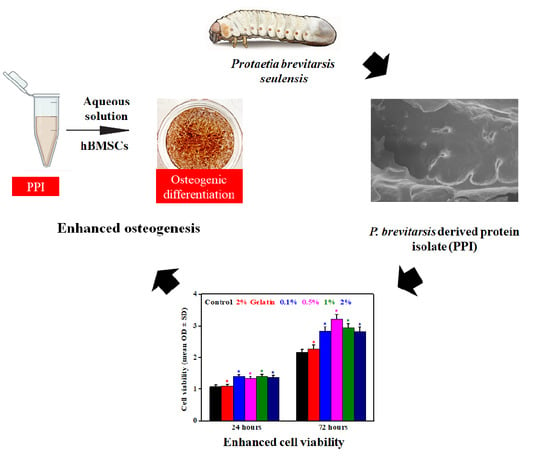
Protaetia brevitarsis seulensis Derived Protein Isolate with Enhanced Osteomodulatory and Antioxidative Property
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
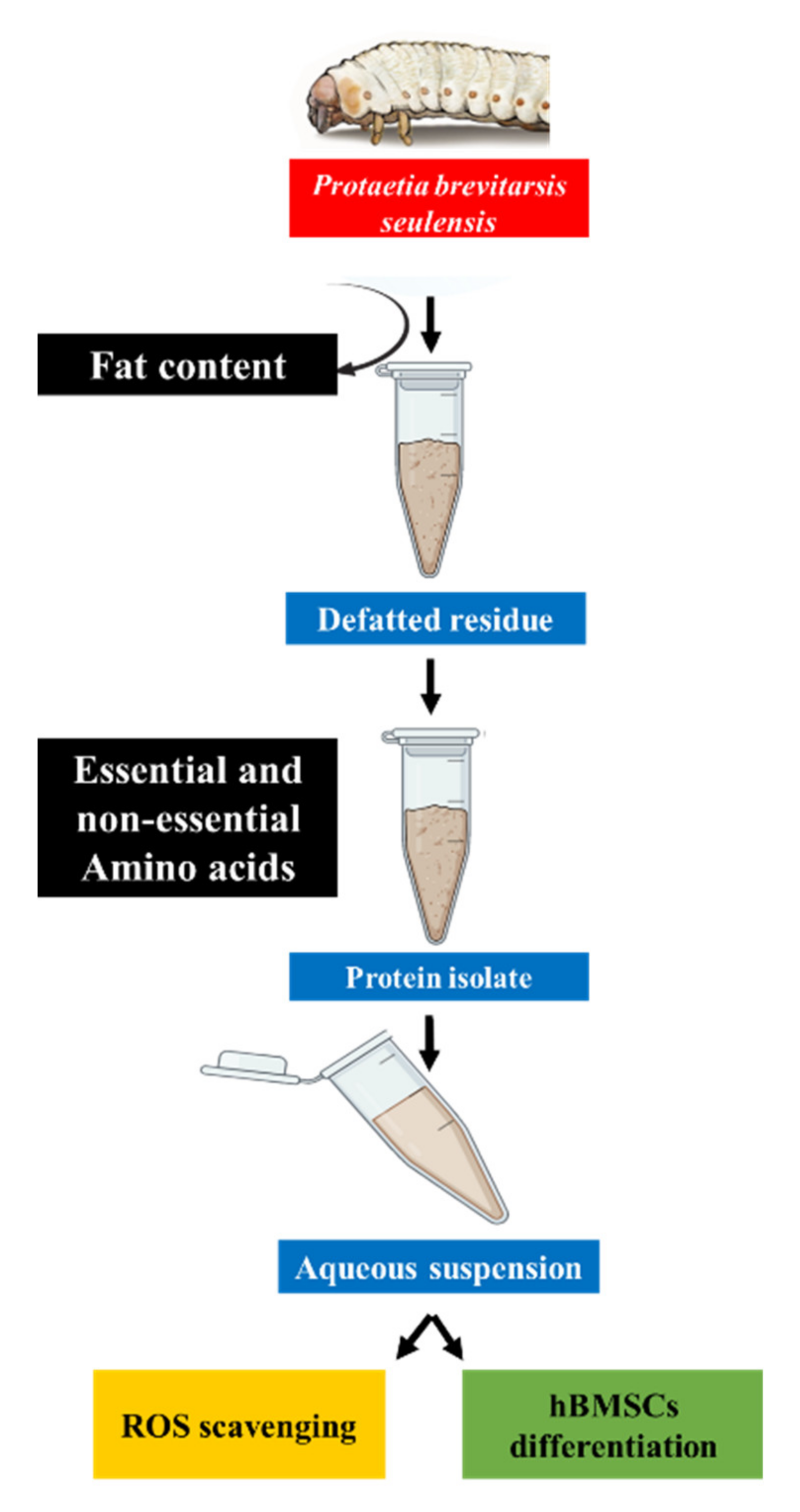
2.2. Defatting and Protein Extraction
2.3. Characterization of the Isolate
2.3.1. Determination of the Nutrient Composition
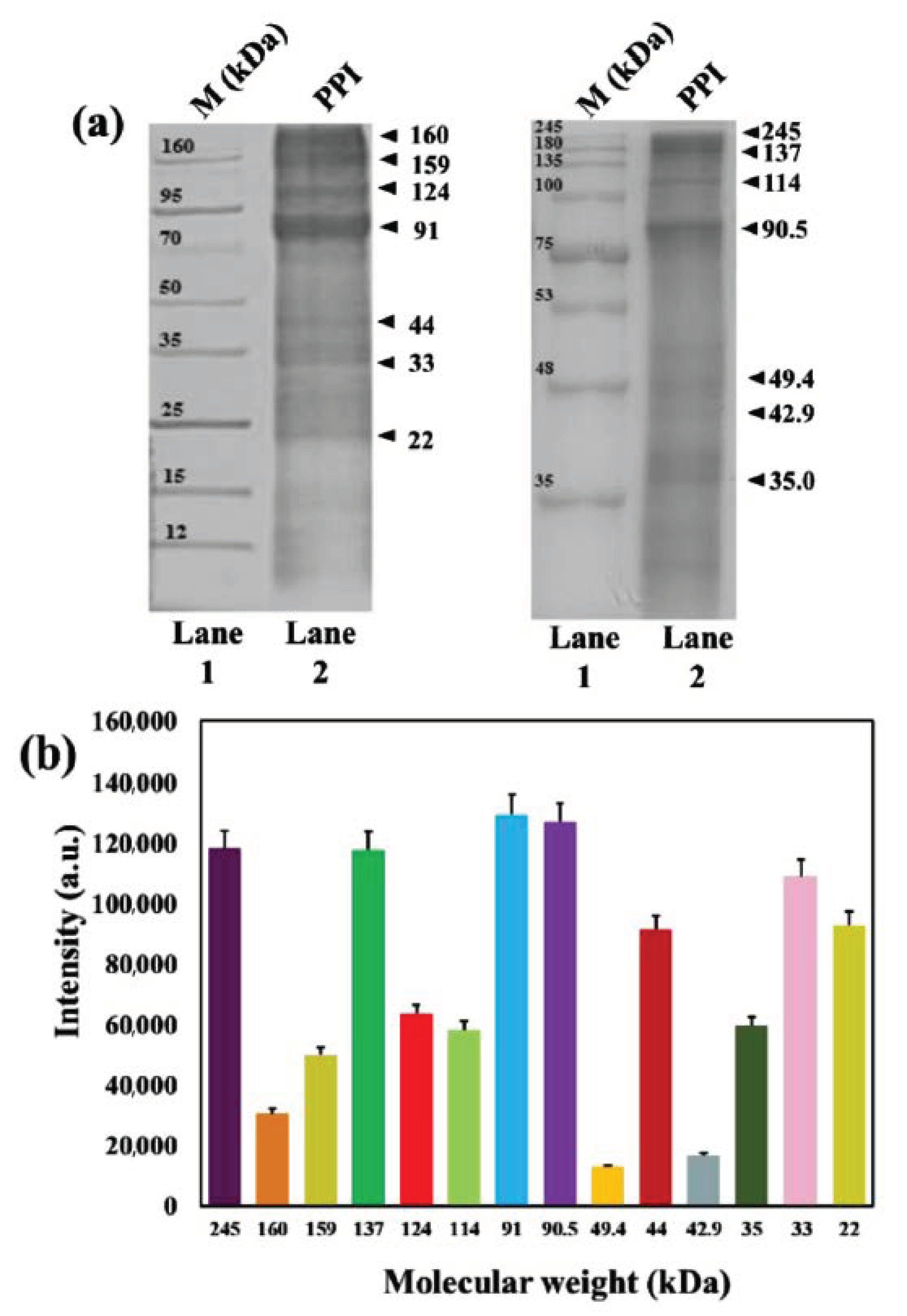
2.3.2. Molecular Weight Determination
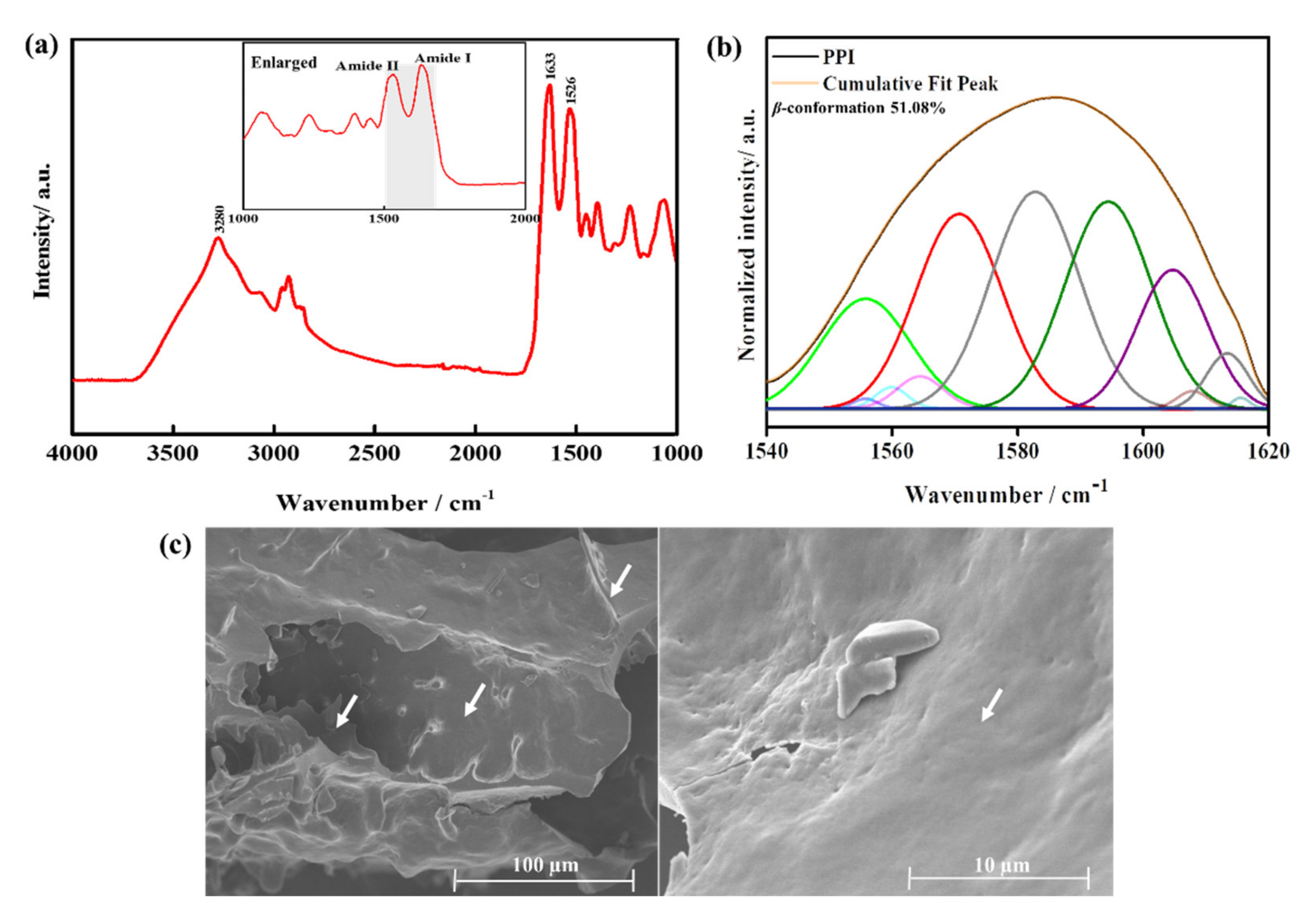
2.3.3. Chemical Composition and Morphological Analysis
2.3.4. DPPH Assay
2.4. Cell Culture
2.5. Cell Viability Assay
2.6. Live-Dead Assay
2.7. Cell Morphology
2.8. Assessment of Reactive Oxygen Species (ROS) Scavenging Property of PPI
2.9. In Vitro Osteogenic Differentiation Study
2.10. RNA Isolation and Real-Time PCR (qRT-PCR) Analysis
2.11. Protein Marker Expression Analysis
2.12. Statistical Analysis
3. Results and Discussion
3.1. Nutritional Composition
3.2. Characterization of the PPI
3.3. Molecular Weight Analysis
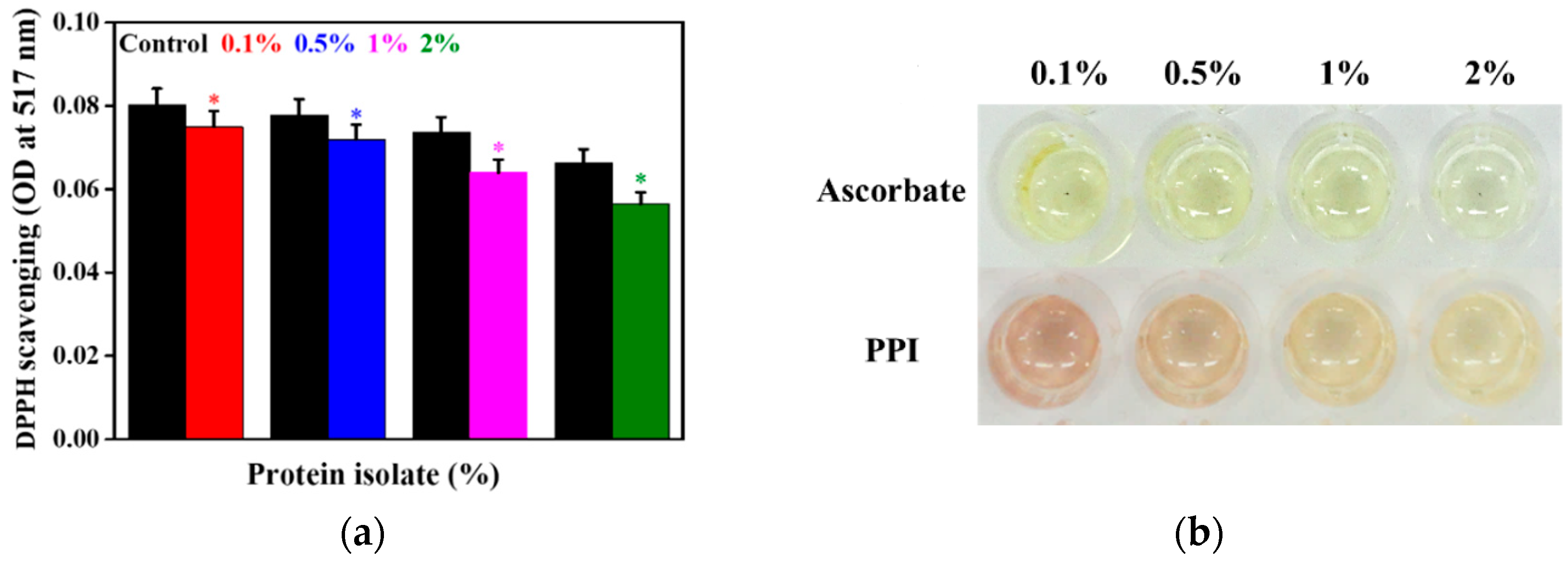
3.4. Antioxidant Activity of the PPI
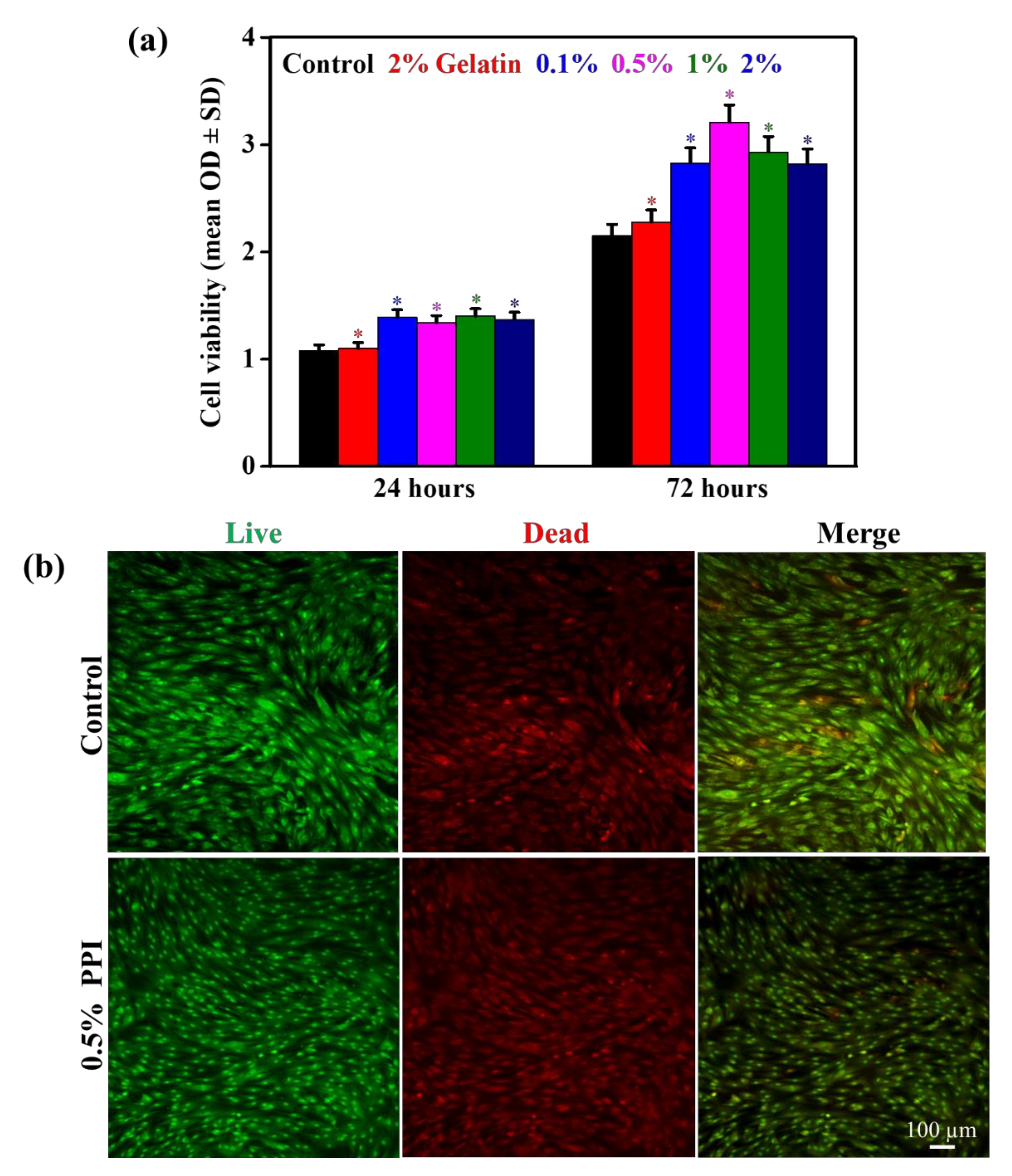
3.5. Cell Viability and Morphology
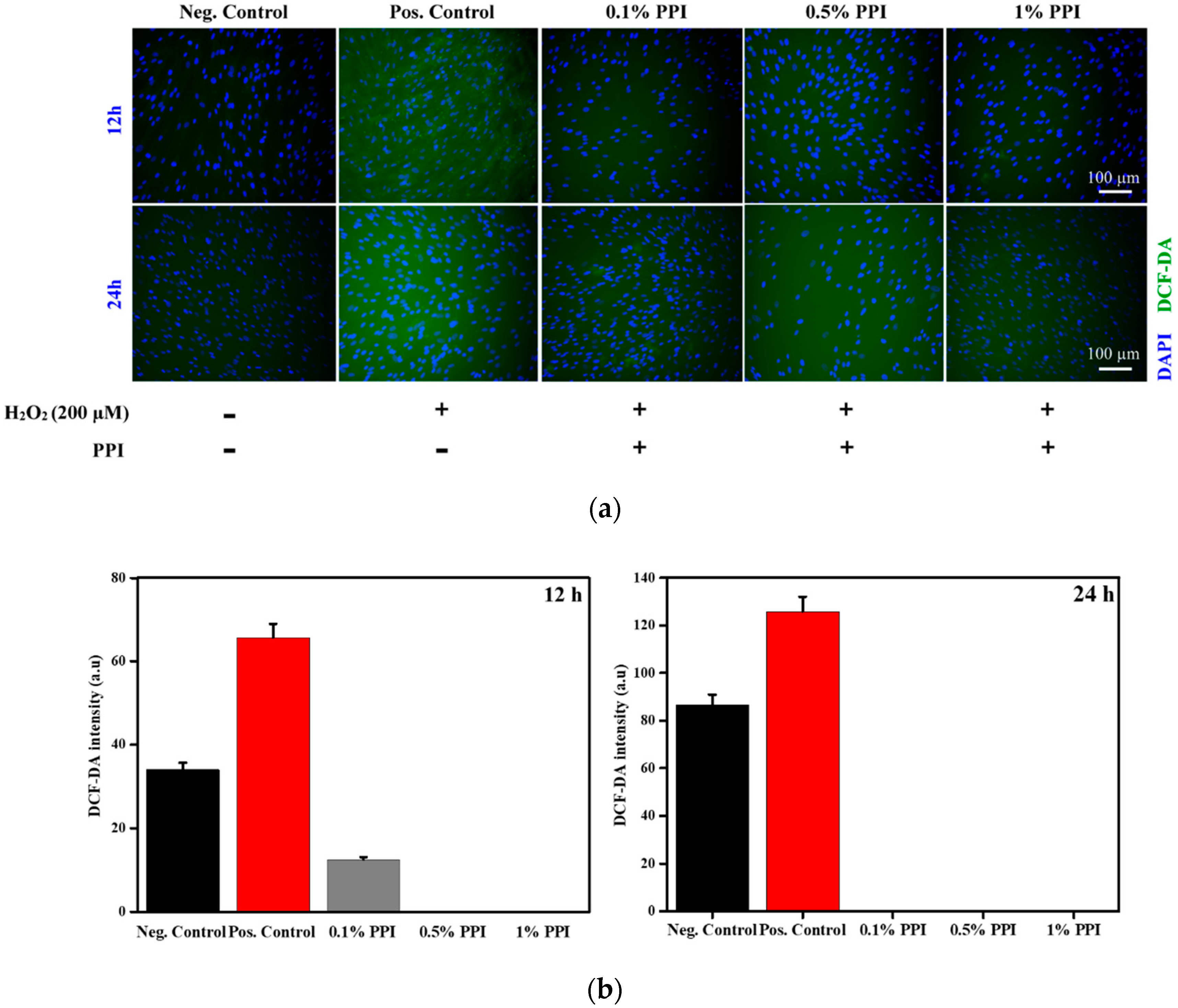
3.6. Intracellular Reactive Oxygen Species (ROS) Scavenging Activity of PPI
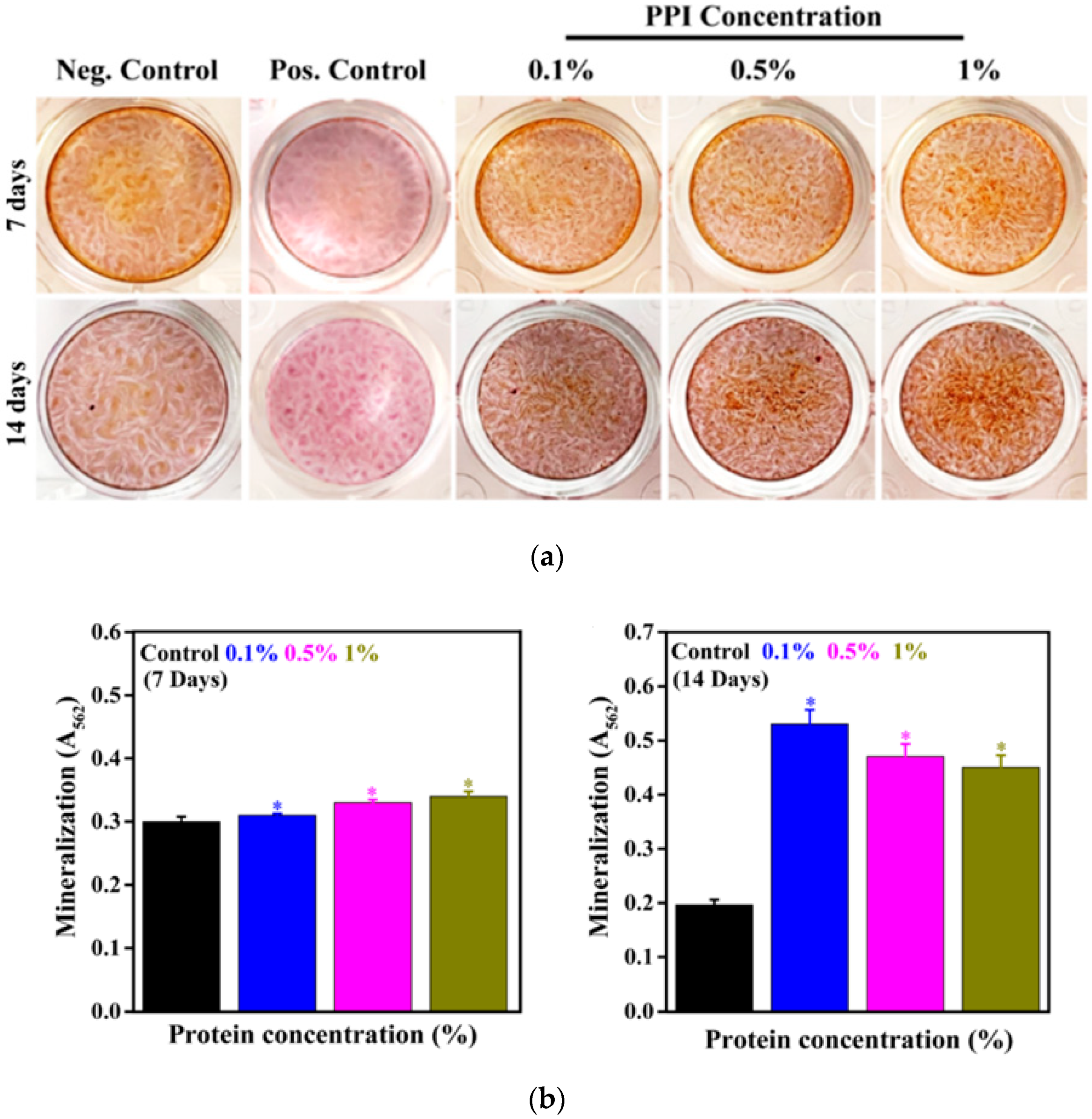
3.7. Mineral Induction in the Presence of PPI
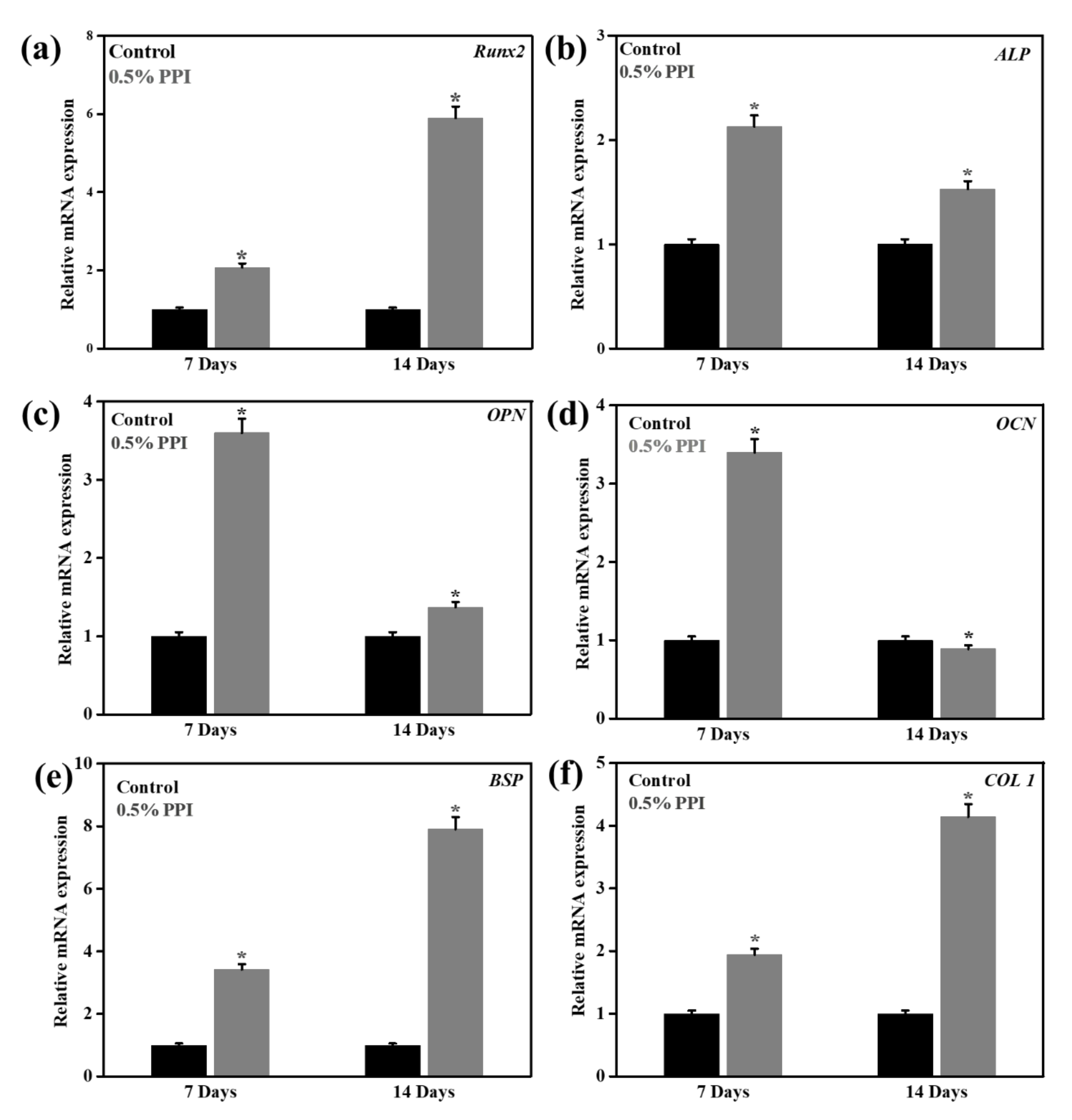
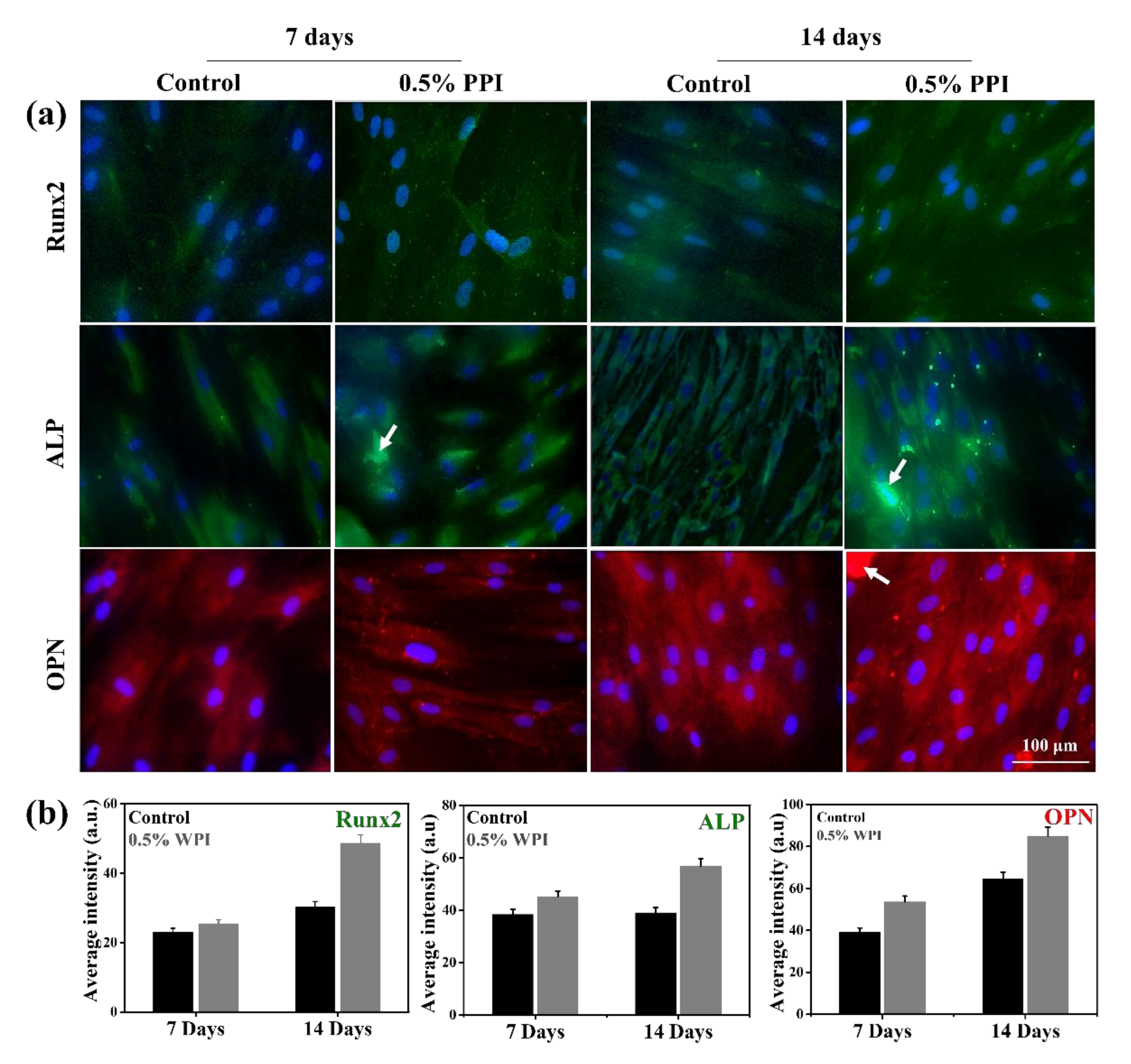
3.8. Gene and Protein Marker Expression
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Shang, F.; Yu, Y.; Liu, S.; Ming, L.; Zhang, Y.; Zhou, Z.; Zhao, J.; Jin, Y. Advancing application of mesenchymal stem cell-based bone tissue regeneration. Bioact. Mater. 2020, 6, 666–683. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.-Y.; Bi, Q.; Zhao, C.; Chen, J.-Y.; Cai, M.-H.; Chen, X.-Y. Recent Advances in Biomaterials for the Treatment of Bone Defects. Organogenesis 2020, 16, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Chen, J.; Liu, S.; Jin, Y. Stem cell-based bone and dental regeneration: A view of microenvironmental modulation. Int. J. Oral Sci. 2019, 11, 23. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Lee, S. A Biomimetic Strategy to Design Biomaterials for In Situ Tissue Regeneration. In In Situ Tissue Regeneration; Elsevier: Amsterdam, The Netherlands, 2016; pp. 185–201. [Google Scholar]
