Chasing Particularities of Guanine- and Cytosine-Rich DNA Strands
Abstract
1. Introduction
2. Results
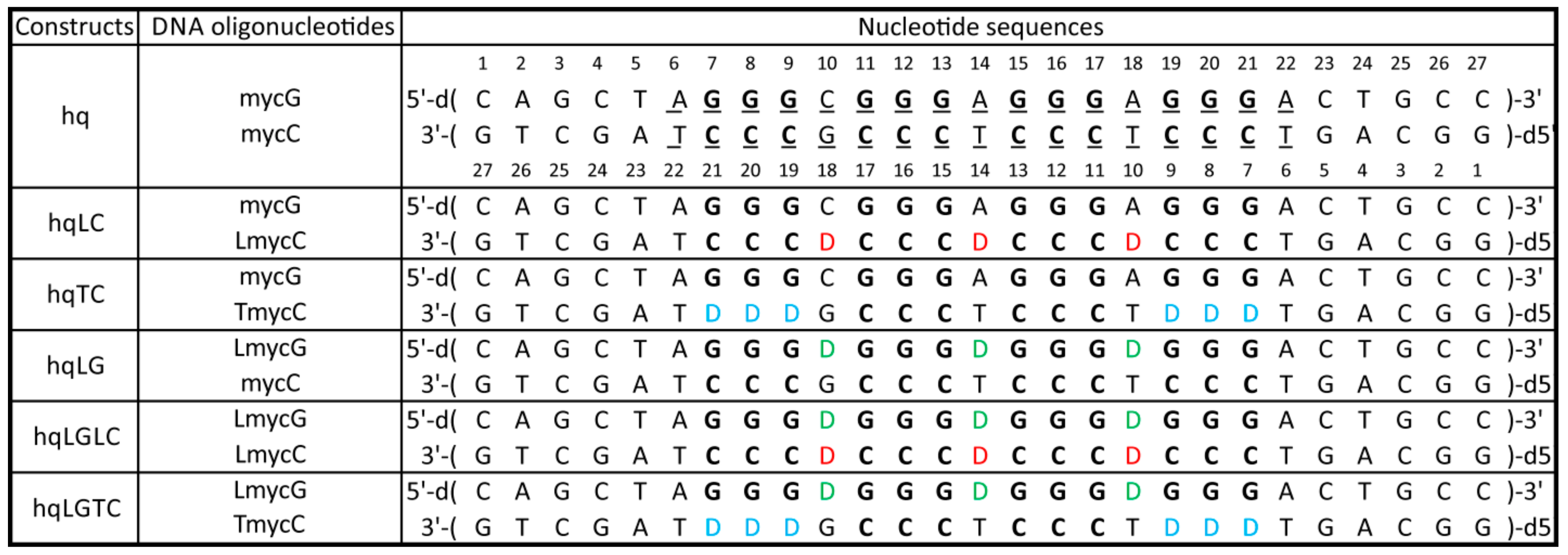
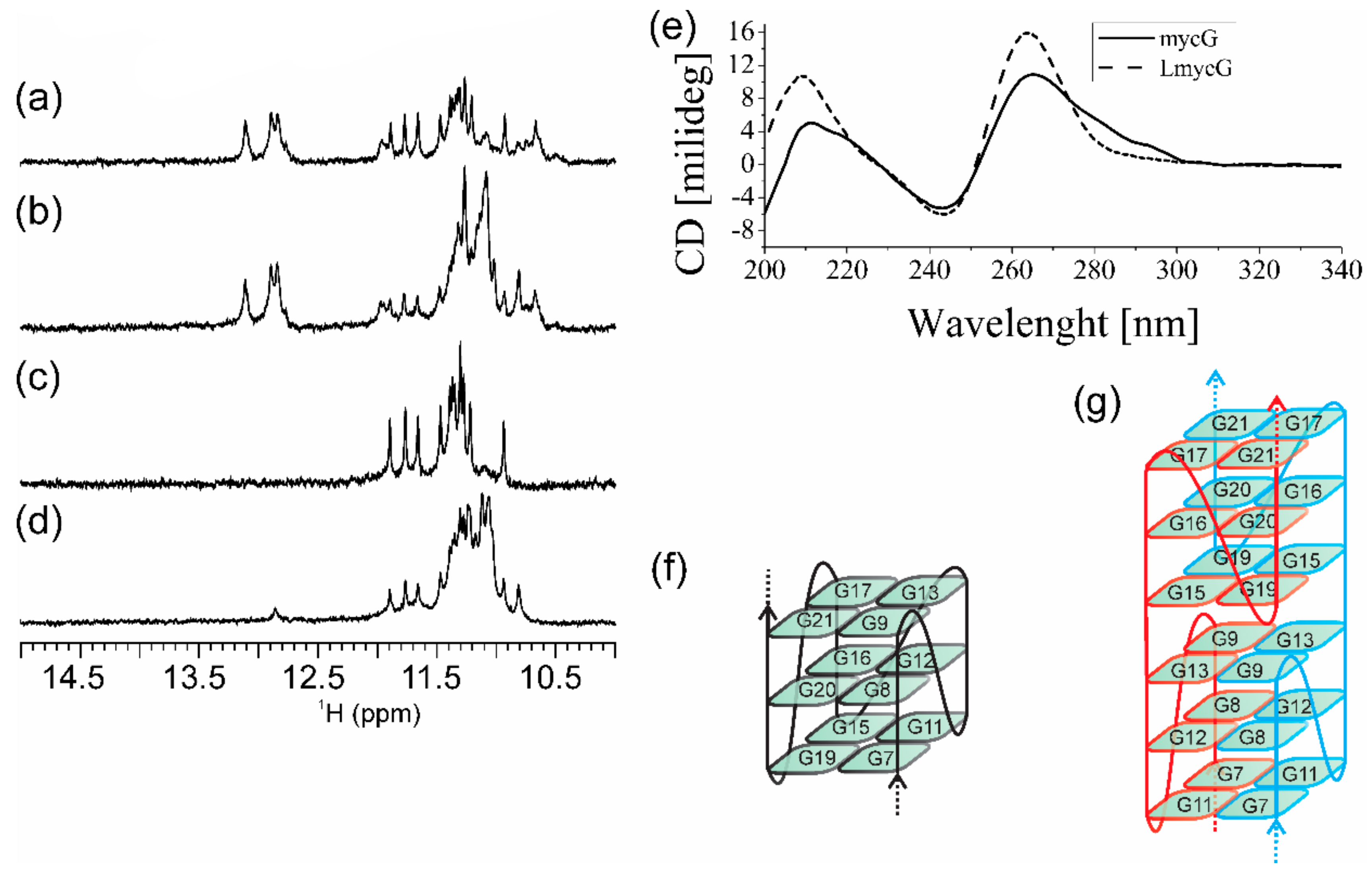
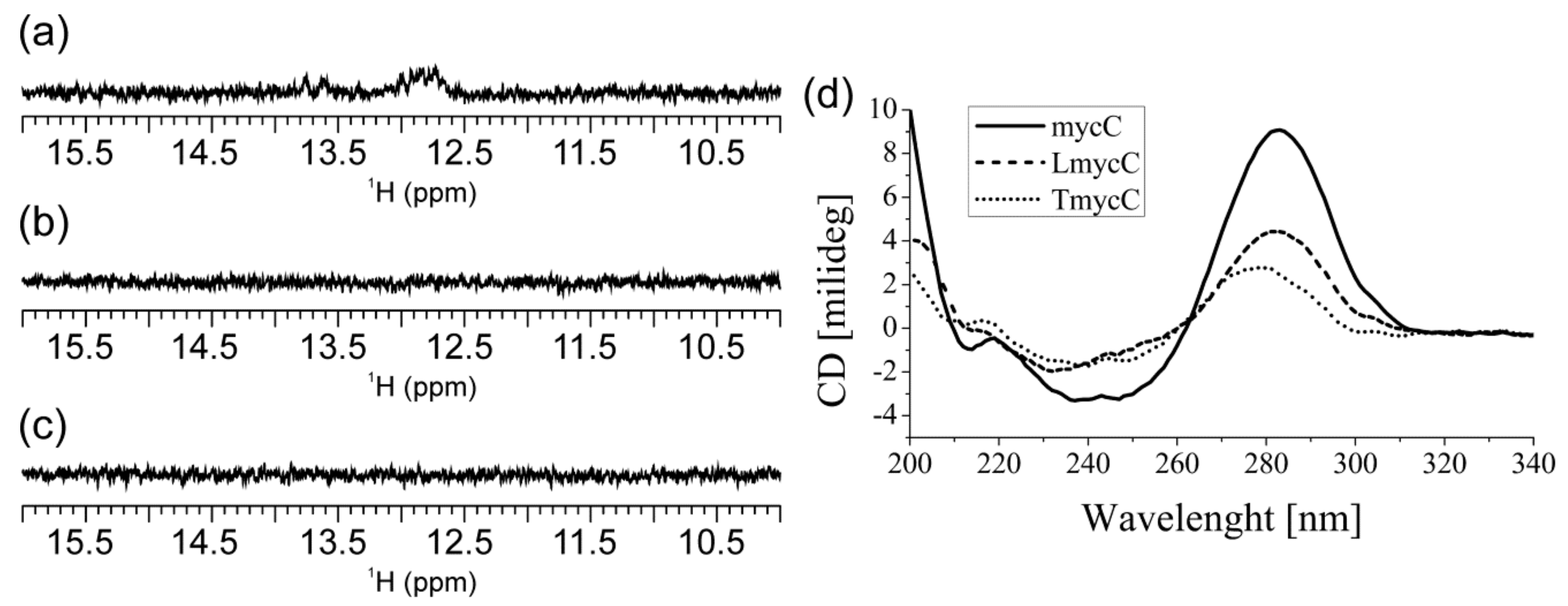
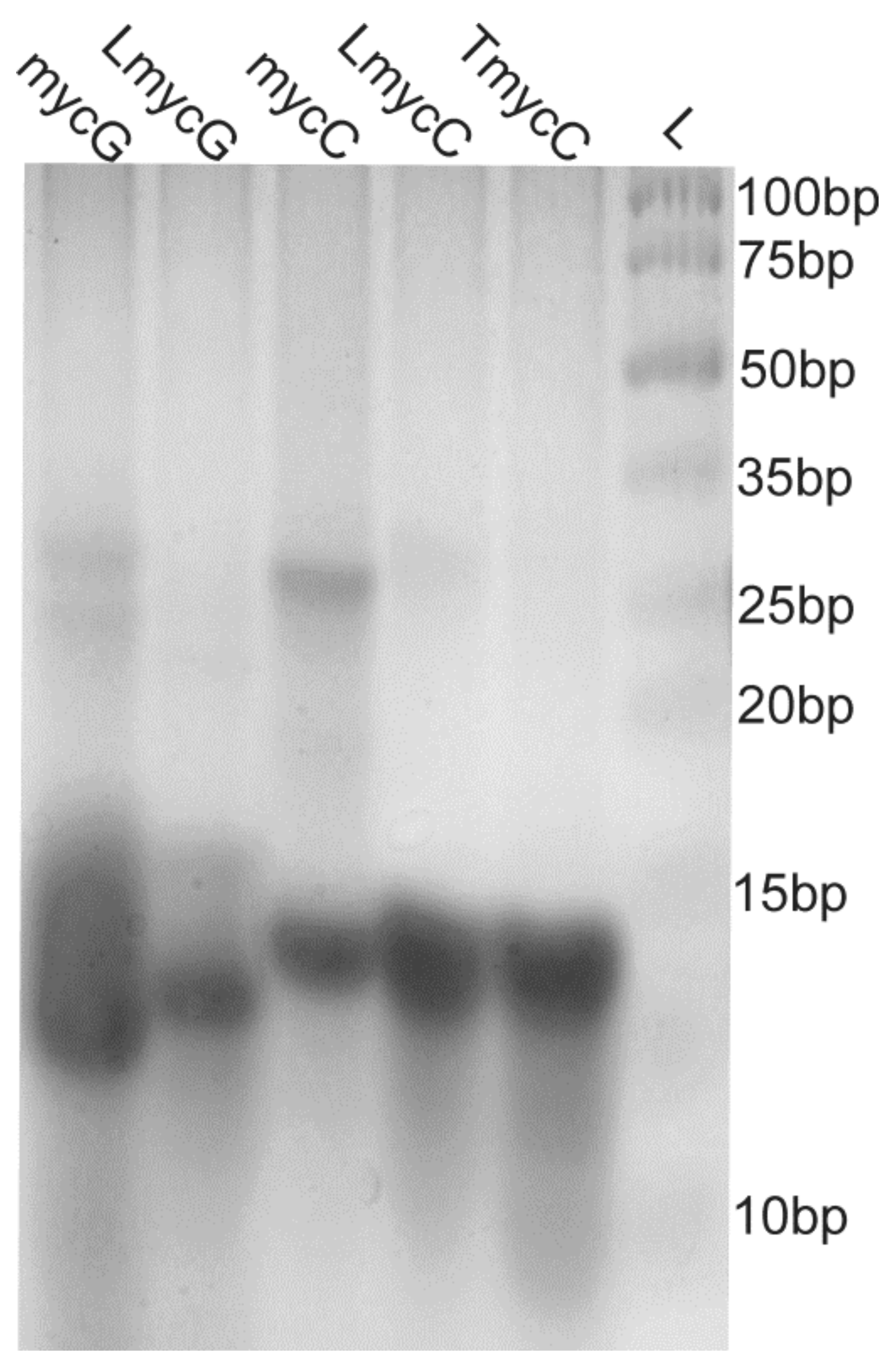
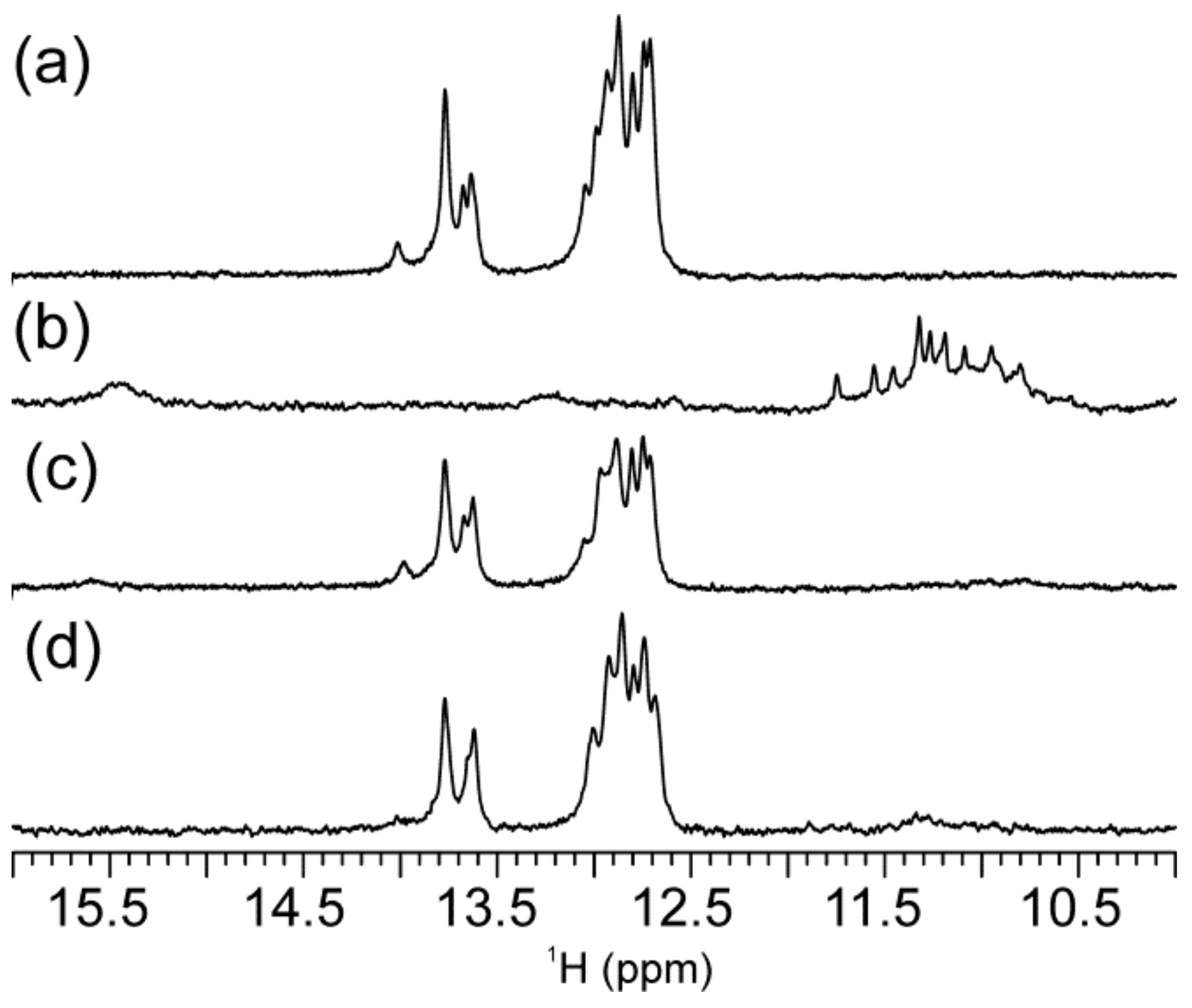
2.1. Abasic Loop Residues Aggravate i-Motif, but Promote G-Quadruplex Formation
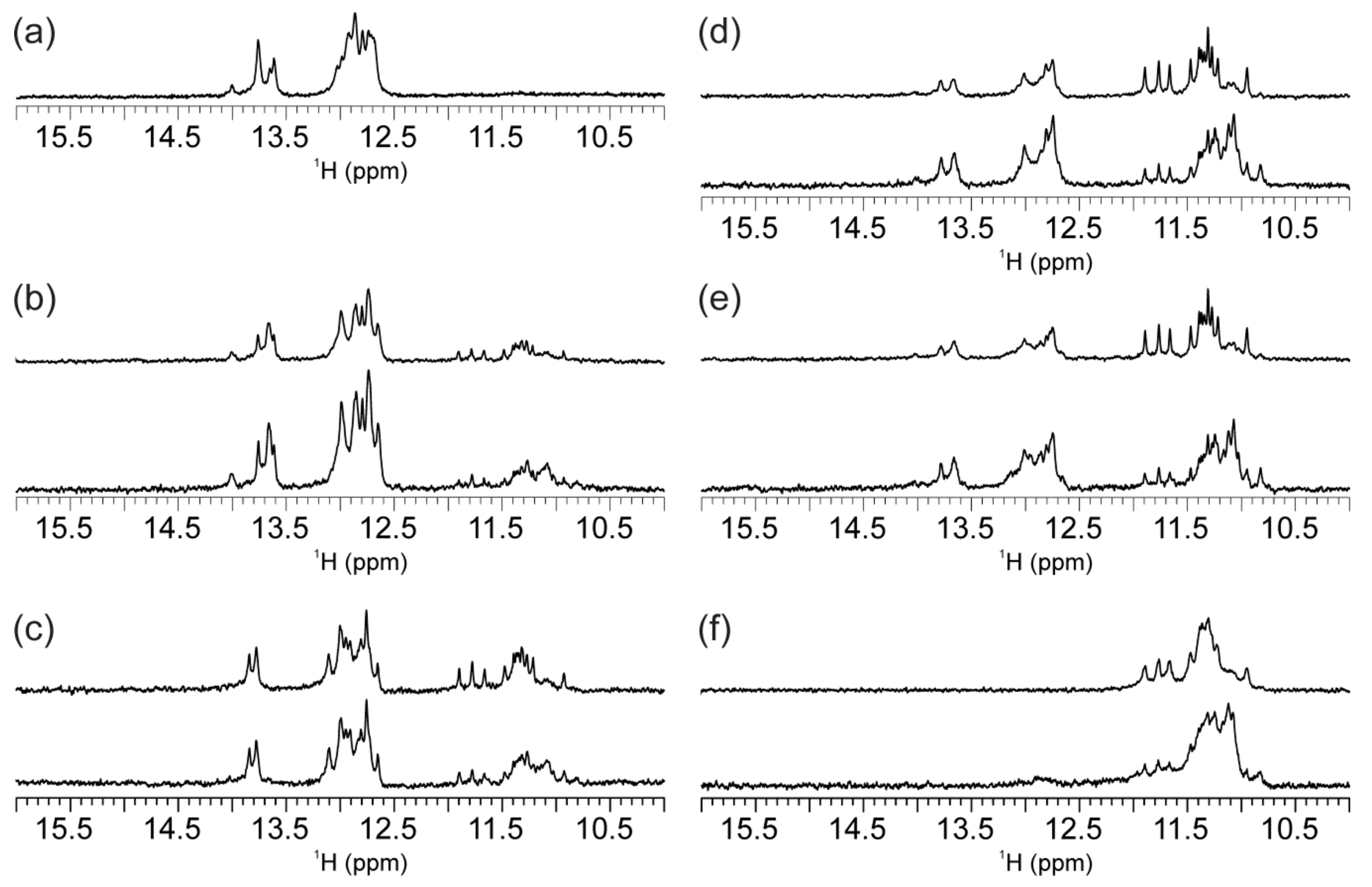
2.2. Individual Oligonucleotide Features Become Apparent Immediately upon Hampering Hybridization between G- and C-rich DNA Strands
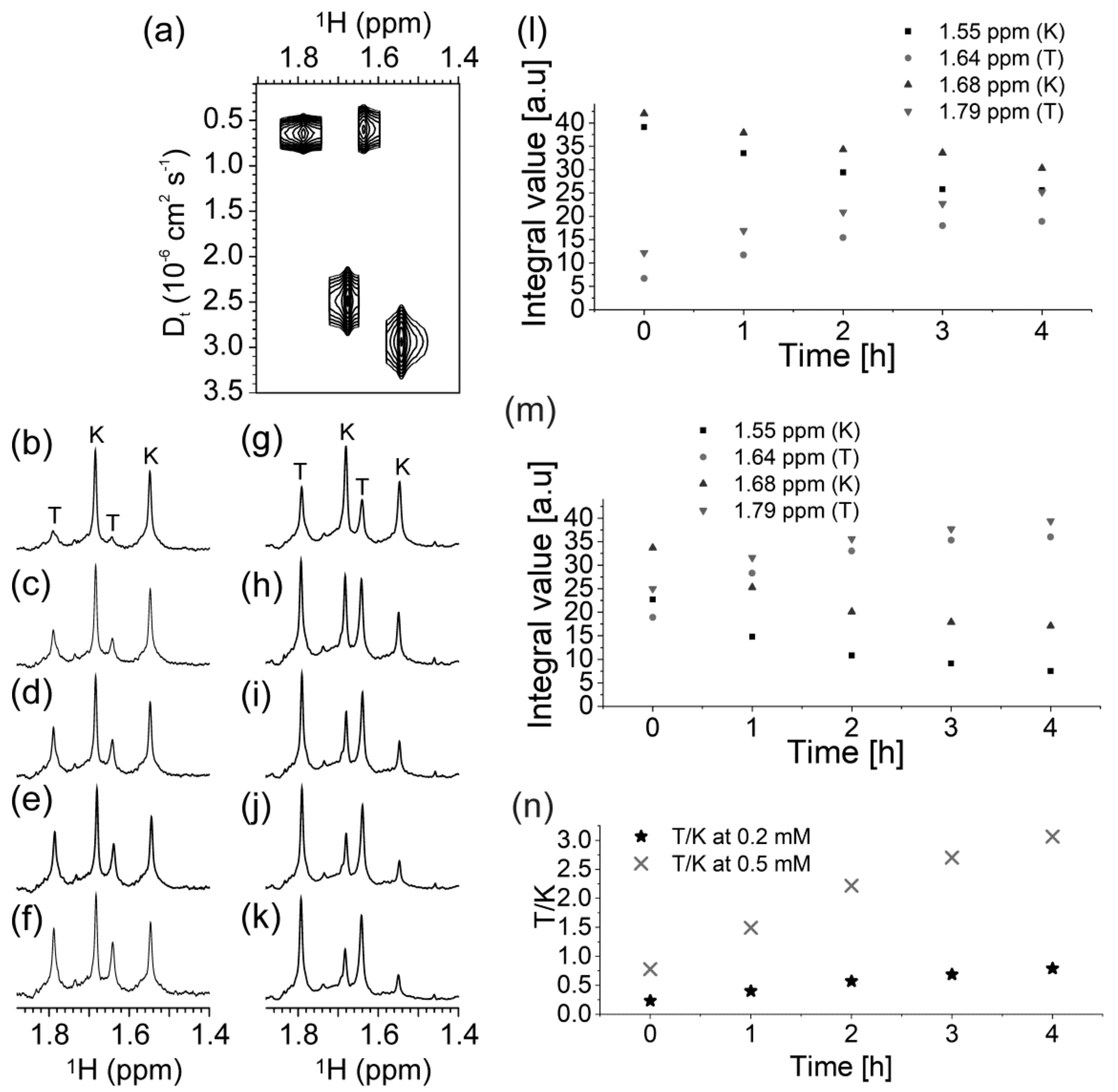
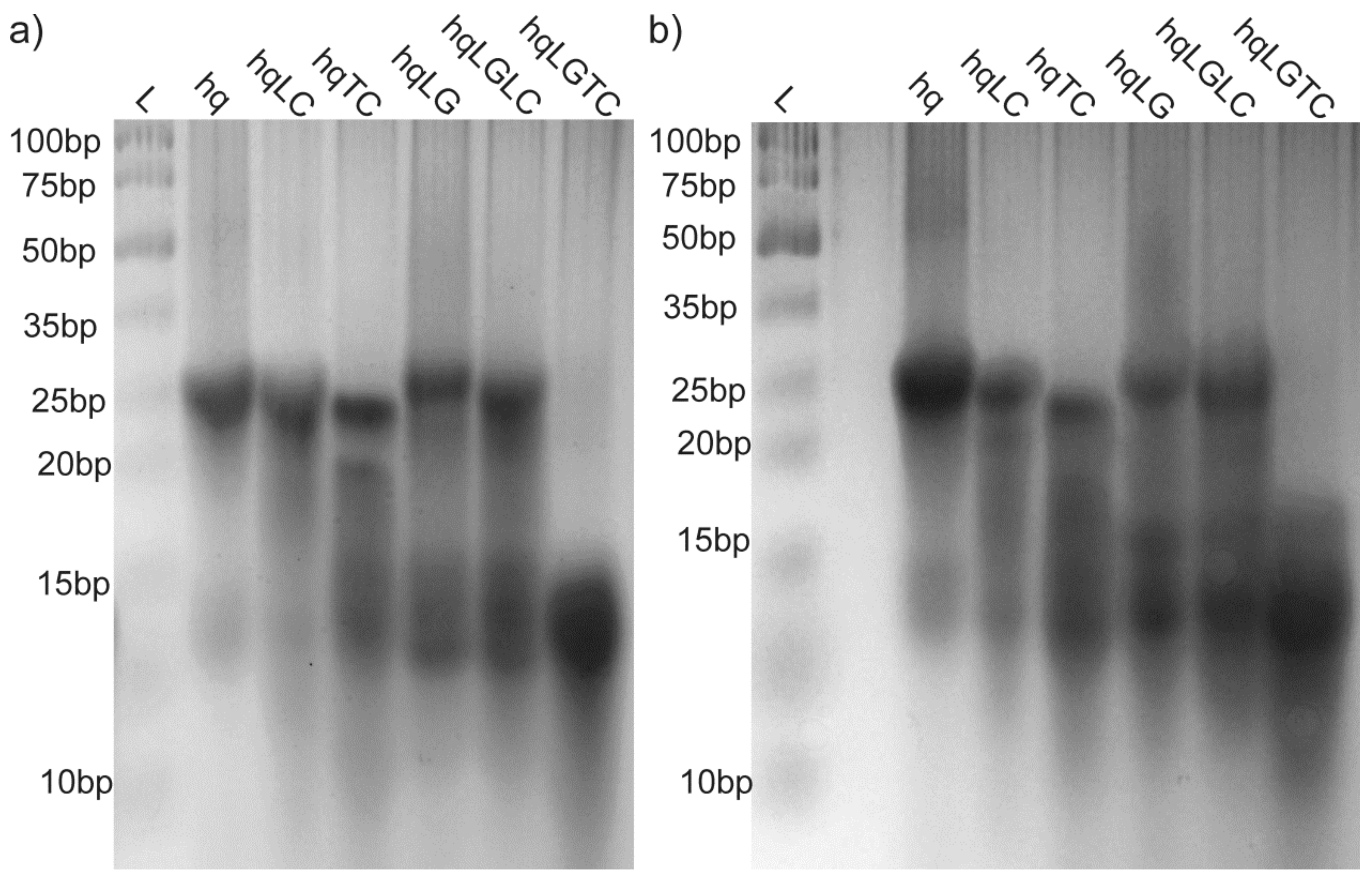
2.3. Presence of Complementary C-Rich Strand Does Not Preclude Transformation of Intra- to Intermolecular G-Quadruplex Species
3. Discussion
4. Materials and Methods
4.1. Oligonucleotides
4.2. NMR Spectroscopy
4.3. CD Spectroscopy
4.4. Native PAGE
4.5 UV Spectroscopy
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Choi, J.; Majima, T. Conformational changes of non-B DNA. Chem. Soc. Rev. 2011, 40, 5893–5909. [Google Scholar] [CrossRef]
- Saini, N.; Zhang, Y.; Usdin, K.; Lobachev, K.S. When secondary comes first-the importance of non-canonical DNA structures. Biochimie 2013, 95, 117–123. [Google Scholar] [CrossRef]
- Sugimoto, N. Noncanonical structures and their thermodynamics of DNA and RNA under molecular crowding: beyond the Watson-Crick double helix. Int. Rev. Cel. Mol. Biol. 2014, 307, 205–273. [Google Scholar]
- Lipps, H.J.; Rhodes, D. G-quadruplex structures: in vivo evidence and function. Trends Cell. Biol. 2009, 19, 414–422. [Google Scholar] [CrossRef] [PubMed]
- Kendrick, S.; Hurley, L.H. The role of G-quadruplex/i-motif secondary structures as cis-acting regulatory elements. Pure Appl. Chem. 2010, 82, 1609–1621. [Google Scholar] [CrossRef] [PubMed]
- Hud, N.V.; Plavec, J. The Role of Cations in Determing Quadruplex Structure and Stability. In Quadruplex Nucleic Acids; Neidle, S., Balasubramanian, S., Eds.; The Royal Society of Chemistry: Cambridge, UK, 2006; pp. 100–130. [Google Scholar]
