Substrate-Specific Activation of α-Secretase by 7-Deoxy-Trans-Dihydronarciclasine Increases Non-Amyloidogenic Processing of β-Amyloid Protein Precursor
Abstract
1. Introduction
2. Results
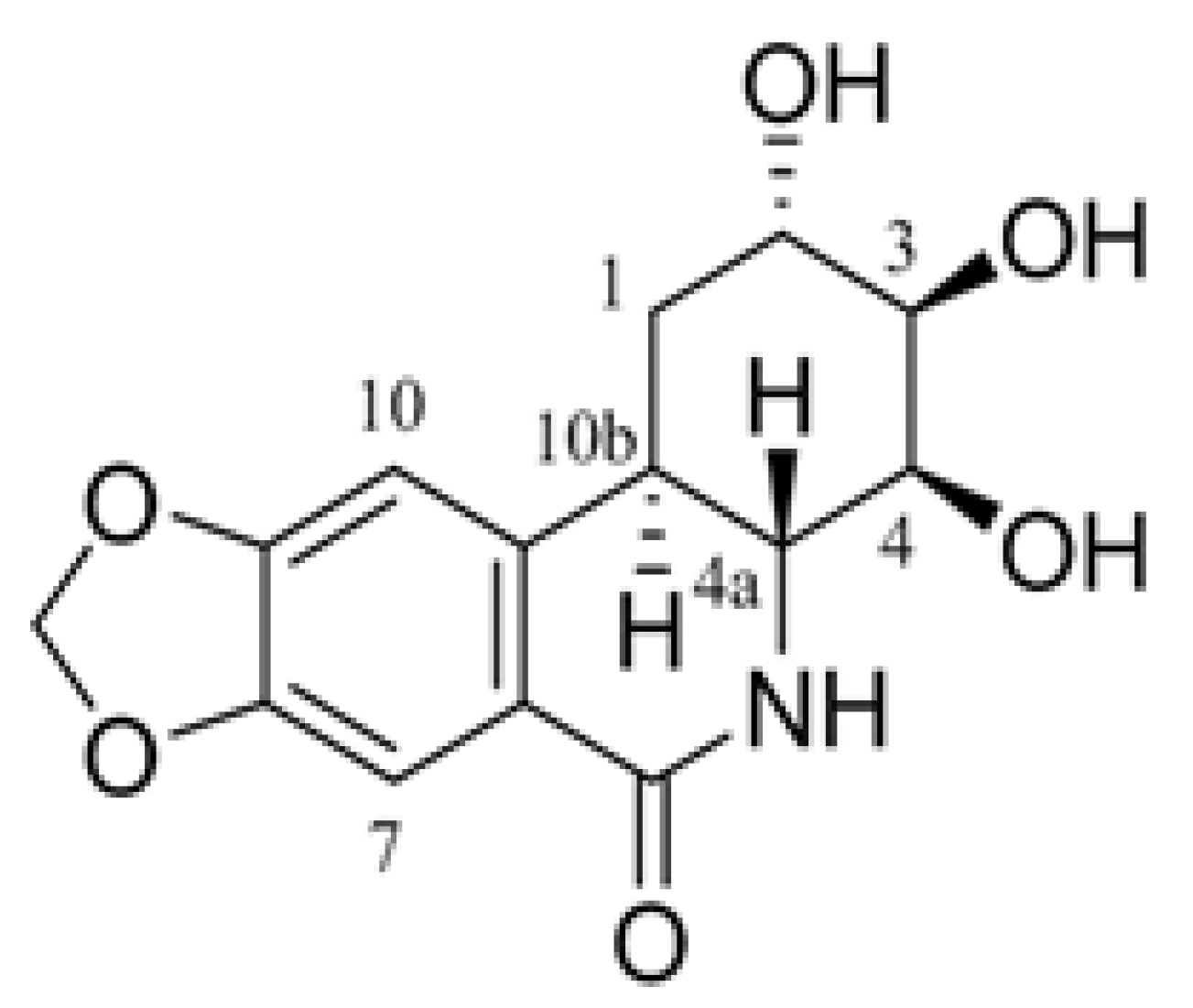
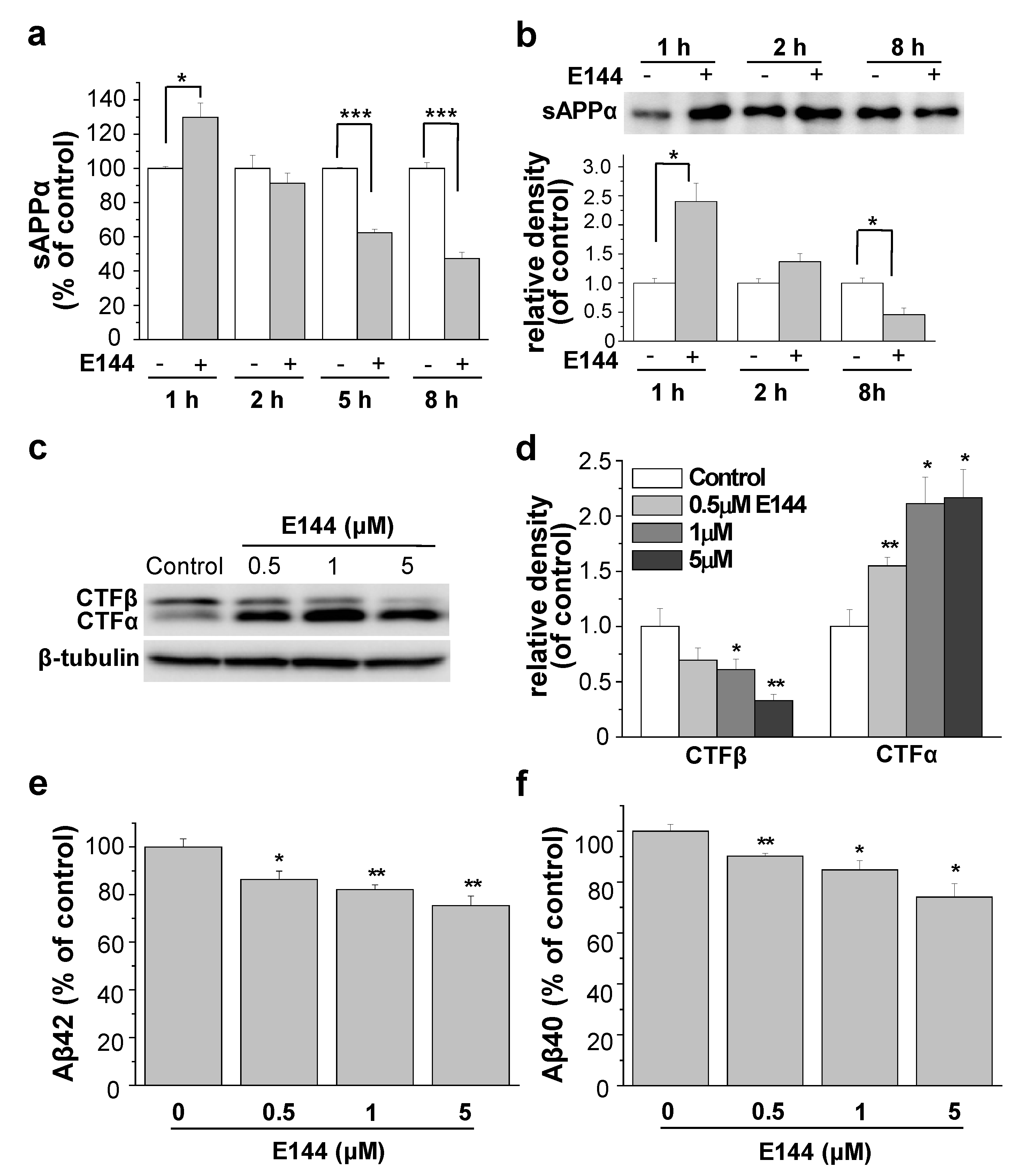
2.1. E144 Increases Secreted sAPPα Level but Decreases Aβ Levels
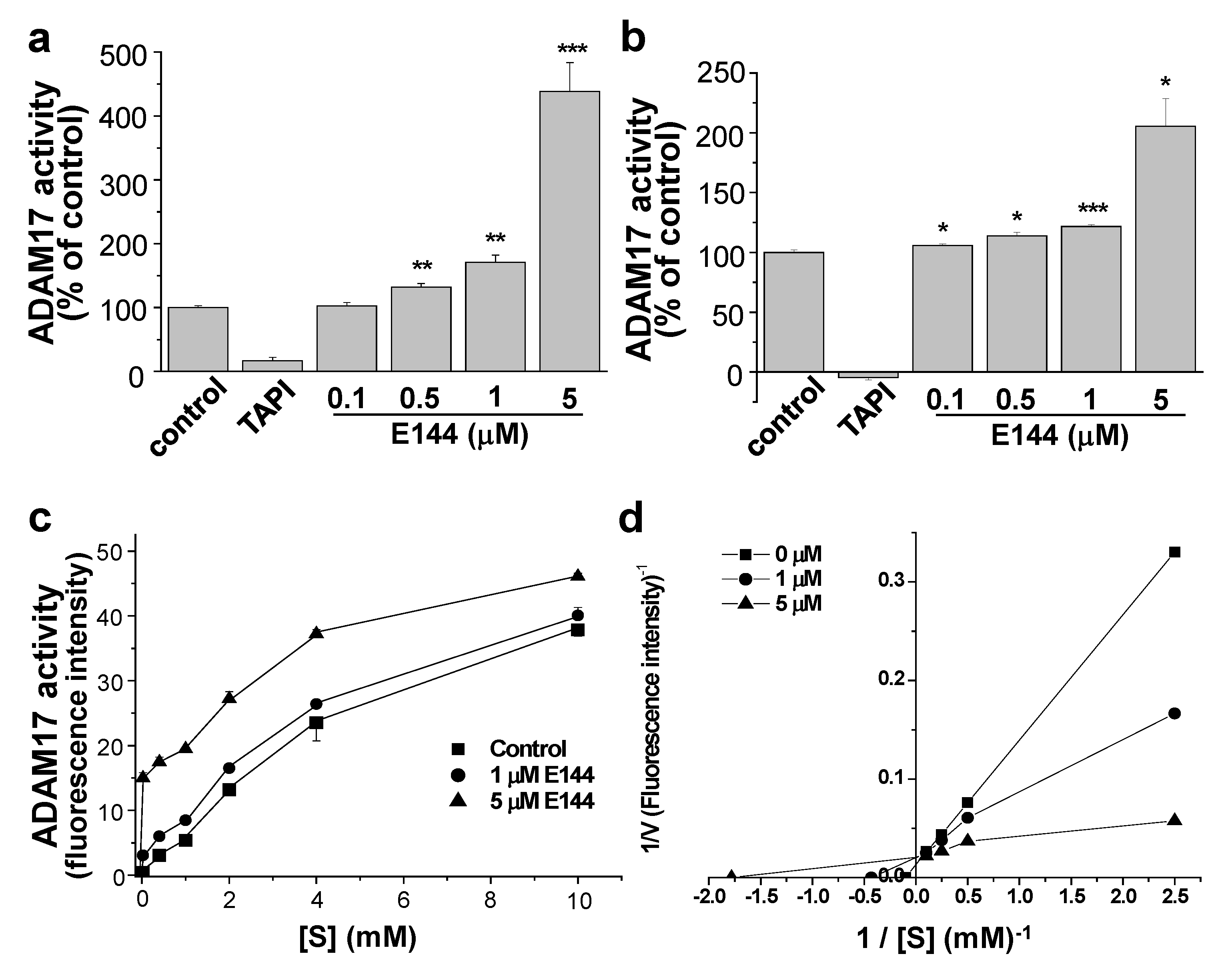
2.2. E144 Directly Activates ADAM17
2.3. E144 Activates ADAM17 in a Substrate-Specific Manner
2.4. E144 Activates ADAM10 in a Substrate-Specific Manner
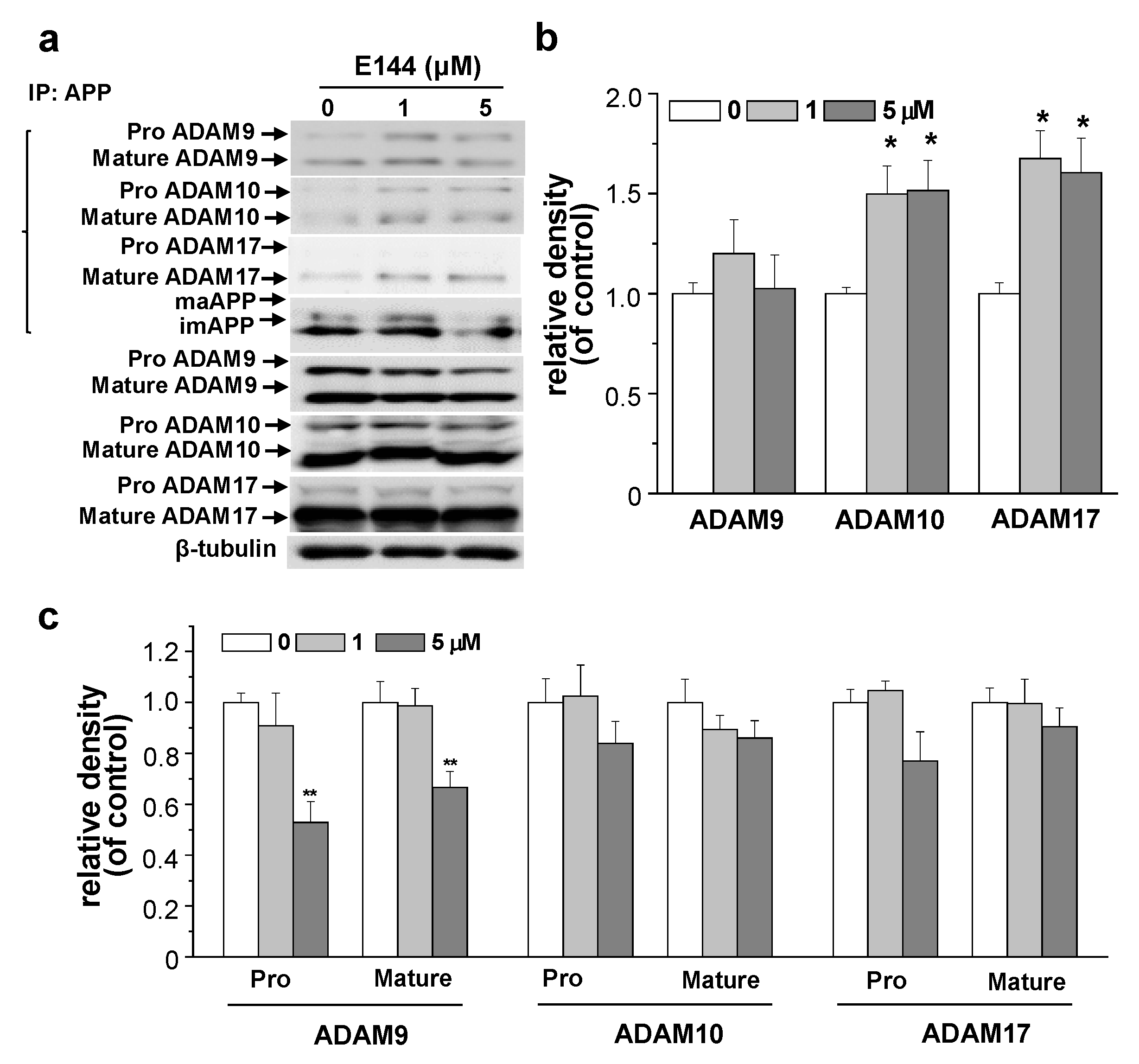
2.5. E144 Increases Interaction of APP with ADAM10 and ADAM17
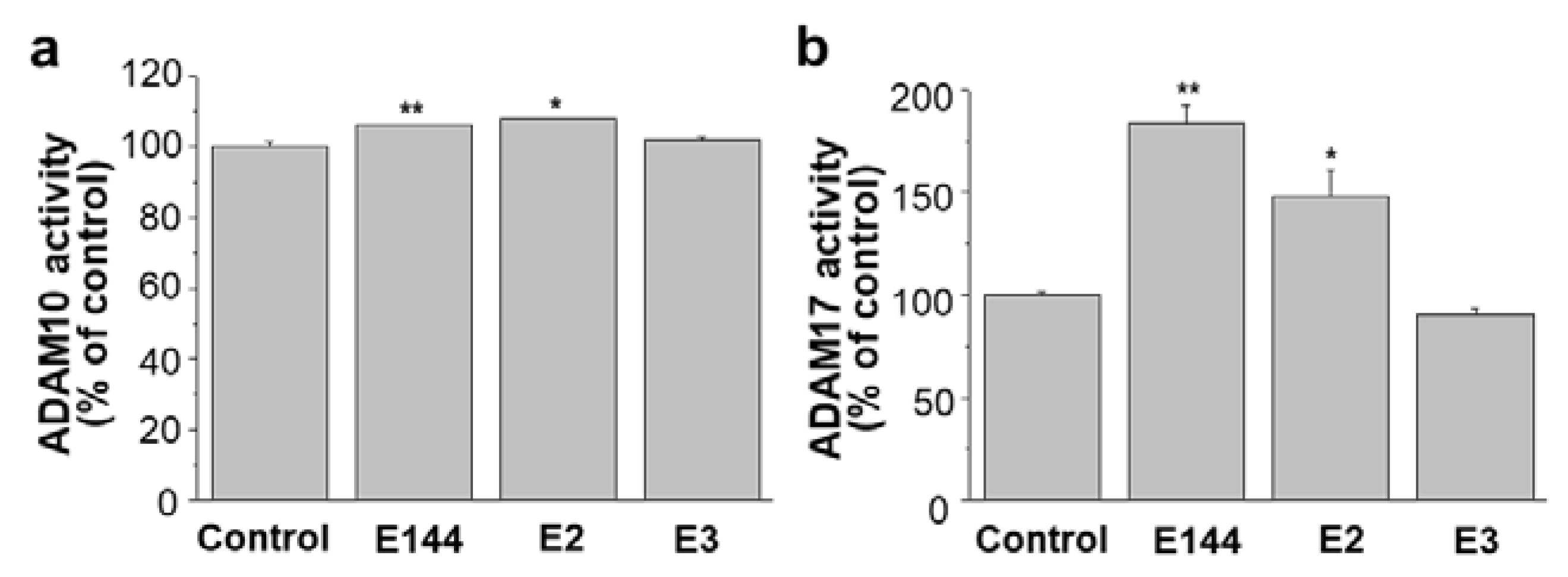
2.6. Differential Effects of Structural Analogues of E144 on ADAM10 and ADAM17
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Experimental Treatments
4.2. sAPPα Immunoprecipitation
4.3. Protein Extraction and Western Blotting
4.4. sAPPα and Aβ Peptide Assay
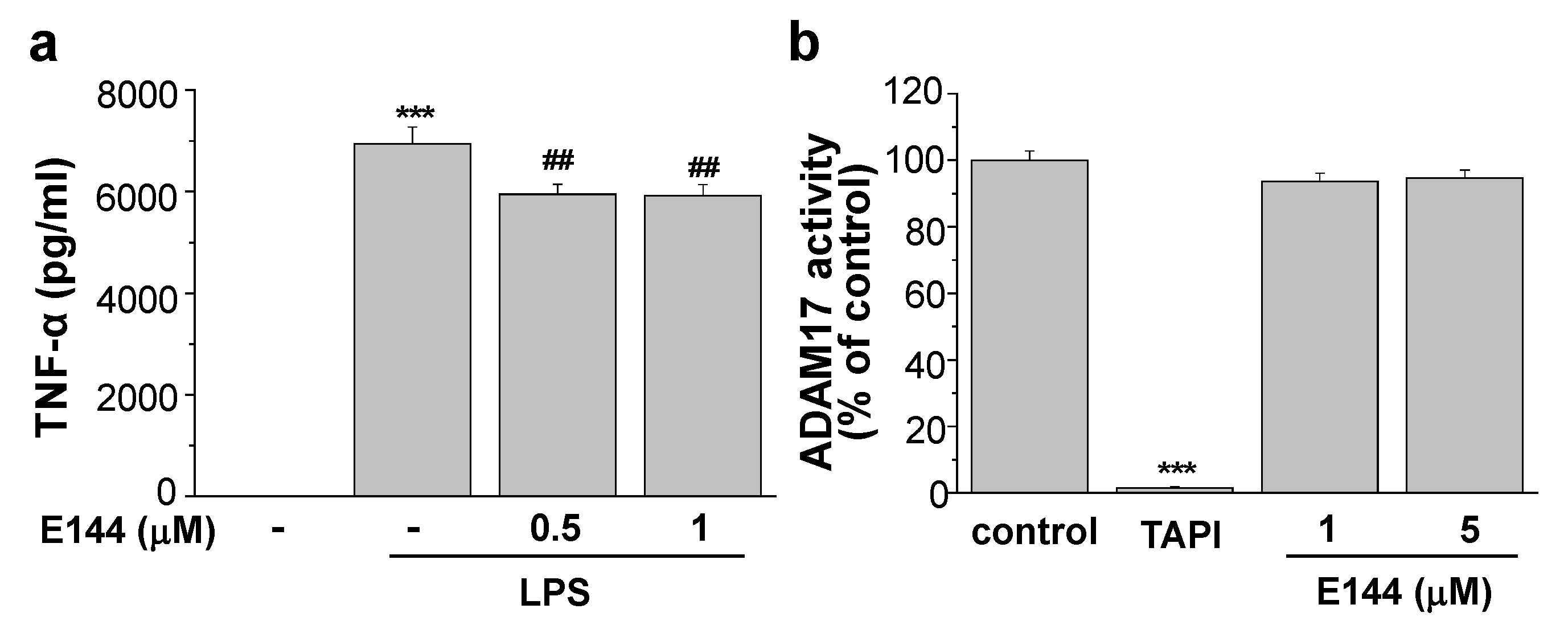
4.5. TNFα Assay
4.6. α-Secretase Activity Assay
4.7. siRNA-Mediated Knockdown of ADAM9, ADAM10, and ADAM17
4.8. Co-Immunoprecipitation
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Selkoe, D.J. Alzheimer’s disease: Genes, proteins, and therapy. Physiol. Rev. 2001, 81, 741–766. [Google Scholar] [CrossRef] [PubMed]
- Tanzi, R.E.; Bertram, L. Twenty Years of the Alzheimer’s Disease Amyloid Hypothesis: A Genetic Perspective. Cell 2005, 120, 545–555. [Google Scholar] [CrossRef]
- Congdon, E.E.; Sigurdsson, E.M. Tau-targeting therapies for Alzheimer disease. Nat. Rev. Neurol. 2018, 14, 399–415. [Google Scholar] [CrossRef]
- Hardy, J.; Selkoe, D.J. The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science 2002, 297, 353–356. [Google Scholar] [CrossRef] [PubMed]
