Chalcones Display Anti-NLRP3 Inflammasome Activity in Macrophages through Inhibition of Both Priming and Activation Steps—Structure-Activity-Relationship and Mechanism Studies
Abstract
1. Introduction
2. Results
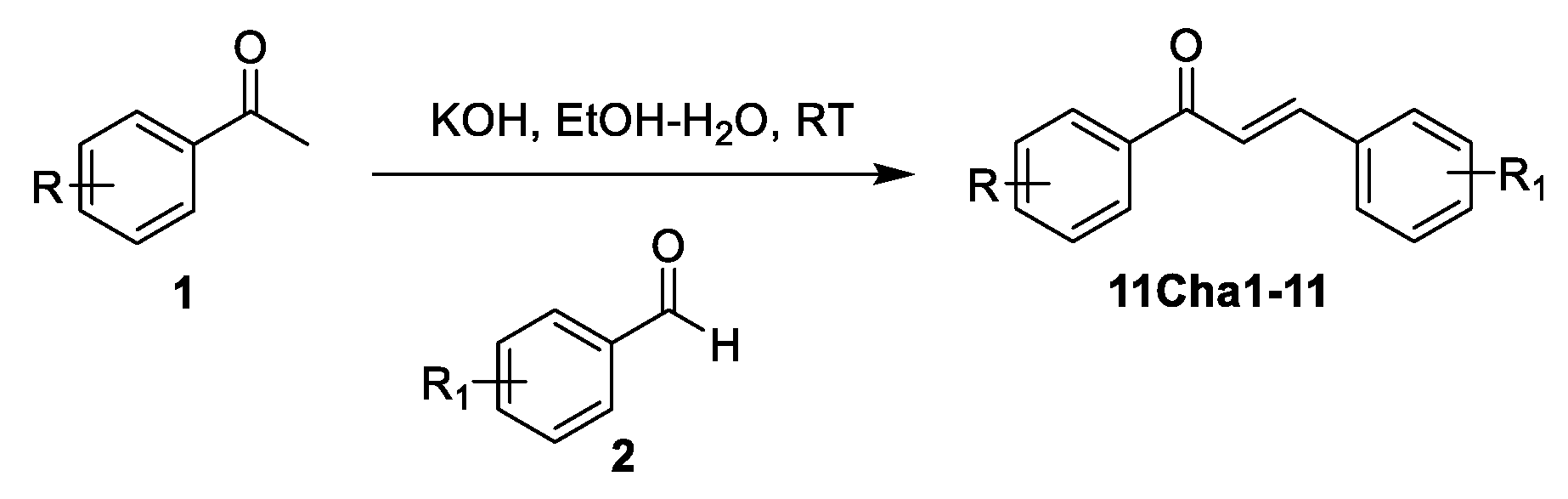
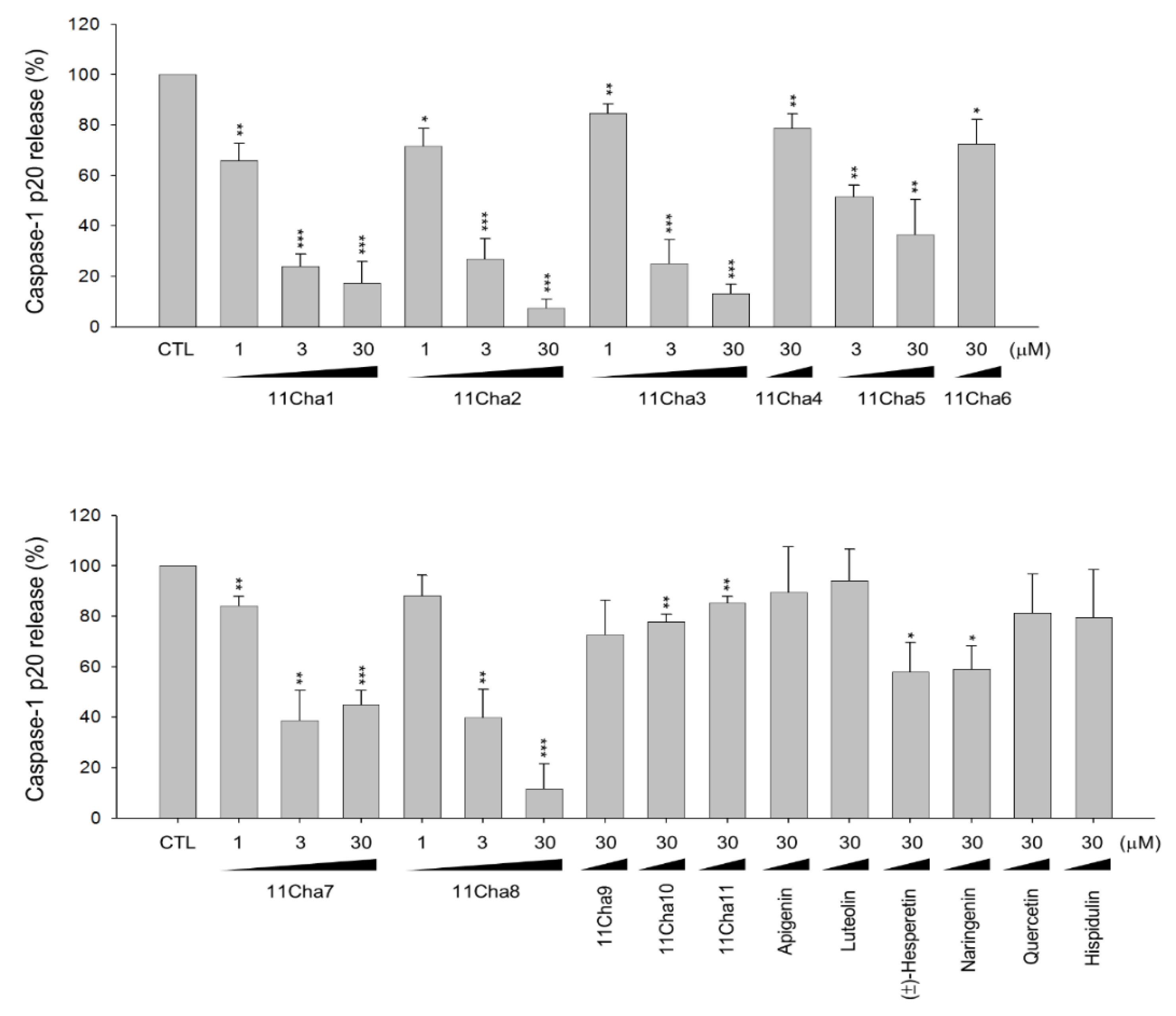
2.1. Detection of Caspase-1 p20 Fragment was Performed to Screen Chalcone Derivatives with Anti-NLRP3 Inflammasome Activity
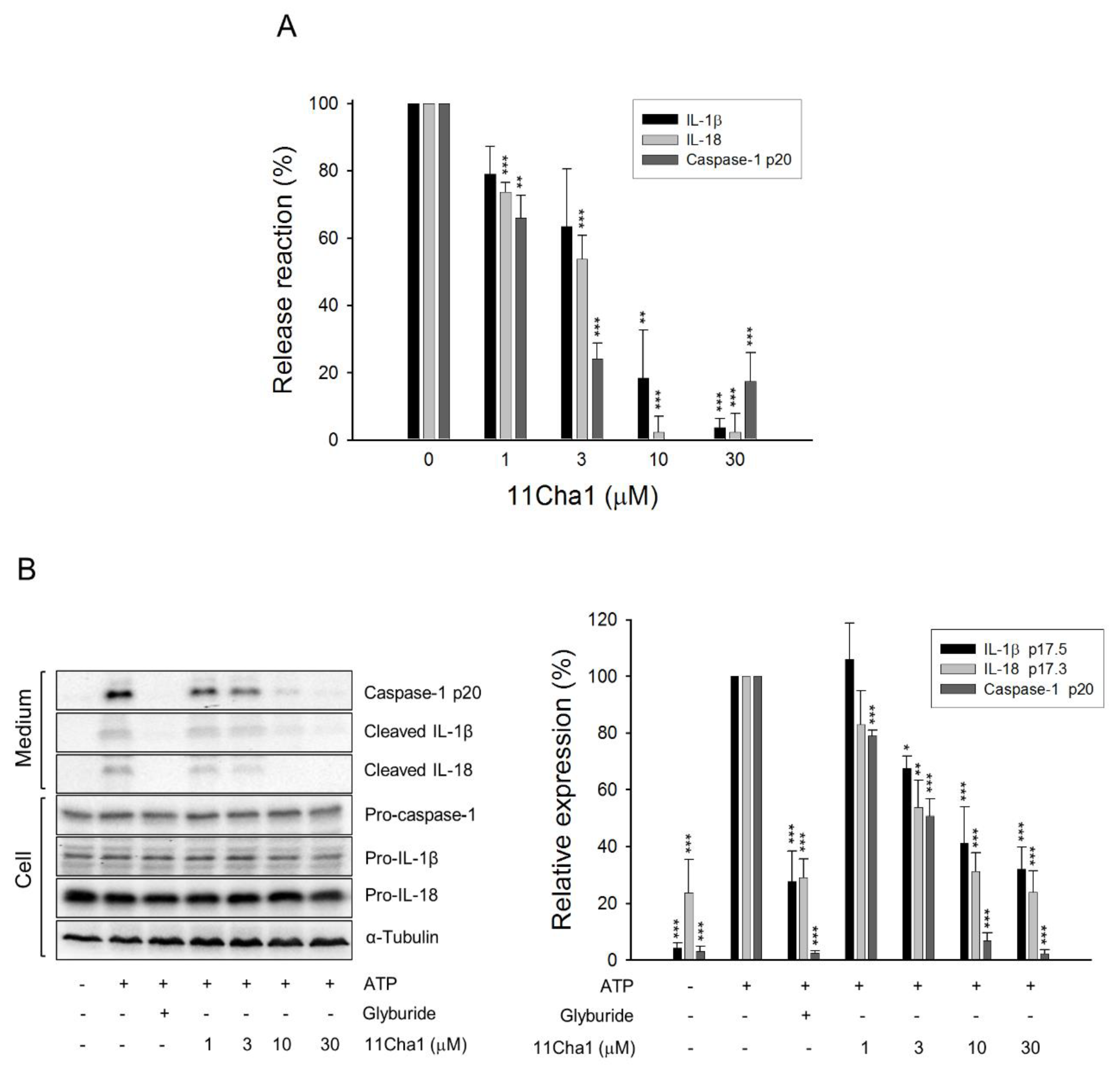
2.2. 11Cha1 Inhibits the Release of Caspase-1 p20 Fragment, IL-1β, and IL-18
2.3. 11Cha1 Inhibits the Activation of NF-ĸB but not MAPKs
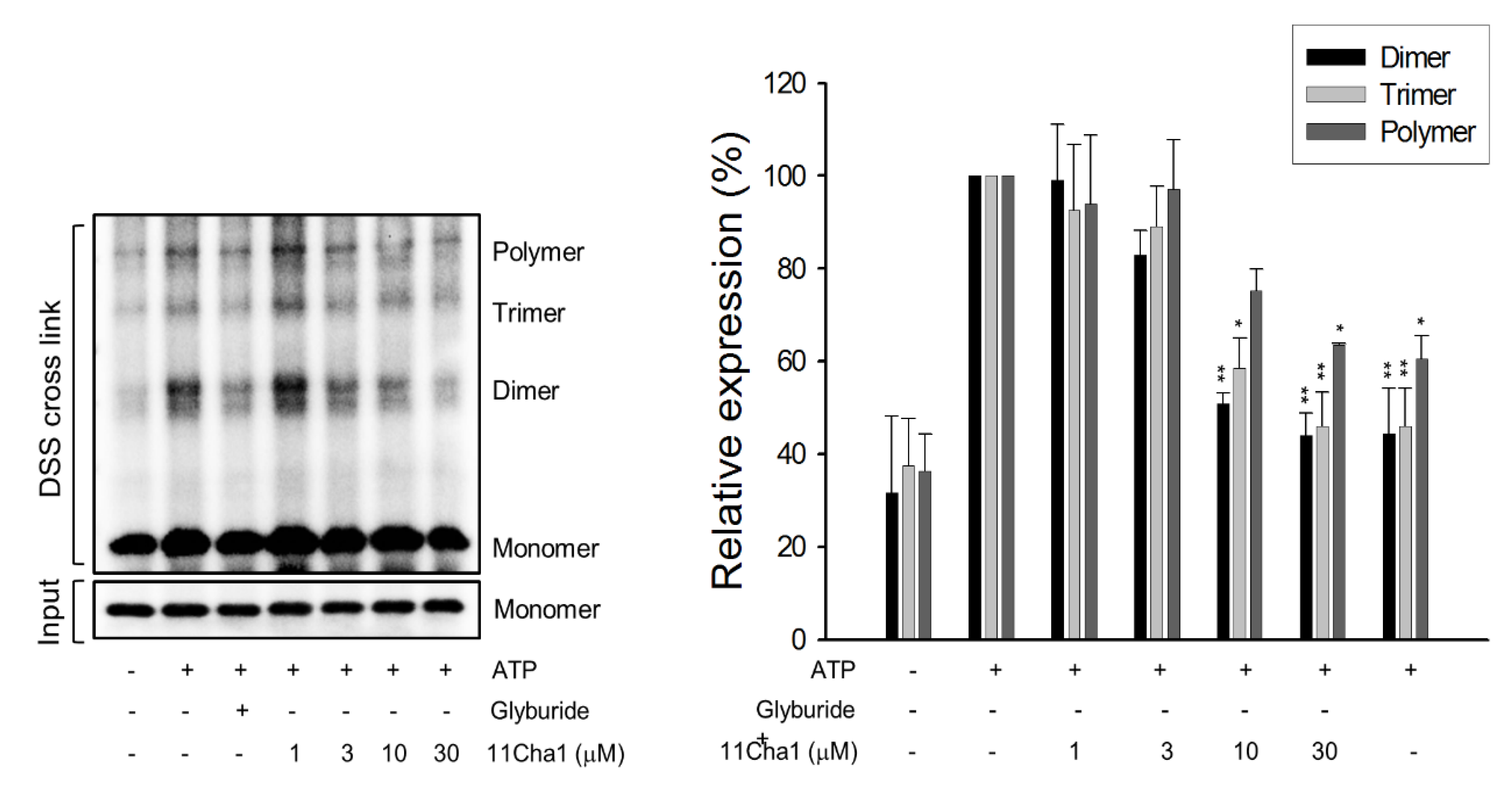
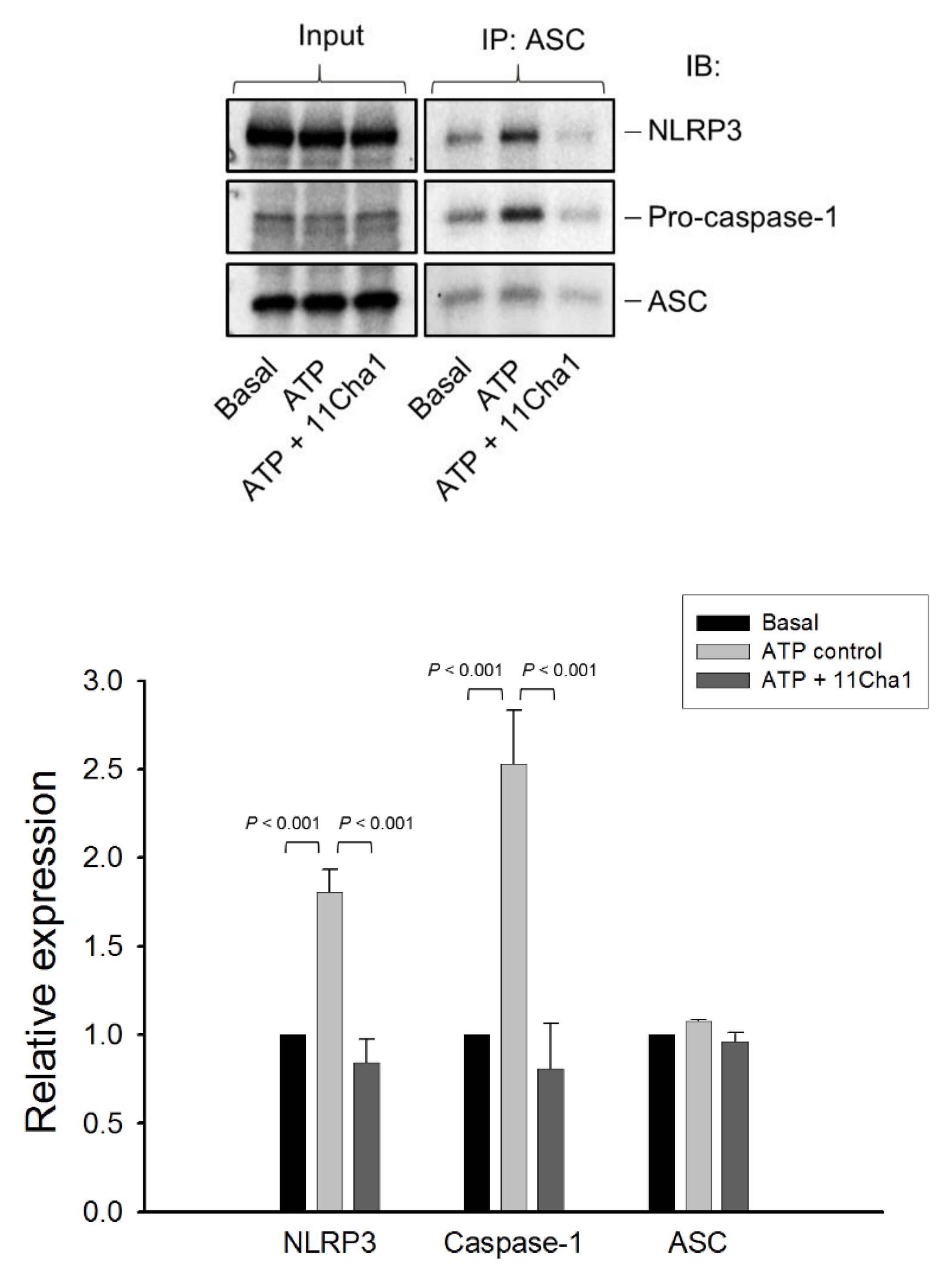
2.4. 11Cha1 Inhibits NLRP3-Dependent ASC Oligomerization and Caspase-1 Activation
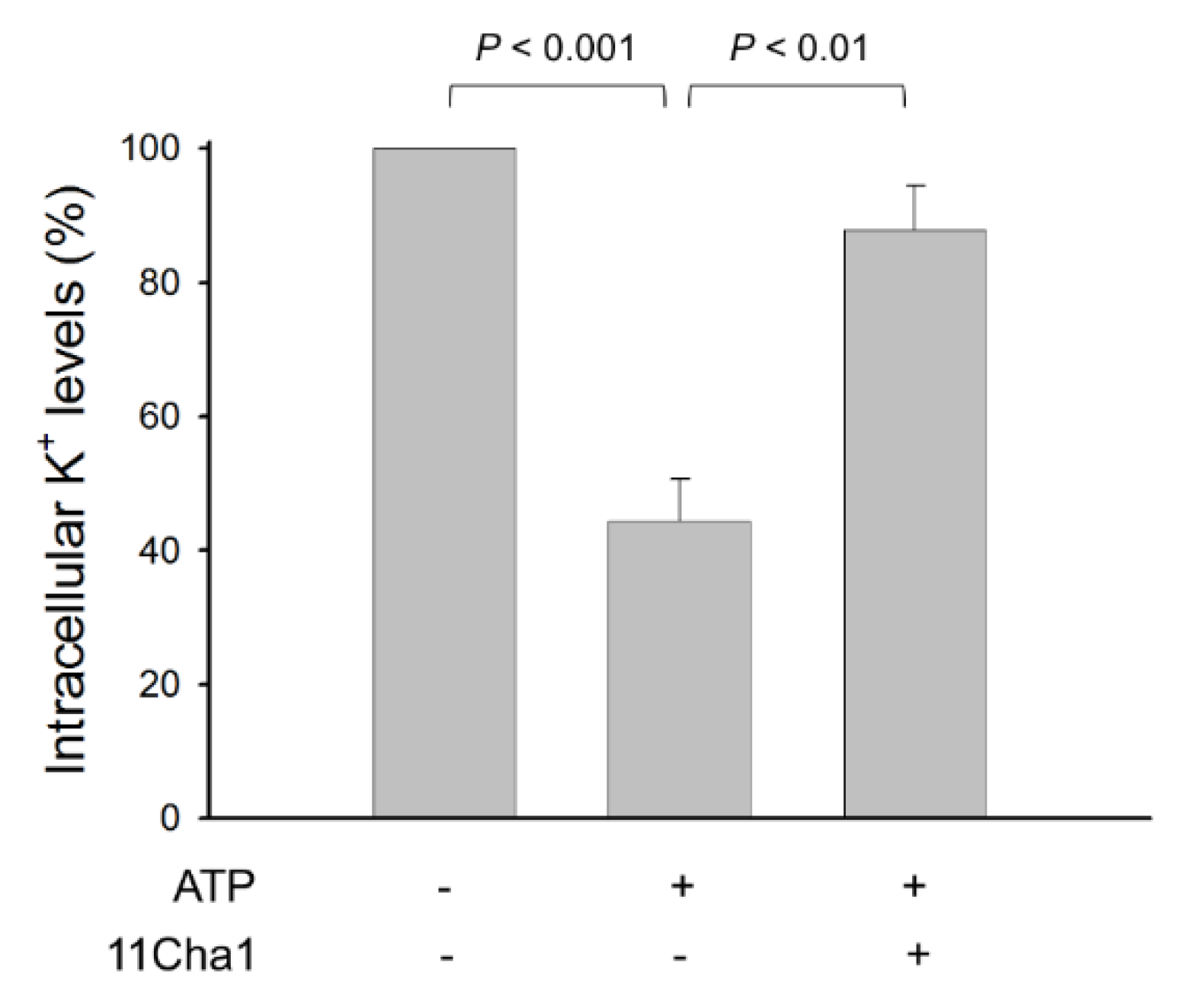
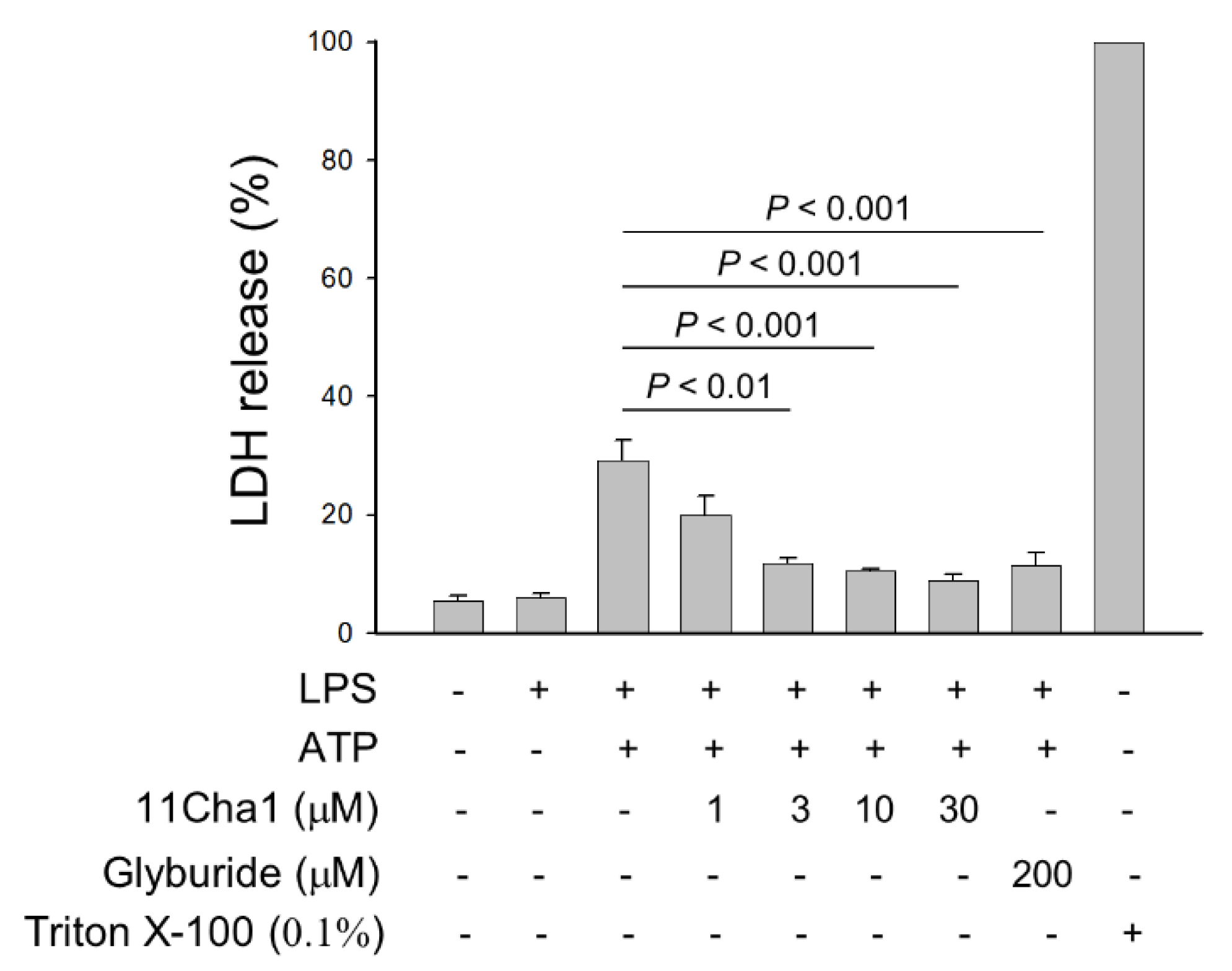
2.5. 11Cha1 Suppresses ATP-Induced K+ Efflux and Pyroptosis in LPS-Primed Macrophages
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Cell Lines and Cell Culture
4.3. MTT Assays
4.4. Cytokine Release
4.5. Western Blotting
4.6. ASC Oligomerization
4.7. Measurement of Intracellular Potassium Content
4.8. Immunoprecipitation Assay
4.9. Lactate Dehydrogenase (LDH) Release
4.10. Data Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ASC | apoptosis-associated speck-like protein containing a CARD |
| ATP | adenosine triphosphate |
| CARD | caspase activation and recruitment domain |
| DAMPs | damage-associated molecular patterns |
| DSS | disuccinimidyl suberate |
| FBS | fetal bovine serum |
| JAK | Janus kinase |
| JNK | c-Jun N-terminal kinase |
| LPS | lipopolysaccharide |
| MAPK | Mitogen-activated protein kinase |
| MK2 | MAPK activated protein kinase 2 |
| mTOR | mammalian target of rapamycin |
| MSU | monosodium urate |
| NF-ĸB | nuclear factor-ĸB |
| NLRP3 | NOD-, LRR- and pyrin domain-containing protein 3 |
| PAMPs | pathogen-associated molecular patterns |
| PI3Ks | phosphoinositide 3-kinases |
| PMA | phorbol 12-myristate 13-acetate |