- Sen, D.; Gilbert, W. A Sodium-Potassium Switch in the Formation of 4-Stranded G4-DNA. Nature 1990, 344, 410–414. [Google Scholar] [CrossRef]
- Dai, J.X.; Carver, M.; Punchihewa, C.; Jones, R.A.; Yang, D.Z. Structure of the Hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: Insights into structure polymorphism of the human telomeric sequence. Nucleic Acids Res. 2007, 35, 4927–4940. [Google Scholar] [CrossRef]
- Gray, R.D.; Chaires, J.B. Kinetics and mechanism of K(+)- and Na(+)-induced folding of models of human telomeric DNA into G-quadruplex structures. Nucleic Acids Res. 2008, 36, 4191–4203. [Google Scholar] [CrossRef]
- Sket, P.; Plavec, J. Tetramolecular DNA Quadruplexes in Solution: Insights into Structural Diversity and Cation Movement. J. Am. Chem. Soc. 2010, 132, 12724–12732. [Google Scholar] [CrossRef]
- Lin, C.T.; Tseng, T.Y.; Wang, Z.F.; Chang, T.C. Structural Conversion of Intramolecular and Intermolecular G-Quadruplexes of bcl2mid: The Effect of Potassium Concentration and Ion Exchange. J. Phys. Chem. B 2011, 115, 2360–2370. [Google Scholar] [CrossRef]
- Day, H.A.; Wright, E.P.; MacDonald, C.J.; Gates, A.J.; Waller, Z.A. Reversible DNA i-motif to hairpin switching induced by copper(ii) cations. Chem. Commun. 2015, 51, 14099–14102. [Google Scholar] [CrossRef] [PubMed]
- Kovanda, A.; Zalar, M.; Sket, P.; Plavec, J.; Rogelj, B. Anti-sense DNA d(GGCCCC)n expansions in C9ORF72 form i-motifs and protonated hairpins. Sci. Rep. 2015, 5, 17944. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, L.; Lienard, H.; Labourier, E.; Djavaheri-Mergny, M.; Lacoste, J.; Leffers, H.; Tazi, J.; Helene, C.; Mergny, J.L. Identification of two human nuclear proteins that recognise the cytosine-rich strand of human telomeres in vitro. Nucleic Acids Res. 2000, 28, 1564–1575. [Google Scholar] [CrossRef] [PubMed]
- Baumann, P.; Cech, T.R. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 2001, 292, 1171–1175. [Google Scholar] [CrossRef] [PubMed]