- Mattson, M.P. Pathways towards and away from Alzheimer’s disease. Nature 2004, 430, 631–639. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Cheung, T.; Cai, X.; Odaka, A.; Otvos, L.; Eckman, C.; Golde, T.; Younkin, S. An increased percentage of long amyloid β protein secreted by familial amyloid β protein precursor (β APP717) mutants. Science 1994, 264, 1336–1340. [Google Scholar] [CrossRef]
- Fernandez, M.A.; Klutkowski, J.A.; Freret, T.; Wolfe, M.S. Alzheimer Presenilin-1 Mutations Dramatically Reduce Trimming of Long Amyloid β-Peptides (Aβ) by γ-Secretase to Increase 42-to-40-Residue Aβ. J. Boil. Chem. 2014, 289, 31043–31052. [Google Scholar] [CrossRef]
- Thinakaran, G.; Koo, E.H. Amyloid precursor protein trafficking, processing, and function. J. Boil. Chem. 2008, 283, 29615–29619. [Google Scholar] [CrossRef]
- Gandy, S. The role of cerebral amyloid β accumulation in common forms of Alzheimer disease. J. Clin. Investig. 2005, 115, 1121–1129. [Google Scholar]
- Lammich, S.; Kojro, E.; Postina, R.; Gilbert, S.; Pfeiffer, R.; Jasionowski, M.; Haass, C.; Fahrenholz, F. Constitutive and regulated α-secretase cleavage of Alzheimer’s amyloid precursor protein by a disintegrin metalloprotease. Proc. Natl. Acad. Sci. USA 1999, 96, 3922–3927. [Google Scholar] [CrossRef]
- Buxbaum, J.D.; Liu, K.N.; Luo, Y.; Slack, J.L.; Stocking, K.L.; Peschon, J.J.; Johnson, R.S.; Castner, B.J.; Cerretti, D.P.; Black, R.A. Evidence that tumor necrosis factor alpha converting enzyme is involved in regulated alpha-secretase cleavage of the Alzheimer amyloid protein precursor. J. Boil. Chem. 1998, 273, 27765–27767. [Google Scholar] [CrossRef] [PubMed]
- Sannerud, R.; Annaert, W. Trafficking, a key player in regulated intramembrane proteolysis. Semin. Cell Dev. Boil. 2009, 20, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Postina, R. Activation of α-secretase cleavage. J. Neurochem. 2011, 120, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Peron, R.; Vatanabe, I.P.; Manzine, P.R.; Camins, A.; Cominetti, M.R. Alpha-Secretase ADAM10 Regulation: Insights into Alzheimer’s Disease Treatment. Pharmaceuticals 2018, 11, 12. [Google Scholar] [CrossRef]