- Di, L.; Shi, Y.-N.; Yan, Y.-M.; Jiang, L.-P.; Hou, B.; Wang, X.-L.; Zuo, Z.-L.; Chen, Y.-B.; Yang, C.-P.; Cheng, Y.-X. Nonpeptide small molecules from the insect Aspongopus chinensis and their neural stem cell proliferation stimulating properties. RSC Adv. 2015, 5, 70985–70991. [Google Scholar] [CrossRef]
- Hirose, Y.; Ohta, E.; Kawai, Y.; Ohta, S. Dorsamin-A’s, glycerolipids carrying a Dehydrophenylalanine Ester Moiety from the seed-eating larvae of the Bruchid Beetle Bruchidius dorsalis. J. Nat. Prod. 2013, 76, 554–558. [Google Scholar] [CrossRef]
- Lee, H.-S.; Ryu, H.-J.; Song, H.-J.; Lee, S.-O.; Lee, H.-S.; Ryu, H.-J.; Song, H.-J.; Lee, S.-W. Enzymatic preparation and antioxidant activities of protein hydrolysates from Protaetia brevitarsis larvae. J. Korean Soc. Food Sci. Nutr. 2017, 46, 1164–1170. [Google Scholar]
- Suh, H.-J.; Kang, S.C. Antioxidant activity of aqueous methanol extracts of Protaetia brevitarsis Lewis (Coleoptera: Scarabaedia) at different growth stages. Nat. Prod. Res. 2012, 26, 510–517. [Google Scholar] [CrossRef]
- Sung, G.A.; Kim, M.H.; Park, S.N. Anti-inflammatory and whitening effects of Protaetia brevitarsis Seulensis extracts by oriental conversion methods. J. Soc. Cosmet. Sci. Korea 2016, 42, 421–432. [Google Scholar] [CrossRef][Green Version]
- Kim, T.-E.; Kim, C.-T.; Sim, H.-J.; Lee, H.-Y.; Kim, S.-O.; Kim, D.-K.; Jo, M.-N.; Lee, S.-J.; Jeon, Y.-D.; Song, Y.-J.; et al. Production of protein hydrolysate from Protaetia brevitarsis seulensis (Kolbe) larvae by enzyme treatment under high pressure. Food Sci. Biotechnol. 2020, 29, 1187–1194. [Google Scholar] [CrossRef]
- Yoon, C.-H.; Jeon, S.-H.; Ha, Y.-J.; Kim, S.-W.; Bang, W.-Y.; Bang, K.-H.; Gal, S.-W.; Kim, I.-S.; Cho, Y.-S. Functional Chemical Components in Protaetia brevitarsis Larvae: Impact of Supplementary Feeds. Food Sci. Anim. Resour. 2020, 40, 461. [Google Scholar] [CrossRef]
- Sim, S.-Y.; Ahn, H.-Y.; Seo, K.-I.; Cho, Y.-S. Physicochemical properties and biological activities of Protaetia brevitarsis seulensis larvae fermented by several kinds of micro-organisms. J. Life Sci. 2018, 28, 827–834. [Google Scholar]