| PMSF | phenylmethylsulfonylfluoride |
| PYD | pyrin domain |
| STAT | signal transducers and activators of transcription |
| TCA | trichloroacetic acid |
| TLR4 | toll-like receptor 4 |
Appendix A
References
- Panche, A.N.; Diwan, A.D.; Chandra, S.R. Flavonoids: An overview. J. Nutr. Sci. 2016, 5, e47. [Google Scholar] [CrossRef] [PubMed]
- Mateeva, N.; Gangapuram, M.; Mazzio, E.; Eyunni, S.; Soliman, K.F.; Redda, K.K. Biological evaluation of synthetic chalcone and flavone derivatives as anti-inflammatory agents. Med. Chem. Res. 2015, 24, 1672–1680. [Google Scholar] [CrossRef]
- Abd, E.L.; Maksoud, A.I.; Taher, R.F.; Gaara, A.H.; Abdelrazik, E.; Keshk, O.S.; Elawdan, K.A.; Morsy, S.E.; Salah, A.; Khalil, H. Selective Regulation of B-Raf Dependent K-Ras/Mitogen-Activated Protein by Natural Occurring Multi-kinase Inhibitors in Cancer Cells. Front. Oncol. 2019, 9, 1220. [Google Scholar]
- Xiao, N.; Qu, J.; He, S.; Huang, P.; Qiao, Y.; Li, G.; Pan, T.; Sui, H.; Zhang, L. Exploring the Therapeutic Composition and Mechanism of Jiang-Suan-Chu-Bi Recipe on Gouty Arthritis Using an Integrated Approach Based on Chemical Profile, Network Pharmacology and Experimental Support Using Molecular Cell Biology. Front. Pharmacol. 2020, 10, 1626. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Li, T.; Liu, Y.; Cai, T.; Yao, W.; Jiang, J.; He, Y.; Shan, L. Hepatoprotective and Anti-Oxidative Effects of Total Flavonoids From Qu Zhi Qiao (Fruit of Citrus Paradisi cv.Changshanhuyou) on Nonalcoholic Steatohepatitis In Vivo and In Vitro Through Nrf2-ARE Signaling Pathway. Front. Pharmacol. 2020, 11, 483. [Google Scholar] [CrossRef] [PubMed]
- Mu, K.; Wang, D.; Kitts, D.D. Molecular Mechanisms That Define Redox Balance Function in Pathogen-Host Interactions-Is There a Role for Dietary Bioactive Polyphenols? Int. J. Mol. Sci. 2019, 20, 6222. [Google Scholar] [CrossRef]