- Kuhn, P.H.; Wang, H.; Dislich, B.; Colombo, A.; Zeitschel, U.; Ellwart, J.W.; Kremmer, E.; Rossner, S.; Lichtenthaler, S.F. ADAM10 is the physiologically relevant, constitutive alpha-secretase of the amyloid precursor protein in primary neurons. EMBO J. 2010, 29, 3020–3032. [Google Scholar] [CrossRef]
- Hsia, H.-E.; Tüshaus, J.; Brummer, T.; Zheng, Y.; Scilabra, S.D.; Lichtenthaler, S.F. Functions of ’A disintegrin and metalloproteases (ADAMs)’ in the mammalian nervous system. Cell. Mol. Life Sci. 2019, 76, 3055–3081. [Google Scholar] [CrossRef]
- Cissé, M.A.; Sunyach, C.; Lefranc-Jullien, S.; Postina, R.; Vincent, B.; Checler, F. The Disintegrin ADAM9 Indirectly Contributes to the Physiological Processing of Cellular Prion by Modulating ADAM10 Activity. J. Boil. Chem. 2005, 280, 40624–40631. [Google Scholar] [CrossRef]
- Cissé, M.; Braun, U.; Leitges, M.; Fisher, A.; Pagès, G.; Checler, F.; Vincent, B. ERK1-independent α-secretase cut of β-amyloid precursor protein via M1 muscarinic receptors and PKCα/ε. Mol. Cell. Neurosci. 2011, 47, 223–232. [Google Scholar] [CrossRef]
- Edwards, D.; Handsley, M.; Pennington, C. The ADAM metalloproteinases. Mol. Asp. Med. 2008, 29, 258–289. [Google Scholar] [CrossRef]
- Black, R.A.; Rauch, C.T.; Kozlosky, C.J.; Peschon, J.J.; Slack, J.L.; Wolfson, M.F.; Castner, B.J.; Stocking, K.L.; Reddy, P.; Srinivasan, S.; et al. A metalloproteinase disintegrin that releases tumour-necrosis factor-α from cells. Nature 1997, 385, 729–733. [Google Scholar] [CrossRef]
- Moss, M.L.; Jin, S.-L.C.; Milla, M.E.; Burkhart, W.; Carter, H.L.; Chen, W.-J.; Clay, W.C.; Didsbury, J.R.; Hassler, D.; Hoffman, C.R.; et al. Cloning of a disintegrin metalloproteinase that processes precursor tumour-necrosis factor-α. Nature 1997, 385, 733–736. [Google Scholar] [CrossRef]