- Uribe, D.J.; Guo, K.X.; Shin, Y.J.; Sun, D. Heterogeneous Nuclear Ribonucleoprotein K and Nucleolin as Transcriptional Activators of the Vascular Endothelial Growth Factor Promoter through Interaction with Secondary DNA Structures. Biochemistry 2011, 50, 3796–3806. [Google Scholar] [CrossRef]
- Paeschke, K.; Capra, J.A.; Zakian, V.A. DNA Replication through G-Quadruplex Motifs Is Promoted by the Saccharomyces cerevisiae Pif1 DNA Helicase. Cell 2011, 145, 678–691. [Google Scholar] [CrossRef]
- Wang, F.; Tang, M.L.; Zeng, Z.X.; Wu, R.Y.; Xue, Y.; Hao, Y.H.; Pang, D.W.; Zhao, Y.; Tan, Z. Telomere- and telomerase-interacting protein that unfolds telomere G-quadruplex and promotes telomere extension in mammalian cells. Proc. Natl. Acad. Sci. USA 2012, 109, 20413–20418. [Google Scholar] [CrossRef]
- Sauer, M.; Paeschke, K. G-quadruplex unwinding helicases and their function in vivo. Biochem. Soc. Trans. 2017, 45, 1173–1182. [Google Scholar] [CrossRef]
- Todd, A.K.; Johnston, M.; Neidle, S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005, 33, 2901–2907. [Google Scholar] [CrossRef]
- Brooks, T.A.; Kendrick, S.; Hurley, L. Making sense of G-quadruplex and i-motif functions in oncogene promoters. Febs J. 2010, 277, 3459–3469. [Google Scholar] [CrossRef]
- Maizels, N.; Gray, L.T. The g4 genome. PLoS Genet. 2013, 9, e1003468. [Google Scholar] [CrossRef] [PubMed]
- Murat, P.; Balasubramanian, S. Existence and consequences of G-quadruplex structures in DNA. Curr. Opin. Genet. Dev. 2014, 25, 22–29. [Google Scholar] [CrossRef]
- Brcic, J.; Plavec, J. Solution structure of a DNA quadruplex containing ALS and FTD related GGGGCC repeat stabilized by 8-bromodeoxyguanosine substitution. Nucleic Acids Res. 2015, 43, 8590–8600. [Google Scholar] [CrossRef] [PubMed]
- Kocman, V.; Plavec, J. A tetrahelical DNA fold adopted by tandem repeats of alternating GGG and GCG tracts. Nat. Commun. 2014, 5, 5831. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kumar, N.; Sahoo, B.; Varun, K.A.S.; Maiti, S. Effect of loop length variation on quadruplex-Watson Crick duplex competition. Nucleic Acids Res. 2008, 36, 4433–4442. [Google Scholar] [CrossRef] [PubMed]