- Baci, D.; Gallazzi, M.; Cascini, C.; Tramacere, M.; De Stefano, D.; Bruno, A.; Noonan, D.M.; Albini, A. Downregulation of Pro-Inflammatory and Pro-Angiogenic Pathways in Prostate Cancer Cells by a Polyphenol-Rich Extract from Olive Mill Wastewater. Int. J. Mol. Sci. 2019, 20, 307. [Google Scholar] [CrossRef]
- Davis, B.K.; Wen, H.; Ting, J.P. The Inflammasome NLRs in Immunity, Inflammation, and Associated Diseases. Annu. Rev. Immunol. 2011, 29, 707–735. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, H.; Kouadir, M.; Song, H.; Shi, F. Recent advances in the mechanisms of NLRP3 inflammasome activation and its inhibitors. Cell Death Dis. 2019, 10, 128. [Google Scholar] [CrossRef]
- Bauernfeind, F.G.; Horvath, G.; Stutz, A.; Alnemri, E.S.; MacDonald, K.; Speert, D.; Fernandes-Alnemri, T.; Wu, J.; Monks, B.G.; Fitzgerald, K.A.; et al. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J. Immunol. 2009, 183, 787–791. [Google Scholar] [CrossRef]
- Franchi, L.; Eigenbrod, T.; Núñez, G. Cutting edge: TNF-alpha mediates sensitization to ATP and silica via the NLRP3 inflammasome in the absence of microbial stimulation. J. Immunol. 2009, 183, 792–796. [Google Scholar] [CrossRef]
- Owona, B.A.; Abia, W.A.; Moundipa, P.F. Natural compounds flavonoids as modulators of inflammasomes in chronic diseases. Int. Immunopharmacol. 2020, 84, 106498. [Google Scholar] [CrossRef] [PubMed]
- Staurengo-Ferrari, L.; Ruiz-Miyazawa, K.W.; Pinho-Ribeiro, F.A.; Fattori, V.; Zaninelli, T.H.; Badaro-Garcia, S.; Borghi, S.M.; Carvalho, T.T.; Alves-Filho, J.C.; Cunha, T.M.; et al. Trans-Chalcone Attenuates Pain and Inflammation in Experimental Acute Gout Arthritis in Mice. Front. Pharmacol. 2018, 9, 1123. [Google Scholar] [CrossRef]
- Woo, J.; Cho, S.; Lee, C.J. Isoliquiritigenin, a chalcone compound, enhances spontaneous inhibitory postsynaptic response. Exp. Neurobiol. 2014, 23, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Honda, H.; Nagai, Y.; Matsunaga, T.; Okamoto, N.; Watanabe, Y.; Tsuneyama, K.; Hayashi, H.; Fujii, I.; Ikutani, M.; Hirai, Y.; et al. Isoliquiritigenin is a potent inhibitor of NLRP3 inflammasome activation and diet-induced adipose tissue inflammation. J. Leukoc. Biol. 2014, 96, 1087–1100. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Wu, G.; Chen, S.; Zhang, L.; Li, F.; Shao, T.; Ren, L.; Chen, S.Y.; Zhang, H.; McClain, C.J.; et al. Chalcone Derivative L6H21 Reduces EtOH + LPS-Induced Liver Injury Through Inhibition of NLRP3 Inflammasome Activation. Alcohol Clin. Exp. Res. 2019, 43, 1662–1671. [Google Scholar] [CrossRef]
- Lv, H.; Yang, H.; Wang, Z.; Feng, H.; Deng, X.; Cheng, G.; Ci, X. Nrf2 signaling and autophagy are complementary in protecting lipopolysaccharide/d-galactosamine-induced acute liver injury by licochalcone A. Cell Death Dis. 2019, 10, 313. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, S.; Cao, H.; Shen, P.; Liu, J.; Fu, Y.; Cao, Y.; Zhang, N. The protective role of phloretin against dextran sulfate sodium-induced ulcerative colitis in mice. Food Funct. 2019, 10, 422–431. [Google Scholar] [CrossRef]
- Yang, G.; Lee, H.E.; Yeon, S.H.; Kang, H.C.; Cho, Y.Y.; Lee, H.S.; Zouboulis, C.C.; Han, S.H.; Lee, J.H.; Lee, J.Y. Licochalcone A attenuates acne symptoms mediated by suppression of NLRP3 inflammasome. Phytother. Res. 2018, 32, 2551–2559. [Google Scholar] [CrossRef]
- Tang, Y.L.; Zheng, X.; Qi, Y.; Pu, X.J.; Liu, B.; Zhang, X.; Li, X.S.; Xiao, W.L.; Wan, C.P.; Mao, Z.W. Synthesis and anti-inflammatory evaluation of new chalcone derivatives bearing bispiperazine linker as IL-1β inhibitors. Bioorg. Chem. 2020, 98, 103748. [Google Scholar] [CrossRef]
- He, Y.; Hara, H.; Núñez, G. Mechanism and Regulation of NLRP3 Inflammasome Activation. Trends Biochem. Sci. 2016, 41, 1012–1021. [Google Scholar] [CrossRef] [PubMed]
- Leu, W.J.; Chen, J.C.; Guh, J.H. Extract from Plectranthus amboinicus Inhibit Maturation and Release of Interleukin 1β Through Inhibition of NF-κB Nuclear Translocation and NLRP3 Inflammasome Activation. Front. Pharmacol. 2019, 10, 573. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-κB signaling in inflammation. Signal Transduct. Target Ther. 2017, 2, 17023. [Google Scholar] [CrossRef] [PubMed]