- Blobel, C.P. ADAMs: key components in EGFR signalling and development. Nat. Rev. Mol. Cell Boil. 2005, 6, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Yu, D.; Bi, Y.; Cao, Y. A disintegrin and metalloprotease 17 promotes microglial cell survival via epidermal growth factor receptor signalling following spinal cord injury. Mol. Med. Rep. 2015, 12, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Sennvik, K.; Fastbom, J.; Blomberg, M.; Wahlund, L.-O.; Winblad, B.; Benedikz, E. Levels of α- and β-secretase cleaved amyloid precursor protein in the cerebrospinal fluid of Alzheimer’s disease patients. Neurosci. Lett. 2000, 278, 169–172. [Google Scholar] [CrossRef]
- Lannfelt, L.; Basun, H.; Wahlund, L.-O.; Rowe, B.A.; Wagner, S.L. Decreased α-secretase-cleaved amyloid precursor protein as a diagnostic marker for Alzheimer’s diseas. Nat. Med. 1995, 1, 829–832. [Google Scholar] [CrossRef]
- Postina, R.; Schroeder, A.; Dewachter, I.; Bohl, J.; Schmitt, U.; Kojro, E.; Prinzen, C.; Endres, K.; Hiemke, C.; Blessing, M.; et al. A disintegrin-metalloproteinase prevents amyloid plaque formation and hippocampal defects in an Alzheimer disease mouse model. J. Clin. Investig. 2004, 113, 1456–1464. [Google Scholar] [CrossRef]
- Kim, M.; Suh, J.; Romano, N.; Truong, M.H.; Mullin, K.; Hooli, B.; Norton, D.; Tesco, G.; Elliott, K.; Wagner, S.L.; et al. Potential late-onset Alzheimer’s disease-associated mutations in the ADAM10 gene attenuate α-secretase activity. Hum. Mol. Genet. 2009, 18, 3987–3996. [Google Scholar] [CrossRef]
- Skovronsky, D.M.; Fath, S.; Lee, V.M.; Milla, M.E. Neuronal localization of the TNFα converting enzyme (TACE) in brain tissue and its correlation to amyloid plaques. J. Neurobiol. 2001, 49, 40–46. [Google Scholar] [CrossRef]