- Choi, B.D.; Wong, N.A.; Auh, J.-H. Defatting and sonication enhances protein extraction from edible insects. Korean J. Food Sci. Anim. Resour. 2017, 37, 955. [Google Scholar] [PubMed]
- AOAC. Official Methods of Analysis of AOAC International, 5th ed.; AOAC International: Rockville, MD, USA, 1999. [Google Scholar]
- Bußler, S.; Rumpold, B.A.; Jander, E.; Rawel, H.M.; Schlüter, O.K. Recovery and techno-functionality of flours and proteins from two edible insect species: Meal worm (Tenebrio molitor) and black soldier fly (Hermetia illucens) larvae. Heliyon 2016, 2, e00218. [Google Scholar] [CrossRef]
- Patel, D.K.; Dutta, S.D.; Hexiu, J.; Ganguly, K.; Lim, K.-T. Bioactive electrospun nanocomposite scaffolds of poly (lactic acid)/cellulose nanocrystals for bone tissue engineering. Int. J. Biol. Macromol. 2020, 162, 1429–1441. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.D.; Patel, D.K.; Seo, Y.-R.; Park, C.-W.; Lee, S.-H.; Kim, J.-W.; Kim, J.; Seonwoo, H.; Lim, K.-T. In Vitro Biocompatibility of Electrospun Poly (ε-Caprolactone)/Cellulose Nanocrystals-Nanofibers for Tissue Engineering. J. Nanomater. 2019, 1–11. [Google Scholar] [CrossRef]
- Chung, M.J.; Lee, S.; Park, Y.-I.; Lee, J.; Kwon, K.H. Neuroprotective effects of phytosterols and flavonoids from Cirsium setidens and Aster scaber in human brain neuroblastoma SK-N-SH cells. Life Sci. 2016, 148, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Gharibi, B.; Hughes, F.J. Effects of medium supplements on proliferation, differentiation potential, and in vitro expansion of mesenchymal stem cells. Stem Cells Transl. Med. 2012, 1, 771–782. [Google Scholar] [CrossRef]
- Huh, J.-E.; Choi, J.-Y.; Shin, Y.-O.; Park, D.-S.; Kang, J.-W.; Nam, D.; Choi, D.-Y.; Lee, J.-D. Arginine enhances osteoblastogenesis and inhibits adipogenesis through the regulation of Wnt and NFATc signaling in human mesenchymal stem cells. Int. J. Mol. Sci. 2014, 15, 13010–13029. [Google Scholar] [CrossRef]
- Choi, K.-M.; Yoon, H.-H.; Seo, Y.-K.; Song, K.-Y.; Kwon, S.-Y.; Lee, H.-S.; Park, Y.-S.; Kim, Y.-J.; Park, J.-K. Effect of essential and nonessential amino acid compositions on the in vitro behavior of human mesenchymal stem cells. Korean J. Chem. Eng. 2007, 24, 1058–1063. [Google Scholar] [CrossRef]