- Brasier, A.R. The nuclear factor-kappaB-interleukin-6 signalling pathway mediating vascular inflammation. Cardiovasc. Res. 2010, 86, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Fann, D.Y.; Lim, Y.A.; Cheng, Y.L.; Lok, K.Z.; Chunduri, P.; Baik, S.H.; Drummond, G.R.; Dheen, S.T.; Sobey, C.G.; Jo, D.G.; et al. Evidence that NF-κB and MAPK Signaling Promotes NLRP Inflammasome Activation in Neurons Following Ischemic Stroke. Mol. Neurobiol. 2018, 55, 1082–1096. [Google Scholar] [CrossRef] [PubMed]
- Schroder, K.; Zhou, R.; Tschopp, J. The NLRP3 inflammasome: A sensor for metabolic danger? Science 2010, 327, 296–300. [Google Scholar] [CrossRef]
- Kelley, N.; Jeltema, D.; Duan, Y.; He, Y. The NLRP3 Inflammasome: An overview of mechanisms of activation and Regulation. Int. J. Mol. Sci. 2019, 20, 3328. [Google Scholar] [CrossRef]
- Muñoz-Planillo, R.; Kuffa, P.; Martínez-Colón, G.; Smith, B.L.; Rajendiran, T.M.; Núñez, G. K⁺ efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity 2013, 38, 1142–1153. [Google Scholar] [CrossRef]
- Xue, Y.; Enosi Tuipulotu, D.; Tan, W.H.; Kay, C.; Man, S.M. Emerging Activators and Regulators of Inflammasomes and Pyroptosis. Trends Immunol. 2019, 40, 1035–1052. [Google Scholar] [CrossRef]
- Marie, J.; Kovacs, D.; Pain, C.; Jouary, T.; Cota, C.; Vergier, B.; Picardo, M.; Taieb, A.; Ezzedine, K.; Cario-André, M. Inflammasome activation and vitiligo/nonsegmental vitiligo progression. Br. J. Dermatol. 2014, 170, 816–823. [Google Scholar] [CrossRef]
- Zhen, Y.; Zhang, H. NLRP3 Inflammasome and Inflammatory Bowel Disease. Front. Immunol. 2019, 10, 276. [Google Scholar] [CrossRef] [PubMed]
