Identification and Characterization of pantocin wh-1, a Novel Cyclic Polypeptide Produced by Pantoea dispersa W18
Abstract
1. Introduction
2. Results
2.1. Isolation and Taxonomical Identification of the Natural Product Producing Strain
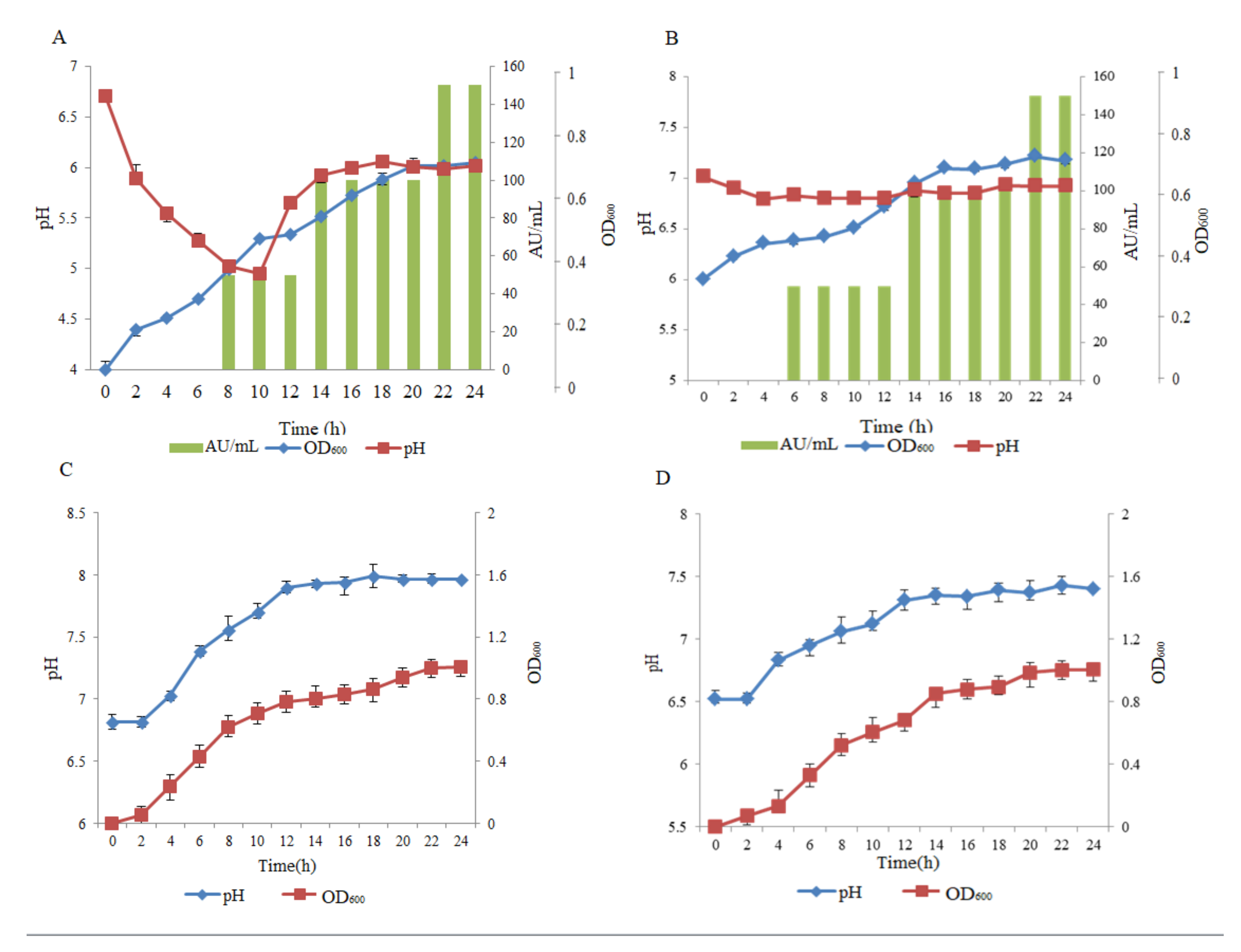
2.2. The Production of Antimicrobial Peptide in Different Media and Physical Conditions
2.3. Purification of pantocin wh-1 and Estimation of the Molecular Weight
2.4. Fragmentation of pantocin wh-1
2.5. Effect of Various Enzymes, Heat, and pH on the Antibacterial Activity of pantocin wh-1
2.6. Antibacterial Spectrum and Time Kill Curve
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. Identification of the pantocin wh-1 Producing Strain
4.3. Purification of pantocin wh-1
4.4. Mass Spectrometry Analysis
4.5. Fragmentation of pantocin wh-1
4.6. Effect of Various Enzymes, Heat, and pH on the Bactericidal Activity of pantocin wh-1
4.7. Detection of Antibacterial Activity
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ortega, M.A.; van der Donk, W.A. New Insights into the Biosynthetic Logic of Ribosomally Synthesized and Post-translationally Modified Peptide Natural Products. Cell Chem. Biol. 2016, 23, 31–44. [Google Scholar] [CrossRef]
- Silver, L.L. Natural products as a source of drug leads to overcome drug resistance. Future Microbiol. 2015, 10, 1711–1718. [Google Scholar] [CrossRef] [PubMed]
- Rutledge, P.J.; Challis, G.L. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol. 2015, 13, 509. [Google Scholar] [CrossRef] [PubMed]
- Milshteyn, A.; Schneider, J.; Brady, S. Mining the Metabiome: Identifying Novel Natural Products from Microbial Communities. Chem. Biol. 2014, 21, 1211–1223. [Google Scholar] [CrossRef] [PubMed]
- Gavini, F.; Mergaert, J.; Beji, A.; Mielcarek, C.; Izard, D.; Kersters, K.; Deley, J. Transfer of Enterobacter-Agglomerans (Beijerinck 1888) Ewing and Fife 1972 to Pantoea Gen-Nov as Pantoea-Agglomerans Comb Nov and Description of Pantoea-Dispersa Sp-Nov. Int. J. Syst. Bacteriol. 1989, 39, 337–345. [Google Scholar] [CrossRef]
- Riggle, J.H.; Klos, E.J. Relationship of Erwinia-Herbicola to Erwinia-Amylovora. Can J Bot. 1972, 50, 1077. [Google Scholar] [CrossRef]
- Chatterjee, A.K.; Gibbins, L.N.; Carpenter, J.A. Some observations on the physiology of Erwinia herbicola and its possible implication as a factor antagonistic to Erwinia amylovora in the “fire-blight” syndrome. Can. J. Microbiol. 1969, 15, 640–642. [Google Scholar] [CrossRef] [PubMed]