- Miura, T.; Thomas, G.J. Structural polymorphism of telomere DNA—Interquadruplex and duplex-quadruplex conversions probed by raman-spectroscopy. Biochemistry 1994, 33, 7848–7856. [Google Scholar] [CrossRef]
- Li, W.; Wu, P.; Ohmichi, T.; Sugimoto, N. Characterization and thermodynamic properties of quadruplex/duplex competition. FEBS Lett. 2002, 526, 77–81. [Google Scholar] [CrossRef]
- Phan, A.T.; Mergny, J.L. Human telomeric DNA: G-quadruplex, i-motif and watson-crick double helix. Nucleic Acids Res. 2002, 30, 4618–4625. [Google Scholar] [CrossRef]
- Ying, L.M.; Green, J.J.; Li, H.T.; Klenerman, D.; Balasubramanian, S. Studies on the structure and dynamics of the human telomeric G quadruplex by single-molecule fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. USA 2003, 100, 14629–14634. [Google Scholar] [CrossRef]
- Li, W.; Miyoshi, D.; Nakano, S.; Sugimoto, N. Structural competition involving G-quadruplex DNA and its complement. Biochemistry 2003, 42, 11736–11744. [Google Scholar] [CrossRef]
- Kumar, N.; Maiti, S. The effect of osmolytes and small molecule on Quadruplex-WC duplex equilibrium: A fluorescence resonance energy transfer study. Nucleic Acids Res. 2005, 33, 6723–6732. [Google Scholar] [CrossRef] [PubMed]
- Jaumot, J.; Eritja, R.; Tauler, R.; Gargallo, R. Resolution of a structural competition involving dimeric G-quadruplex and its C-rich complementary strand. Nucleic Acids Res. 2006, 34, 206–216. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Risitano, A.; Fox, K.R. Stability of intramolecular DNA quadruplexes: Comparison with DNA duplexes. Biochemistry 2003, 42, 6507–6513. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.G.; Xie, X.; Li, P.Z.; Zhao, J.Y.; Huang, L.L.; Zhou, X. Equilibrious Strand Exchange Promoted by DNA Conformational Switching. Sci. Rep. 2013, 3, 1121. [Google Scholar] [CrossRef]
- Rangan, A.; Fedoroff, O.Y.; Hurley, L.H. Induction of duplex to G-quadruplex transition in the c-myc promoter region by a small molecule. J. Biol. Chem. 2001, 276, 4640–4646. [Google Scholar] [CrossRef]