- Pellegrini, C.; Fornai, M.; Antonioli, L.; Blandizzi, C.; Calderone, V. Phytochemicals as novel therapeutic strategies for NLRP3 inflammasome-related neurological, metabolic, and inflammatory diseases. Int. J. Mol. Sci. 2019, 20, 2876. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Yang, Q.; Chen, J.; Yu, C.; Zhang, L.; Zhou, W.; Chen, M. Salvianolic acid A ameliorates early-stage atherosclerosis development by inhibiting NLRP3 inflammasome activation in Zucker diabetic fatty rats. Molecules 2020, 25, 1089. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Akira, S. Signaling to NF-kappaB by Toll-like receptors. Trends Mol. Med. 2007, 13, 460–469. [Google Scholar] [CrossRef]
- Song, N.; Liu, Z.S.; Xue, W.; Bai, Z.F.; Wang, Q.Y.; Dai, J.; Liu, X.; Huang, Y.J.; Cai, H.; Zhan, X.Y.; et al. NLRP3 phosphorylation is an essential priming event for inflammasome activation. Mol. Cell 2017, 68, 185–197. [Google Scholar] [CrossRef]
- Ghonime, M.G.; Shamaa, O.R.; Das, S.; Eldomany, R.A.; Fernandes-Alnemri, T.; Alnemri, E.S.; Gavrilin, M.A.; Wewers, M.D. Inflammasome priming by lipopolysaccharide is dependent upon ERK signaling and proteasome function. J. Immunol. 2014, 192, 3881–3888. [Google Scholar] [CrossRef]
- Song, N.; Li, T. Regulation of NLRP3 Inflammasome by Phosphorylation. Front. Immunol. 2018, 9, 2305. [Google Scholar] [CrossRef]
- Yeung, Y.T.; Aziz, F.; Guerrero-Castilla, A.; Arguelles, S. Signaling pathways in inflammation and anti-inflammatory therapies. Curr. Pharm. Des. 2018, 24, 1449–1484. [Google Scholar] [CrossRef]
- Wang, C.; Hockerman, S.; Jacobsen, E.J.; Alippe, Y.; Selness, S.R.; Hope, H.R.; Hirsch, J.L.; Mnich, S.J.; Saabye, M.J.; Hood, W.F.; et al. Selective inhibition of the p38α MAPK-MK2 axis inhibits inflammatory cues including inflammasome priming signals. J. Exp. Med. 2018, 215, 1315–1325. [Google Scholar] [CrossRef]
- Abais, J.M.; Xia, M.; Zhang, Y.; Boini, K.M.; Li, P.L. Redox regulation of NLRP3 inflammasomes: ROS as trigger or effector? Antioxid. Redox Signal. 2015, 22, 1111–1129. [Google Scholar] [CrossRef]
- Bauernfeind, F.; Bartok, E.; Rieger, A.; Franchi, L.; Núñez, G.; Hornung, V. Cutting edge: Reactive oxygen species inhibitors block priming, but not activation, of the NLRP3 inflammasome. J. Immunol. 2011, 187, 613–617. [Google Scholar] [CrossRef] [PubMed]