- Bisi, D.C.; Lampe, D.J. Secretion of anti-Plasmodium effector proteins from a natural Pantoea agglomerans isolate by using PelB and HlyA secretion signals. Appl. Env. Microbiol. 2011, 77, 4669–4675. [Google Scholar] [CrossRef]
- Pusey, P.L.; Stockwell, V.O.; Reardon, C.L.; Smits, T.H.; Duffy, B. Antibiosis activity of Pantoea agglomerans biocontrol strain E325 against Erwinia amylovora on apple flower stigmas. Phytopathology 2011, 101, 1234–1241. [Google Scholar] [CrossRef]
- Brady, S.F.; Wright, S.A.; Lee, J.C.; Sutton, A.E.; Zumoff, C.H.; Wodzinski, R.S.; Beer, S.V.; Clardy, J. Pantocin B, an antibiotic from Erwinia herbicola discovered by heterologous expression of cloned genes. J. Am. Chem. Soc. 1999, 121, 11912–11913. [Google Scholar] [CrossRef]
- Jin, M.; Fischbach, M.A.; Clardy, J. A biosynthetic gene cluster for the acetyl-CoA carboxylase inhibitor andrimid. J. Am. Chem. Soc. 2006, 128, 10660–10661. [Google Scholar] [CrossRef] [PubMed]
- Sammer, U.F.; Volksch, B.; Mollmann, U.; Schmidtke, M.; Spiteller, P.; Spiteller, M.; Spiteller, D. 2-Amino-3-(Oxirane-2,3-Dicarboxamido)-Propanoyl-Valine, an Effective Peptide Antibiotic from the Epiphyte Pantoea agglomerans 48b/90. Appl. Env. Microb. 2009, 75, 7710–7717. [Google Scholar] [CrossRef] [PubMed]
- Giddens, S.R.; Feng, Y.J.; Mahanty, H.K. Characterization of a novel phenazine antibiotic gene cluster in Erwinia herbicola Eh1087. Mol. Microbiol. 2002, 45, 769–783. [Google Scholar] [CrossRef] [PubMed]
- Aydin, M.; Lucht, N.; Konig, W.A.; Lupp, R.; Jung, G.; Winkelmann, G. Structure Elucidation of the Peptide Antibiotics Herbicolin-a and Herbicolin-B. Liebigs. Ann. Chem. 1985, 1985, 2285–2300. [Google Scholar] [CrossRef]
- Gavrish, E.; Sit, C.S.; Cao, S.; Kandror, O.; Spoering, A.; Peoples, A.; Ling, L.; Fetterman, A.; Hughes, D.; Bissell, A.; et al. Lassomycin, a ribosomally synthesized cyclic peptide, kills mycobacterium tuberculosis by targeting the ATP-dependent protease ClpC1P1P2. Chem. Biol. 2014, 21, 509–518. [Google Scholar] [CrossRef]
- Parish, T. Targeting mycobacterial proteolytic complexes with natural products. Chem. Biol. 2014, 21, 437–438. [Google Scholar] [CrossRef][Green Version]
- Gao, W.; Kim, J.Y.; Anderson, J.R.; Akopian, T.; Hong, S.; Jin, Y.Y.; Kandror, O.; Kim, J.W.; Lee, I.A.; Lee, S.Y.; et al. The cyclic peptide ecumicin targeting ClpC1 is active against Mycobacterium tuberculosis in vivo. Antimicrob. Agents Ch. 2015, 59, 880–889. [Google Scholar] [CrossRef]
- Sass, P.; Josten, M.; Famulla, K.; Schiffer, G.; Sahl, H.G.; Hamoen, L.; Brotz-Oesterhelt, H. Antibiotic acyldepsipeptides activate ClpP peptidase to degrade the cell division protein FtsZ. Proc. Natl. Acad. Sci. USA 2011, 108, 17474–17479. [Google Scholar] [CrossRef]