- Kumar, N.; Basundra, R.; Maiti, S. Elevated polyamines induce c-MYC overexpression by perturbing quadruplexWC duplex equilibrium. Nucleic Acids Res. 2009, 37, 3321–3331. [Google Scholar] [CrossRef]
- Zhang, J.L.; Fu, Y.; Zheng, L.; Li, W.; Li, H.; Sun, Q.; Xiao, Y.; Geng, F. Natural isoflavones regulate the quadruplexduplex competition in human telomeric DNA. Nucleic Acids Res. 2009, 37, 2471–2482. [Google Scholar] [CrossRef]
- Zhang, Z.J.; He, X.W.; Yuan, G. Regulation of the equilibrium between G-quadruplex and duplex DNA in promoter of human c-myc oncogene by a pyrene derivative. Int. J. Biol. Macromol. 2011, 49, 1173–1176. [Google Scholar] [CrossRef]
- Shalaby, T.; Fiaschetti, G.; Nagasawa, K.; Shin-ya, K.; Baumgartner, M.; Grotzer, M. G-Quadruplexes as Potential Therapeutic Targets for Embryonal Tumors. Molecules 2013, 18, 12500–12537. [Google Scholar] [CrossRef]
- Monchaud, D.; Teulade-Fichou, M.P. A hitchhiker’s guide to G-quadruplex ligands. Org. Biomol. Chem. 2008, 6, 627–636. [Google Scholar] [CrossRef]
- Monchaud, D.; Granzhan, A.; Saettel, N.; Guedin, A.; Mergny, J.L.; Teulade-Fichou, M.P. “One ring to bind them all”-part I: The efficiency of the macrocyclic scaffold for g-quadruplex DNA recognition. J. Nucl. Acids 2010, 2010, 525862. [Google Scholar] [CrossRef] [PubMed]
- Shirude, P.S.; Okumus, B.; Ying, L.M.; Ha, T.; Balasubramanian, S. Single-molecule conformational analysis of G-quadruplex formation in the promoter DNA duplex of the proto-oncogene C-kit. J. Am. Chem. Soc. 2007, 129, 7484–7485. [Google Scholar] [CrossRef] [PubMed]
- Dhakal, S.; Yu, Z.B.; Konik, R.; Cui, Y.X.; Koirala, D.; Mao, H.B. G-Quadruplex and i-Motif Are Mutually Exclusive in ILPR Double-Stranded DNA. Biophys. J. 2012, 102, 2575–2584. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.S.; Qu, X.G.; Trent, J.O.; Chaires, J.B. Tiny telomere DNA. Nucleic Acids Res. 2002, 30, 2307–2315. [Google Scholar] [CrossRef] [PubMed]