- Slack, B.E.; Ma, L.K.; Seah, C.C. Constitutive shedding of the amyloid precursor protein ectodomain is up-regulated by tumour necrosis factor-α converting enzyme. Biochem. J. 2001, 357, 787–794. [Google Scholar] [CrossRef]
- Merlos-Suárez, A.; Fernández-Larrea, J.; Reddy, P.; Baselga, J.; Arribas, J. Pro-tumor necrosis factor-α processing activity is tightly controlled by a component that does not affect notch processing. J. Boil. Chem. 1998, 273, 24955–24962. [Google Scholar] [CrossRef]
- Qian, M.; Shen, X.; Wang, H. The Distinct Role of ADAM17 in APP Proteolysis and Microglial Activation Related to Alzheimer’s Disease. Cell. Mol. Neurobiol. 2015, 36, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Park, Y.; Chun, Y.S.; Cha, J.W.; Kwon, H.C.; Oh, M.S.; Chung, S.; Yang, H.O. Effect of Lycoris chejuensis and its active components on experimental models of Alzheimer’s disease. J. Agric. Food Chem. 2015, 63, 6979–6988. [Google Scholar] [CrossRef] [PubMed]
- Chun, Y.S.; Zhang, L.; Li, H.; Park, Y.; Chung, S.; Yang, H.O. 7-Deoxy-trans-dihydronarciclasine Reduces β-Amyloid and Ameliorates Memory Impairment in a Transgenic Model of Alzheimer’s Disease. Mol. Neurobiol. 2018, 55, 8953–8964. [Google Scholar] [CrossRef] [PubMed]
- Citron, M.; Oltersdorf, T.; Haass, C.; McConlogue, L.; Hung, A.Y.; Seubert, P.; Vigo-Pelfrey, C.; Lieberburg, I.; Selkoe, D.J. Mutation of the β-amyloid precursor protein in familial Alzheimer’s disease increases β-protein production. Nature 1992, 360, 672–674. [Google Scholar] [CrossRef] [PubMed]
- Pruessmeyer, J.; Ludwig, A. The good, the bad and the ugly substrates for ADAM10 and ADAM17 in brain pathology, inflammation and cancer. Semin. Cell Dev. Boil. 2009, 20, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Pei, G. Visualization of Alzheimer’s Disease Related α-/β-/γ-Secretase Ternary Complex by Bimolecular Fluorescence Complementation Based Fluorescence Resonance Energy Transfer. Front. Mol. Neurosci. 2018, 11, 431. [Google Scholar] [CrossRef]
