memory in aging and mild cognitive impairment. Arch. Neurol. 2003, 60, 729–736. [Google Scholar] [CrossRef] [PubMed]
- Condello, C.; Schain, A.; Grutzendler, J. Multicolor time-stamp reveals the dynamics and toxicity of amyloid deposition. Sci. Rep. 2011, 1, 19. [Google Scholar] [CrossRef] [PubMed]
- Yan, P.; Bero, A.W.; Cirrito, J.R.; Xiao, Q.; Hu, X.; Wang, Y.; Gonzales, E.; Holtzman, D.M.; Lee, J.M. Characterizing the appearance and growth of amyloid plaques in APP/PS1 mice. J. Neurosci. 2009, 29, 10706–10714. [Google Scholar] [CrossRef]
- Bode, D.C.; Freeley, M.; Nield, J.; Palma, M.; Viles, J.H. Amyloid-beta oligomers have a profound detergent-like effect on lipid membrane bilayers, imaged by atomic force and electron microscopy. J. Biol. Chem. 2019, 294, 7566–7572. [Google Scholar] [CrossRef]
Sample Availability: CRANAD-28 is available from the authors. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ran, K.; Yang, J.; Nair, A.V.; Zhu, B.; Ran, C. CRANAD-28: A Robust Fluorescent Compound for Visualization of Amyloid Beta Plaques. Molecules 2020, 25, 863. https://doi.org/10.3390/molecules25040863
Ran K, Yang J, Nair AV, Zhu B, Ran C. CRANAD-28: A Robust Fluorescent Compound for Visualization of Amyloid Beta Plaques. Molecules. 2020; 25(4):863. https://doi.org/10.3390/molecules25040863
Chicago/Turabian StyleRan, Kathleen, Jing Yang, Anil V. Nair, Biyue Zhu, and Chongzhao Ran. 2020. "CRANAD-28: A Robust Fluorescent Compound for Visualization of Amyloid Beta Plaques" Molecules 25, no. 4: 863. https://doi.org/10.3390/molecules25040863
APA StyleRan, K., Yang, J., Nair, A. V., Zhu, B., & Ran, C. (2020). CRANAD-28: A Robust Fluorescent Compound for Visualization of Amyloid Beta Plaques. Molecules, 25(4), 863. https://doi.org/10.3390/molecules25040863