- Dutta, K.; Fujimoto, T.; Inoue, M.; Miyoshi, D.; Sugimoto, N. Development of new functional nanostructures consisting of both DNA duplex and quadruplex. Chem. Commun. 2010, 46, 7772–7774. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Amrane, S.; Korkut, D.N.; Bourdoncle, A.; He, H.Z.; Ma, D.L.; Mergny, J.L. Combination of i-Motif and G-Quadruplex Structures within the Same Strand: Formation and Application. Angew. Chem. Int. Ed. 2013, 52, 7742–7746. [Google Scholar] [CrossRef] [PubMed]
- Endo, M.; Xing, X.; Zhou, X.; Emura, T.; Hidaka, K.; Tuesuwan, B.; Sugiyama, H. Single-Molecule Manipulation of the Duplex Formation and Dissociation at the G-Quadruplex/i-Motif Site in the DNA Nanostructure. ACS Nano 2015, 9, 9922–9929. [Google Scholar] [CrossRef]
- Lim, K.W.; Phan, A.T. Structural basis of DNA quadruplex-duplex junction formation. Angew. Chem. Int. Ed. 2013, 52, 8566–8569. [Google Scholar] [CrossRef]
- Lim, K.W.; Jenjaroenpun, P.; Low, Z.J.; Khong, Z.J.; Ng, Y.S.; Kuznetsov, V.A.; Phan, A.T. Duplex stem-loop-containing quadruplex motifs in the human genome: A combined genomic and structural study. Nucleic Acids Res. 2015, 43, 5630–5646. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, H.H.; Zheng, K.W.; Hao, Y.H.; Tan, Z. DNA G-quadruplex formation in response to remote downstream transcription activity: long-range sensing and signal transducing in DNA double helix. Nucleic Acids Res. 2013, 14, 7144–7152. [Google Scholar] [CrossRef]
- Konig, S.L.; Huppert, J.L.; Sigel, R.K.; Evans, A.C. Distance-dependent duplex DNA destabilization proximal to G-quadruplex/i-motif sequences. Nucleic Acids Res. 2013, 41, 7453–7461. [Google Scholar] [CrossRef] [PubMed]
- Trajkovski, M.; Webba da Silva, M.; Plavec, J. Unique Structural Features of Interconverting Monomeric and Dimeric G-Quadruplexes Adopted by a Sequence from the Intron of the N-myc Gene. J. Am. Chem. Soc. 2012, 134, 4132–4141. [Google Scholar] [CrossRef] [PubMed]
- Manzini, G.; Yathindra, N.; Xodo, L.E. Evidence for Intramolecularly Folded I-DNA Structures in Biologically Relevant Ccc-Repeat Sequences. Nucleic Acids Res. 1994, 22, 4634–4640. [Google Scholar] [CrossRef] [PubMed]