- Ollinger, J.; O’Malley, T.; Kesicki, E.A.; Odingo, J.; Parish, T. Validation of the Essential ClpP Protease in Mycobacterium tuberculosis as a Novel Drug Target. J. Bacteriol. 2012, 194, 663–668. [Google Scholar] [CrossRef]
- Hartkoorn, R.C.; Sala, C.; Neres, J.; Pojer, F.; Magnet, S.; Mukherjee, R.; Uplekar, S.; Boy-Rottger, S.; Altmann, K.H.; Cole, S.T. Towards a new tuberculosis drug: Pyridomycin-nature’s isoniazid. Embo. Mol. Med. 2012, 4, 1032–1042. [Google Scholar] [CrossRef]
- Kremer, L.; Douglas, J.D.; Baulard, A.R.; Morehouse, C.; Guy, M.R.; Alland, D.; Dover, L.G.; Lakey, J.H.; Jacobs, W.R.; Brennan, P.J.; et al. Thiolactomycin and related analogues as novel anti-mycobacterial agents targeting KasA and KasB condensing enzymes in Mycobacterium tuberculosis. J. Biol. Chem. 2000, 275, 16857–16864. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, E.K.; Riwanto, M.; Sambandamurthy, V.; Roggo, S.; Miault, C.; Zwingelstein, C.; Krastel, P.; Noble, C.; Beer, D.; Rao, S.P.S.; et al. The Natural Product Cyclomarin Kills Mycobacterium Tuberculosis by Targeting the ClpC1 Subunit of the Caseinolytic Protease. Angew. Chem. Int. Ed. 2011, 50, 5889–5891. [Google Scholar] [CrossRef] [PubMed]
- Vasudevan, D.; Rao, S.P.S.; Noble, C.G. Structural Basis of Mycobacterial Inhibition by Cyclomarin A. J. Biol. Chem. 2013, 288, 30883–30891. [Google Scholar] [CrossRef] [PubMed]
- Ling, L.L.; Schneider, T.; Peoples, A.J.; Spoering, A.L.; Engels, I.; Conlon, B.P.; Mueller, A.; Schaberle, T.F.; Hughes, D.E.; Epstein, S.; et al. A new antibiotic kills pathogens without detectable resistance. Nature 2015, 517, 455. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Huang, H.; Xie, Y.; Liu, Z.; Zhao, J.; Zhang, C.; Jia, Y.; Zhang, Y.; Zhang, H.; Zhang, T.; et al. Biosynthesis of ilamycins featuring unusual building blocks and engineered production of enhanced anti-tuberculosis agents. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Boglárka, S.; Eniko, W.; László, M.; Tibor, M. Evaluation of the Biolog system for the identification of certain closely related Pasteurella species. Diagn. Microbiol. Infect. Dis. 2011, 71, 6–11. [Google Scholar]
- Masuda, Y.; Ono, H.; Kitagawa, H.; Ito, H.; Mu, F.; Sawa, N.; Zendo, T.; Sonomoto, K. Identification and characterization of leucocyclicin Q, a novel cyclic bacteriocin produced by Leuconostoc mesenteroides TK41401. Appl. Environ. Microbiol. 2011, 77, 8164–8170. [Google Scholar] [CrossRef]