- Yu, Y.; Newman, H.; Shen, L.; Sharma, D.; Hu, G.; Mirando, A.j.; Zhang, H.; Knudsen, E.; Zhang, G.-F.; Hilton, M.J. Glutamine metabolism regulates proliferation and lineage allocation in skeletal stem cells. Cell Metab. 2019, 29, 966–978.e4. [Google Scholar] [CrossRef]
- Huang, Y.-C.; Li, Z.; Li, J.; Lyu, F.-J. Interaction between Stem Cells and the Microenvironment for Musculoskeletal Repair; Hindawi: London, UK, 2020. [Google Scholar]
- Onak, G.; Sen, M.; Horzum, N.; Ercan, U.K.; Yarali, Z.B.; Garipcan, B.; Karaman, O. Aspartic and Glutamic Acid Templated Peptides Conjugation on Plasma Modified Nanofibers for Osteogenic Differentiation of Human Mesenchymal Stem Cells: A Comparative Study. Sci. Rep. 2018, 8, 17620. [Google Scholar] [CrossRef] [PubMed]
- Ding, K.-H.; Cain, M.; Davis, M.; Bergson, C.; McGee-Lawrence, M.; Perkins, C.; Hardigan, T.; Shi, X.; Zhong, Q.; Xu, J.; et al. Amino acids as signaling molecules modulating bone turnover. Bone 2018, 115, 15–24. [Google Scholar] [CrossRef] [PubMed]
- El Refaey, M.; Zhong, Q.; Ding, K.-H.; Shi, X.-M.; Xu, J.; Bollag, W.B.; Hill, W.D.; Chutkan, N.; Robbins, R.; Nadeau, H.; et al. Impact of dietary aromatic amino acids on osteoclastic activity. Calcif. Tissue Int. 2014, 95, 174–182. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Arrondo, J.L.R.; Muga, A.; Castresana, J.; Goni, F.M. Quantitative studies of the structure of proteins in solution by Fourier-transform infrared spectroscopy. Prog. Biophys. Mol. Biol. 1993, 59, 23–56. [Google Scholar] [CrossRef]
- Nandiyanto, A.B.D.; Oktiani, R.; Ragadhita, R. How to read and interpret FTIR spectroscope of organic material. Indones. J. Sci. Technol. 2019, 4, 97–118. [Google Scholar] [CrossRef]
- Barth, A. Infrared spectroscopy of proteins. Biochim. Biophys. Acta BBA Bioenerg. 2007, 1767, 1073–1101. [Google Scholar] [CrossRef]
- Lorenz-Fonfria, V.A. Infrared Difference Spectroscopy of Proteins: From Bands to Bonds. Chem. Rev. 2020, 120, 3466–3576. [Google Scholar] [CrossRef]