- Bryan, T.M.; Baumann, P. G-quadruplexes: From guanine gels to chemotherapeutics. Mol. Biotechnol. 2011, 49, 198–208. [Google Scholar] [CrossRef]
- Smirnov, I.V.; Shafer, R.H. Electrostatics dominate quadruplex stability. Biopolymers 2007, 85, 91–101. [Google Scholar] [CrossRef]
- Nakano, S.; Miyoshi, D.; Sugimoto, N. Effects of Molecular Crowding on the Structures, Interactions, and Functions of Nucleic Acids. Chem. Rev. 2014, 114, 2733–2758. [Google Scholar] [CrossRef]
- Trajkovski, M.; Endoh, T.; Tateishi-Karimata, H.; Ohyama, T.; Tanaka, S.; Plavec, J.; Sugimoto, N. Pursuing origins of (poly)ethylene glycol-induced G-quadruplex structural modulations. Nucleic Acids Res. 2018, 46, 4301–4315. [Google Scholar] [CrossRef]
- Cui, Y.; Kong, D.; Ghimire, C.; Xu, C.; Mao, H. Mutually Exclusive Formation of G-Quadruplex and i-Motif is a General Phenomenon Governed by Steric Hindrance in Duplex DNA. Biochemistry 2016, 55, 2291–2299. [Google Scholar] [CrossRef]
- Karsisiotis, A.I.; Hessari, N.M.; Novellino, E.; Spada, G.P.; Randazzo, A.; Webba da Silva, M. Topological Characterization of Nucleic Acid G-Quadruplexes by UV Absorption and Circular Dichroism. Angew. Chem. Int. Ed. 2011, 50, 10645–10648. [Google Scholar] [CrossRef]
- Webba da Silva, M.; Trajkovski, M.; Sannohe, Y.; Hessari, N.M.; Sugiyama, H.; Plavec, J. Design of a G-Quadruplex Topology through Glycosidic Bond Angles. Angew. Chem. Int. Ed. 2009, 48, 9167–9170. [Google Scholar] [CrossRef]
- Cevec, M.; Plavec, J. Role of loop residues and cations on the formation and stability of dimeric DNA G-quadruplexes. Biochemistry 2005, 44, 15238–15246. [Google Scholar] [CrossRef] [PubMed]
- Kuryavyi, V.; Phan, A.T.; Patel, D.J. Solution structures of all parallel-stranded monomeric and dimeric G-quadruplex scaffolds of the human c-kit2 promoter. Nucleic Acids Res. 2010, 38, 6757–6773. [Google Scholar] [CrossRef] [PubMed]
- Do, N.Q.; Phan, A.T. Monomer-Dimer Equilibrium for the 5′-5′ Stacking of Propeller-Type Parallel-Stranded G-Quadruplexes: NMR Structural Study. Chem. Eur. J. 2012, 18, 14752–14759. [Google Scholar] [CrossRef] [PubMed]