- Detlev, S.; Anja, R.; Martin, S.; Peter, H.; Jochen, F.; Armin, H. A novel MALDI LIFT-TOF/TOF mass spectrometer for proteomics. Anal. Bioanal. Chem. 2003, 376, 952–965. [Google Scholar]
- Romy, S.; Joachim, V.; Anto, B.; Zhiyuan, W.; Yueqiu, H.; Kristin, D.; Torsten, S.; Stefanie, H.; Peter, L.; Rainer, B. Amylocyclicin, a novel circular bacteriocin produced by Bacillus amyloliquefaciens FZB42. J. Bacteriol. 2014, 196, 1842–1852. [Google Scholar]
- Batdorj, B.; Dalgalarrondo, M.; Choiset, Y.; Pedroche, J.; Métro, F.; Prévost, H.; J-M, C.; Haertlé, T. Purification and characterization of two bacteriocins produced by lactic acid bacteria isolated from Mongolian airag. J. Appl. Microbiol. 2010, 101, 837–848. [Google Scholar] [CrossRef]
- Lash, B.W.; Mysliwiec, T.H.; Gourama, H. Detection and partial characterization of a broad-range bacteriocin produced by Lactobacillus plantarum (ATCC 8014). Food Microbiol. 2005, 22, 199–204. [Google Scholar] [CrossRef]
- Miao, J.Y.; Guo, H.X.; Ou, Y.W.; Liu, G.; Fang, X.; Liao, Z.L.; Ke, C.; Chen, Y.J.; Zhao, L.C.; Cao, Y. Purification and characterization of bacteriocin F1, a novel bacteriocin produced by Lactobacillus paracasei subsp tolerans FX-6 from Tibetan kefir, a traditional fermented milk from Tibet, China. Food Control. 2014, 42, 48–53. [Google Scholar] [CrossRef]
- Tianyu, Z.; Si-Yang, L.; Nuermberger, E.L. Autoluminescent Mycobacterium tuberculosis for rapid, real-time, non-invasive assessment of drug and vaccine efficacy. Plos ONE 2012, 7, e29774. [Google Scholar]
- Zhang, T.Y.; Li, S.Y.; Converse, P.J.; Almeida, D.V.; Grosset, J.H.; Nuermberger, E.L. Using Bioluminescence To Monitor Treatment Response in Real Time in Mice with Mycobacterium ulcerans Infection. Antimicrob. Agents Ch. 2011, 55, 56–61. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds pantocin wh-1 are available from the authors. |



| Strain | Name | Structure | Ref. |
|---|---|---|---|
| P. agglomerans EH318 | pantocin B | (R)-N-[((S)-2-amino-propanoylamino)-methyl]-2-methanesulfonyl-succinamic acid | [10] |
| pantocin A | unknown | ||
| P. agglomerans Eh335 | andrimid | 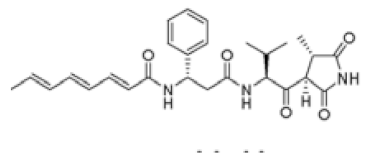 | [11] |
| P. agglomerans C9-1/48b/90 | 2-amino-3-(oxirane-2, 3-dicarboxamido)-propanoyl-valine | [12] | |
| P. agglomerans C9-1 | herbicolin O | unknown | |
| herbicolin I | unknown | ||
| P. agglomerans Eh1087 | phenazine | d-alanylgriseoluteic acid (AGA) | [13] |
| E. herbicola 112Y | 112Y | unknown | |
| E. herbicola A 111 | herbicolin A | (R)-3-OH-Cl4-DH-Abu-l-Thr-d-aThr-d-Leu-Gly-d-Glx-Gly-l-Me-aThr-L-Arg | [14] |