- Jackson, M.; Mantsch, H.H. The use and misuse of FTIR spectroscopy in the determination of protein structure. Crit. Rev. Biochem. Mol. Biol. 1995, 30, 95–120. [Google Scholar] [CrossRef]
- Litvinov, R.I.; Faizullin, D.A.; Zuev, Y.F.; Weisel, J.W. The α-helix to β-sheet transition in stretched and compressed hydrated fibrin clots. Biophys. J. 2012, 103, 1020–1027. [Google Scholar] [CrossRef]
- Collier, J.H.; Rudra, J.S.; Gasiorowski, J.Z.; Jung, J.P. Multi-component extracellular matrices based on peptide self-assembly. Chem. Soc. Rev. 2010, 39, 3413–3424. [Google Scholar] [CrossRef]
- Ami, D.; Lavatelli, F.; Rognoni, P.; Palladini, G.; Raimondi, S.; Giorgetti, S.; Monti, L.; Doglia, S.M.; Natalello, A.; Merlini, G. In situ characterization of protein aggregates in human tissues affected by light chain amyloidosis: A FTIR microspectroscopy study. Sci. Rep. 2016, 6, 29096. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.-K.; Yong, H.H.; Chun, H.H.; Lee, M.-A.; Kim, Y.B.; Choi, Y.S. Changes of amino acid composition and protein technical functionality of edible insects by extracting steps. J. Asia-Pac. Entomol. 2020, 23, 298–305. [Google Scholar] [CrossRef]
- Pownall, T.L.; Udenigwe, C.C.; Aluko, R.E. Amino acid composition and antioxidant properties of pea seed (Pisum sativum L.) enzymatic protein hydrolysate fractions. J. Agric. Food Chem. 2010, 58, 4712–4718. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.K.; Tsai, M.L.; Huang, J.R.; Chen, R.H. In vitro antioxidant activities of low-molecular-weight polysaccharides with various functional groups. J. Agric. Food Chem. 2009, 57, 2699–2704. [Google Scholar] [CrossRef]
- Ranathunga, S.; Rajapakse, N.; Kim, S.-K. Purification and characterization of antioxidative peptide derived from muscle of conger eel (Conger myriaster). Eur. Food Res. Technol. 2006, 222, 310–315. [Google Scholar] [CrossRef]
- Yang, R.; Li, X.; Lin, S.; Zhang, Z.; Chen, F. Identification of novel peptides from 3 to 10 kDa pine nut (Pinus koraiensis) meal protein, with an exploration of the relationship between their antioxidant activities and secondary structure. Food Chem. 2017, 219, 311–320. [Google Scholar] [CrossRef]