- Seals, D.F.; Courtneidge, S.A. The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev. 2003, 17, 7–30. [Google Scholar] [CrossRef]
- Mattson, M.P. Cellular actions of β-amyloid precursor protein and its soluble and fibrillogenic derivatives. Physiol. Rev. 1997, 77, 1081–1132. [Google Scholar] [CrossRef]
- Kögel, N.; Deller, T.; Behl, C. Roles of amyloid precursor protein family members in neuroprotection, stress signaling and aging. Exp. Brain Res. 2011, 217, 471–479. [Google Scholar] [CrossRef]
- Guo, Q.; Robinson, N.; Mattson, M.P. Secreted β-amyloid precursor protein counteracts the proapoptotic action of mutant presenilin-1 by activation of NF-kappaB and stabilization of calcium homeostasis. J. Boil. Chem. 1998, 273, 12341–12351. [Google Scholar] [CrossRef]
- Isacson, O. Alzheimer’s disease and Down’s syndrome: roles of APP, trophic factors and ACh. Trends Neurosci. 2002, 25, 79–84. [Google Scholar] [CrossRef]
- Postina, R. A Closer Look at α-Secretase. Curr. Alzheimer Res. 2008, 5, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Arduise, C.; Abache, T.; Li, L.; Billard, M.; Chabanon, A.; Ludwig, A.; Mauduit, P.; Boucheix, C.; Rubinstein, E.; Le Naour, F. Tetraspanins regulate ADAM10-mediated cleavage of TNF-α and epidermal growth factor. J. Immunol. 2008, 181, 7002–7013. [Google Scholar] [CrossRef] [PubMed]
- Horiuchi, K.; Le Gall, S.; Schulte, M.; Yamaguchi, T.; Reiss, K.; Murphy, G.; Toyama, Y.; Hartmann, D.; Saftig, P.; Blobel, C.P. Substrate Selectivity of Epidermal Growth Factor-Receptor Ligand Sheddases and their Regulation by Phorbol Esters and Calcium Influx. Mol. Boil. Cell 2007, 18, 176–188. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Gu, M.Y.; Zhang, L.J.; Jeon, H.J.; Cho, Y.B.; Yang, H.O. 7-Deoxy-trans-dihydronarciclasine isolated from Lycoris chejuensis inhibits neuroinflammation in experimental models. J. Agric. Food Chem. 2019, 67, 9796–9804. [Google Scholar] [CrossRef] [PubMed]