| herbicolin B | d-Abu-l-Thr-d-aThr-d-Leu-Gly-d-Gln-Gly-N-Me-l-aThr-l-Arg |
| Method | Vol(ml) | Total Activity (AU) 1 | Yield (%) | Total Protein (mg) 2 | Purification (fold) |
|---|---|---|---|---|---|
| Supernatant | 2000 | 1.60 × 106 | 100 | 16,520 | 1 |
| SP-Sepharose | 350 | 5 × 105 | 80 | 3537 | 75 |
| Octyl-Sepharose | 120 | 5 × 105 | 80 | 623 | 212 |
| RP-HPLC | 4 | 2.3 × 105 | 23 | 1.52 | 963 |
| Treatment | Molecular Mass of Fragment (Da) | Amino Acid Sequence |
|---|---|---|
| 0.6 M HCl | 851.94 | YVFGYGF |
| BNPS-skatole | 1163.3 | FGYGFNCAVW |
| Trypsin | 848.04 | MWCYVF |
| 1190.43 | WVMWCYVFG | |
| Merge | 1927.76 | —VMWCYVFGYGFNCAVW— |
| Compound | Source | Strategy | Target | MIC (μM) | Ref. |
|---|---|---|---|---|---|
| Lassomycin | Lentzea kentuckyensis sp. | In situ cultivation and prolonged incubation | ClpC1 | 0.42–1.57 | [15,16] |
| Ecumicin | Nonomuraea sp., strain MJM5123 | High-throughput screening and stress of nutrient limitation | ClpC1 | 0.13–0.3 | [17] |
| Acyldepsipeptides | Streptococcus hawaiiensis | Stress of nutrient limitation | ClpP | 31–65 | [18,19] |
| Pyridomycin | Dactylosporangium fulvum | Genetic manipulation | InhA | 0.72–1.44 | [20] |
| Thiolactomycin | Nocardia sp. | Genetic manipulation | β-ketoacyl-ACP synthase | 92.5 | [21] |
| Cyclomarin A | Streptomyces sp. | Stress of nutrient limitation | ClpC1 | 0.3–2.5 | [22,23] |
| Teixobactin | Eleftheria terrae | ichip | lipid II | 0.125 | [24] |
| Ilamycins | Streptomyces sp. SCSIO16 | Gene inactivation and isotope labeled | — | 0.0098 | [25] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teng, T.; Li, X.; Zhang, L.; Li, Y. Identification and Characterization of pantocin wh-1, a Novel Cyclic Polypeptide Produced by Pantoea dispersa W18. Molecules 2020, 25, 485. https://doi.org/10.3390/molecules25030485
Teng T, Li X, Zhang L, Li Y. Identification and Characterization of pantocin wh-1, a Novel Cyclic Polypeptide Produced by Pantoea dispersa W18. Molecules. 2020; 25(3):485. https://doi.org/10.3390/molecules25030485
Chicago/Turabian StyleTeng, Tieshan, Xianghui Li, Lei Zhang, and Yanzhang Li. 2020. "Identification and Characterization of pantocin wh-1, a Novel Cyclic Polypeptide Produced by Pantoea dispersa W18" Molecules 25, no. 3: 485. https://doi.org/10.3390/molecules25030485
APA StyleTeng, T., Li, X., Zhang, L., & Li, Y. (2020). Identification and Characterization of pantocin wh-1, a Novel Cyclic Polypeptide Produced by Pantoea dispersa W18. Molecules, 25(3), 485. https://doi.org/10.3390/molecules25030485