- Van Bruggen, R.; Köker, M.Y.; Jansen, M.; van Houdt, M.; Roos, D.; Kuijpers, T.W.; van den Berg, T.K. Human NLRP3 inflammasome activation is Nox1-4 independent. Blood 2010, 115, 5398–5400. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Zeng, M.Y.; Yang, D.; Motro, B.; Núñez, G. NEK7 is an essential mediator of NLRP3 activation downstream of potassium efflux. Nature 2016, 530, 354–357. [Google Scholar] [CrossRef] [PubMed]
- Groß, C.J.; Mishra, R.; Schneider, K.S.; Médard, G.; Wettmarshausen, J.; Dittlein, D.C.; Shi, H.; Gorka, O.; Koenig, P.A.; Fromm, S.; et al. K+efflux-independent NLRP3 inflammasome activation by small molecules targeting mitochondria. Immunity 2016, 45, 761–773. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Tian, Y.; Yin, Q. AIM2 Inflammasome Assembly and Signaling. Adv. Exp. Med. Biol. 2019, 1172, 143–155. [Google Scholar] [PubMed]

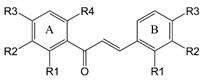 | 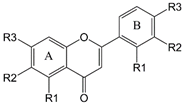 | 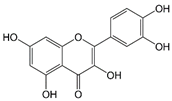 | ||||||
| Chalcones | Flavonoids | Quercetin | ||||||
| A ring | B ring | |||||||
| Chalcones | −R1 | −R2 | −R3 | −R4 | −R1 | −R2 | −R3 | IC50 (μM) |
| 11Cha1 | −OH | −H | −OCH3 | −OCH3 | −H | −H | −COOH | 1.5 |
| 11Cha2 | −OH | −H | −OMOM a | −OMOM a | −H | −H | −COOH | 1.7 |
| 11Cha3 | −H | −H | −OCH3 | −H | −H | −Br | −OH | 1.9 |
| 11Cha4 | −H | −H | −OCH3 | −H | −H | −H | −I | >30 |
| 11Cha5 | −H | −H | −OH | −H | −H | −H | −OCH2COOCH3 | 3.8 |
| 11Cha6 | −OMOM a | −OCH3 | −OMOM a | −OH | −H | −H | −OBn b | >30 |
| 11Cha7 | −H | −H | −OBn b | −H | −H | −H | −OH | 2.3 |
| 11Cha8 | −H | −H | −H | −H | −OCH2COOCH2CH3 | −H | −H | 2.4 |
| 11Cha9 | −OBn b | −I | −OMOM a | −OH | −H | −H | −OBn b | >30 |
| 11Cha10 | −OBn b | −OCH3 | −OH | −OH | −H | −H | −OBn b | >30 |
| 11Cha11 | −OBn b | −OCD3 | −OH | −OH | −H | −H | −OBn b | >30 |
| Flavonoids | −R1 | −R2 | −R3 | −R1 | −R2 | −R3 | Release (%) | IC50 (μM) |