- Ahn, M.Y.; Joo, H.J.; Kim, J.S.; Yeon, Y.; Ryu, H.Y.; Choi, B.G.; Song, K.S.; Kim, S.H.; Park, M.K.; Jo, Y.Y. Toxicity assessment of Gryllus bimaculatus (a type of cricket) glycosaminoglycan. Toxicol. Res. 2020, 36, 319–328. [Google Scholar] [CrossRef]
- Young Ahn, M.; Bae, H.J.; Kim, I.S.; Yoo, E.J.; Kwack, S.J.; Kim, H.S.; Kim, D.H.; Ryu, K.S.; Lee, H.S.; Kim, J.W.; et al. Genotoxic evaluation of the biocomponents of the cricket, Gryllus bimaculatus, using three mutagenicity tests. J. Toxicol. Environ. Health Part A 2005, 68, 2111–2118. [Google Scholar] [CrossRef]
- Srinroch, C.; Srisomsap, C.; Chokchaichamnakit, D.; Punyarit, P.; Phiriyangkul, P. Identification of novel allergen in edible insect, Gryllus bimaculatus and its cross-reactivity with Macrobrachium spp. allergens. Food Chem. 2015, 184, 160–166. [Google Scholar] [CrossRef]
- Lutolf, M.P.; Gilbert, P.M.; Blau, H.M. Designing materials to direct stem-cell fate. Nature 2009, 462, 433–441. [Google Scholar] [CrossRef]
- Nita, M.; Grzybowski, A. The role of the reactive oxygen species and oxidative stress in the pathomechanism of the age-related ocular diseases and other pathologies of the anterior and posterior eye segments in adults. Oxidative Med. Cell. Longev. 2016, 2016, 3164734. [Google Scholar] [CrossRef] [PubMed]
- Hwang, D.; Goo, T.-W.; Yun, E.-Y. In Vitro Protective Effect of Paste and Sauce Extract Made with Protaetia brevitarsis Larvae on HepG2 Cells Damaged by Ethanol. Insects 2020, 11, 494. [Google Scholar] [CrossRef] [PubMed]
- Douglas, T.E.; Vandrovcova, M.; Krocilova, N.; Keppler, J.K.; Zarubova, J.; Skirtach, A.G.; Bacakova, L. Application of whey protein isolate in bone regeneration: Effects on growth and osteogenic differentiation of bone-forming cells. J. Dairy Sci. 2018, 101, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Karadjian, M.; Senger, A.S.; Esser, C.; Wilkesmann, S.; Heller, R.; Fellenberg, J.; Simon, R.; Westhauser, F. Human Platelet Lysate Can Replace Fetal Calf Serum as a Protein Source to Promote Expansion and Osteogenic Differentiation of Human Bone-Marrow-Derived Mesenchymal Stromal Cells. Cells 2020, 9, 918. [Google Scholar] [CrossRef] [PubMed]
- Hansamuit, K.; Osathanon, T.; Suwanwela, J. Effect of Jagged1 on the expression of genes in regulation of osteoblast differentiation and bone mineralization ontology in human dental pulp and periodontal ligament cells. J. Oral Biol. Craniofacial Res. 2020, 10, 233–237. [Google Scholar] [CrossRef] [PubMed]