- Karthick, C.; Periyasamy, S.; Jayachandran, K.S.; Anusuyadevi, M. Intrahippocampal Administration of Ibotenic Acid Induced Cholinergic Dysfunction via NR2A/NR2B Expression: Implications of Resveratrol against Alzheimer Disease Pathophysiology. Front. Mol. Neurosci. 2016, 9, 555. [Google Scholar] [CrossRef]
- Sathya, M.; Moorthi, P.; Premkumar, P.; Kandasamy, M.; Jayachandran, K.S.; Anusuyadevi, M. Resveratrol Intervenes Cholesterol- and Isoprenoid-Mediated Amyloidogenic Processing of AβPP in Familial Alzheimer’s Disease. J. Alzheimer’s Dis. 2017, 60, S3–S23. [Google Scholar] [CrossRef]
- Menon, V.P.; Sudheer, A.R. Antioxidant and anti-inflammatory properties of curcumin. Kidney Dev. Dis. 2007, 595, 105–125. [Google Scholar]
- Narasingappa, R.B.; Javagal, M.R.; Pullabhatla, S.; Htoo, H.H.; Rao, J.K.; Hernandez, J.-F.; Govitrapong, P.; Vincent, B. Corrigendum to “Activation of α-secretase by curcumin-aminoacid conjugates” [Biochem. Biophys. Res. Commun. 424 (2012) 691–696]. Biochem. Biophys. Res. Commun. 2012, 426, 665. [Google Scholar] [CrossRef]
- Tippmann, F.; Hundt, J.; Schneider, A.; Endres, K.; Fahrenholz, F. Up-regulation of the alpha-secretase ADAM10 by retinoic acid receptors and acitretin. FASEB J. 2009, 23, 1643–1654. [Google Scholar] [CrossRef]
- Endres, K.; Fahrenholz, F.; Lotz, J.; Hiemke, C.; Teipel, S.; Lieb, K.; Tuscher, O.; Fellgiebel, A. Increased CSFAPPs-alpha levels in patients with Alzheimer disease treated with acitretin. Neurology 2014, 83, 1930–1935. [Google Scholar] [CrossRef] [PubMed]
- Brummer, T.; Müller, S.A.; Pan-Montojo, F.; Yoshida, F.; Fellgiebel, A.; Tomita, T.; Endres, K.; Lichtenthaler, S.F. Nr CAM is a marker for substrate-selective activation of ADAM 10 in Alzheimer’s disease. EMBO Mol. Med. 2019, 11, e9695. [Google Scholar] [CrossRef] [PubMed]