- Kuryavyi, V.; Cahoon, L.A.; Seifert, H.S.; Patel, D.J. RecA-Binding pilE G4 Sequence Essential for Pilin Antigenic Variation Forms Monomeric and 5′ End-Stacked Dimeric Parallel G-Quadruplexes. Structure 2012, 20, 2090–2102. [Google Scholar] [CrossRef]
- Boan, F.; Gomez-Marquez, J. In Vitro Recombination Mediated by G-Quadruplexes. Chembiochem 2010, 11, 331–334. [Google Scholar] [CrossRef]
- Wanrooij, P.H.; Uhler, J.P.; Shi, Y.H.; Westerlund, F.; Falkenberg, M.; Gustafsson, C.M. A hybrid G-quadruplex structure formed between RNA and DNA explains the extraordinary stability of the mitochondrial R-loop. Nucleic Acids Res. 2012, 40, 10334–10344. [Google Scholar] [CrossRef]
- Zheng, K.W.; Xiao, S.; Liu, J.Q.; Zhang, J.Y.; Hao, Y.H.; Tan, Z. Co-transcriptional formation of DNA:RNA hybrid G-quadruplex and potential function as constitutional cis element for transcription control. Nucleic Acids Res. 2013, 41, 5533–5541. [Google Scholar] [CrossRef]
- Xu, Y.; Ishizuka, T.; Yang, J.; Ito, K.; Katada, H.; Komiyama, M.; Hayashi, T. Oligonucleotide Models of Telomeric DNA and RNA Form a Hybrid G-quadruplex Structure as a Potential Component of Telomeres. J. Biol. Chem. 2012, 287, 41787–41796. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trajkovski, M.; Plavec, J. Chasing Particularities of Guanine- and Cytosine-Rich DNA Strands. Molecules 2020, 25, 434. https://doi.org/10.3390/molecules25030434
Trajkovski M, Plavec J. Chasing Particularities of Guanine- and Cytosine-Rich DNA Strands. Molecules. 2020; 25(3):434. https://doi.org/10.3390/molecules25030434
Chicago/Turabian StyleTrajkovski, Marko, and Janez Plavec. 2020. "Chasing Particularities of Guanine- and Cytosine-Rich DNA Strands" Molecules 25, no. 3: 434. https://doi.org/10.3390/molecules25030434
APA StyleTrajkovski, M., & Plavec, J. (2020). Chasing Particularities of Guanine- and Cytosine-Rich DNA Strands. Molecules, 25(3), 434. https://doi.org/10.3390/molecules25030434