- Kirkham, G.; Cartmell, S. Genes and proteins involved in the regulation of osteogenesis. Top. Tissue Eng. 2007, 3, 1–22. [Google Scholar]
- Komori, T. Regulation of proliferation, differentiation and functions of osteoblasts by Runx2. Int. J. Mol. Sci. 2019, 20, 1694. [Google Scholar] [CrossRef]
- Vimalraj, S. Alkaline Phosphatase: Structure, Expression and its Function in Bone Mineralization. Gene 2020, 754, 144855. [Google Scholar] [CrossRef]
- Morinobu, M.; Ishijima, M.; Rittlimg, S.R.; Tsuji, K.; Yamamoto, H.; Nifuji, A.; Denhardt, D.T.; Noda, M. Osteopontin expression in osteoblasts and osteocytes during bone formation under mechanical stress in the calvarial suture in vivo. J. Bone Miner. Res. 2003, 18, 1706–1715. [Google Scholar] [CrossRef]
- Thurner, P.J.; Chen, C.G.; Martin, S.I.; Sun, L.; Harman, A.; Porter, A.; Ager, J.W., III; Ritchie, R.O.; Alliston, T. Osteopontin deficiency increases bone fragility but preserves bone mass. Bone 2010, 46, 1564–1573. [Google Scholar] [CrossRef]
- Rittling, S.R.; Matsumoto, H.N.; Mckee, M.D.; Nanci, A.; An, X.R.; Novick, K.E.; Kowalski, A.J.; Noda, M.; Denhardt, D.T. Mice lacking osteopontin show normal development and bone structure but display altered osteoclast formation in vitro. J. Bone Miner. Res. 1998, 13, 1101–1111. [Google Scholar] [CrossRef] [PubMed]
- Malaval, L.; Wade-Gueye, N.M.; Boudiffa, M.; Fei, J.; Zirngibl, R.; Chen, F.; Laroche, N.; Roux, J.P.; Burt-Pichat, B.; Duboeuf, F.; et al. Bone sialoprotein plays a functional role in bone formation and osteoclastogenesis. J. Exp. Med. 2008, 205, 1145–1153. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Bank, R.A.; Tekoppele, J.M.; Agrawal, C.M. The role of collagen in determining bone mechanical properties. J. Orthop. Res. 2001, 19, 1021–1026. [Google Scholar] [CrossRef]

| Genes | GenBank Accession No. | Sequences (5′ to 3′) |
|---|---|---|
| β-actin | NM_031144 | ACCCGCGAGTACAACCTTCT CTTCTGACCCATACCCACCA |
| Runx2 | NM_001146038 | CGCACGACAACCGCACCAT CAGCACGGAGCACAGGAAGTT |
| BSP | L09555 | AACTTTTATGTCCCCCGTTGA TGGACTGGAAACCGTTTCAGA |