- Aragão, A.Z.B.; Nogueira, M.L.C.; Granato, D.C.; Simabuco, F.M.; Honorato, R.V.; Hoffman, Z.; Yokoo, S.; Laurindo, F.R.M.; Squina, F.M.; Zeri, A.C.M.; et al. Identification of Novel Interaction between ADAM17 (a Disintegrin and Metalloprotease 17) and Thioredoxin-1. J. Boil. Chem. 2012, 287, 43071–43082. [Google Scholar] [CrossRef] [PubMed]
- Peiretti, F.; Deprez-Beauclair, P.; Bonardo, B.; Aubert, H.; Juhan-Vague, I.; Nalbone, G. Identification of SAP97 as an intracellular binding partner of TACE. J. Cell Sci. 2003, 116, 1949–1957. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chun, Y.S.; Cho, Y.Y.; Kwon, O.H.; Zhao, D.; Yang, H.O.; Chung, S. Substrate-Specific Activation of α-Secretase by 7-Deoxy-Trans-Dihydronarciclasine Increases Non-Amyloidogenic Processing of β-Amyloid Protein Precursor. Molecules 2020, 25, 646. https://doi.org/10.3390/molecules25030646
Chun YS, Cho YY, Kwon OH, Zhao D, Yang HO, Chung S. Substrate-Specific Activation of α-Secretase by 7-Deoxy-Trans-Dihydronarciclasine Increases Non-Amyloidogenic Processing of β-Amyloid Protein Precursor. Molecules. 2020; 25(3):646. https://doi.org/10.3390/molecules25030646
Chicago/Turabian StyleChun, Yoon Sun, Yoon Young Cho, Oh Hoon Kwon, Dong Zhao, Hyun Ok Yang, and Sungkwon Chung. 2020. "Substrate-Specific Activation of α-Secretase by 7-Deoxy-Trans-Dihydronarciclasine Increases Non-Amyloidogenic Processing of β-Amyloid Protein Precursor" Molecules 25, no. 3: 646. https://doi.org/10.3390/molecules25030646
APA StyleChun, Y. S., Cho, Y. Y., Kwon, O. H., Zhao, D., Yang, H. O., & Chung, S. (2020). Substrate-Specific Activation of α-Secretase by 7-Deoxy-Trans-Dihydronarciclasine Increases Non-Amyloidogenic Processing of β-Amyloid Protein Precursor. Molecules, 25(3), 646. https://doi.org/10.3390/molecules25030646