| Control | 100±0 | |||||||
| Apigenin | −OH | −H | −OH | −H | −H | −OH | 89.5±18.1c | >30 |
| Luteolin | −OH | −H | −OH | −H | −OH | −OH | 94.2±12.4 c | >30 |
| (±)−Hesperetin | −OH | −H | −OH | −H | −OH | −OCH3 | 58.1±11.6 c | >30 |
| Naringenin | −OH | −H | −OH | −H | −H | −OH | 59.0±9.3 c | >30 |
| Hispidulin | −OH | −OCH3 | −OH | −H | −H | −OH | 79.6±19.1c | >30 |
| Quercetin | 81.3±15.6 c | >30 | ||||||
Sample Availability: Samples of the compounds are not available from the authors. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leu, W.-J.; Chu, J.-C.; Hsu, J.-L.; Du, C.-M.; Jiang, Y.-H.; Hsu, L.-C.; Huang, W.-J.; Guh, J.-H. Chalcones Display Anti-NLRP3 Inflammasome Activity in Macrophages through Inhibition of Both Priming and Activation Steps—Structure-Activity-Relationship and Mechanism Studies. Molecules 2020, 25, 5960. https://doi.org/10.3390/molecules25245960
Leu W-J, Chu J-C, Hsu J-L, Du C-M, Jiang Y-H, Hsu L-C, Huang W-J, Guh J-H. Chalcones Display Anti-NLRP3 Inflammasome Activity in Macrophages through Inhibition of Both Priming and Activation Steps—Structure-Activity-Relationship and Mechanism Studies. Molecules. 2020; 25(24):5960. https://doi.org/10.3390/molecules25245960
Chicago/Turabian StyleLeu, Wohn-Jenn, Jung-Chun Chu, Jui-Ling Hsu, Chi-Min Du, Yi-Huei Jiang, Lih-Ching Hsu, Wei-Jan Huang, and Jih-Hwa Guh. 2020. "Chalcones Display Anti-NLRP3 Inflammasome Activity in Macrophages through Inhibition of Both Priming and Activation Steps—Structure-Activity-Relationship and Mechanism Studies" Molecules 25, no. 24: 5960. https://doi.org/10.3390/molecules25245960
APA StyleLeu, W.-J., Chu, J.-C., Hsu, J.-L., Du, C.-M., Jiang, Y.-H., Hsu, L.-C., Huang, W.-J., & Guh, J.-H. (2020). Chalcones Display Anti-NLRP3 Inflammasome Activity in Macrophages through Inhibition of Both Priming and Activation Steps—Structure-Activity-Relationship and Mechanism Studies. Molecules, 25(24), 5960. https://doi.org/10.3390/molecules25245960