| ALP | NM_007431 | CCAACTCTTTTGTGCCAGAGA GGCTACATTGGTGTTGAGCTTTT |
| OPN | J04765 | TGAAACGAGTCAGCTGGATG TGAAATTCATGGCTGTGGAA |
| COL1 | NM007742 | GCTCCTCTTAGGGGCCACT CCACGTCTCACCATTGGGG |
| Sample | Crude Protein (%, DM) | Crude Fat (%, DM) | Crude Ash (%, DM) | Carbohydrate * (%, DM) |
|---|---|---|---|---|
| P. brevitarsis protein isolate | 77.52 ± 0.31 a | 0.52 ± 0.14 c | 4.07 ± 0.35 b | 17.89 |
| Essential Amino Acid | Contents (g/100 g Sample) | Non-Essential Amino Acid | Contents (g/100g Sample) |
|---|---|---|---|
| Isoleucine | 3.76 | Aspartic acid | 7.48 |
| Leucine | 5.92 | Serine | 3.39 |
| Lysine | 5.31 | Glutamic acid | 9.68 |
| Methionine | 1.47 | Proline | 5.44 |
| Phenylalanine | 3.93 | Glycine | 3.22 |
| Tyrosine | 5.24 | Alanine | 3.61 |
| Threonine | 3.63 | Cysteine | 1.54 |
| Valine | 4.12 | Arginine | 4.33 |
| Histidine | 2.26 | Non-essential A.A | 38.69 |
| Tryptophan | 1.07 | ||
| Essential A.A | 36.71 |
Sample Availability: Samples of the Protaetia brevitarsis derived protein isolate (PPI) is available from the authors. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ganguly, K.; Jeong, M.-S.; Dutta, S.D.; Patel, D.K.; Cho, S.-J.; Lim, K.-T. Protaetia brevitarsis seulensis Derived Protein Isolate with Enhanced Osteomodulatory and Antioxidative Property. Molecules 2020, 25, 6056. https://doi.org/10.3390/molecules25246056
Ganguly K, Jeong M-S, Dutta SD, Patel DK, Cho S-J, Lim K-T. Protaetia brevitarsis seulensis Derived Protein Isolate with Enhanced Osteomodulatory and Antioxidative Property. Molecules. 2020; 25(24):6056. https://doi.org/10.3390/molecules25246056
Chicago/Turabian StyleGanguly, Keya, Min-Soo Jeong, Sayan Deb Dutta, Dinesh K. Patel, Seong-Jun Cho, and Ki-Taek Lim. 2020. "Protaetia brevitarsis seulensis Derived Protein Isolate with Enhanced Osteomodulatory and Antioxidative Property" Molecules 25, no. 24: 6056. https://doi.org/10.3390/molecules25246056
APA StyleGanguly, K., Jeong, M.-S., Dutta, S. D., Patel, D. K., Cho, S.-J., & Lim, K.-T. (2020). Protaetia brevitarsis seulensis Derived Protein Isolate with Enhanced Osteomodulatory and Antioxidative Property. Molecules, 25(24), 6056. https://doi.org/10.3390/molecules25246056