Figure 1.
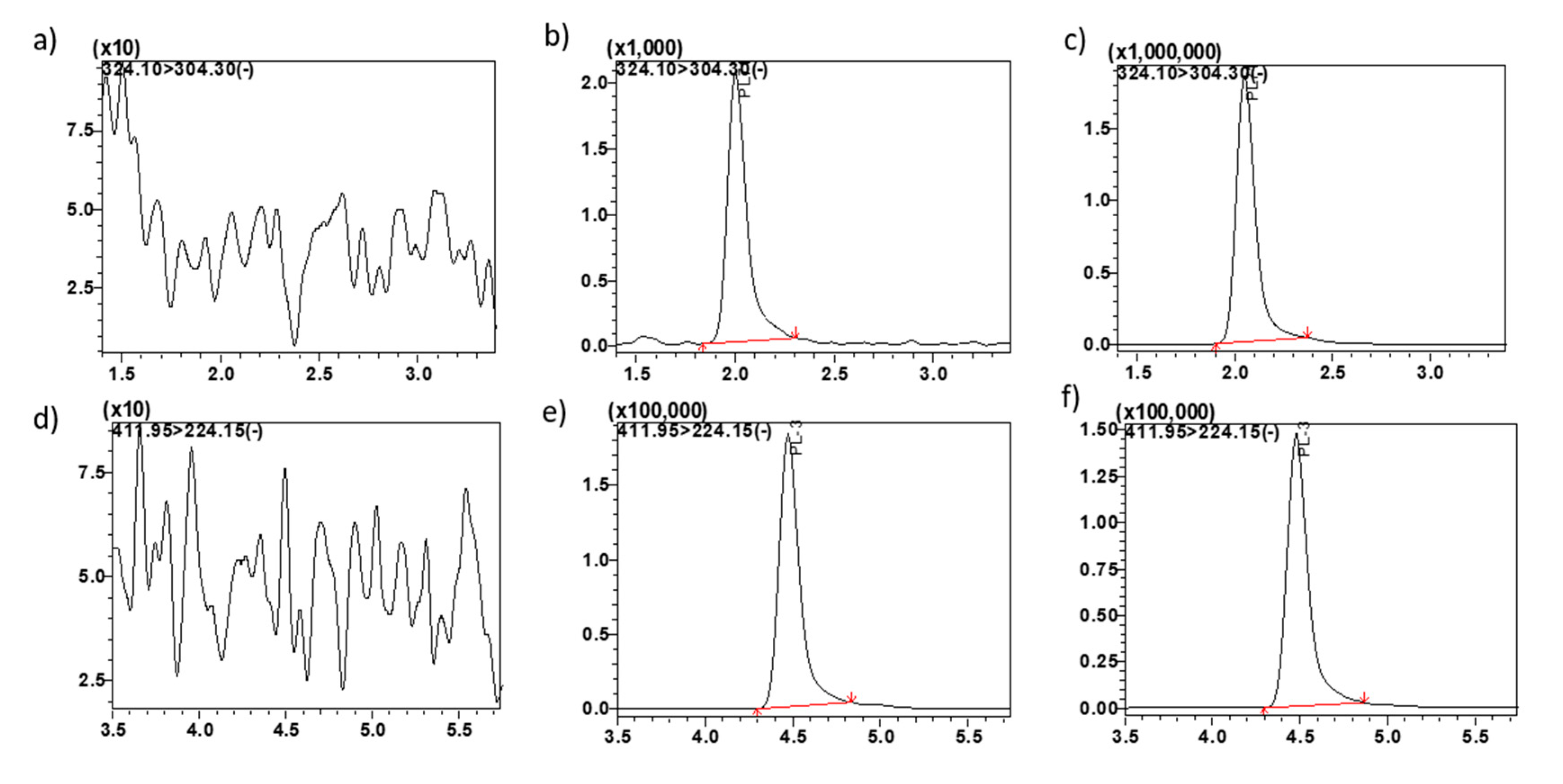
Representative MRM ion-chromatograms. (a) blank mouse plasma using the conditions for MP1 detection, (b) MP1 spiked in mouse plasma at LQC 0.6 ng/mL, (c) mouse plasma from pre-clinical study sample at 0.5 h time point showing MP1, (d) blank plasma using the conditions for PL3 (IS) detection, (e) PL3 (IS) spiked in plasma 0.5 µg/mL, (f) plasma from pre-clinical study sample at 0.5 h time point spiked with IS showing PL3.
Figure 1.
Representative MRM ion-chromatograms. (a) blank mouse plasma using the conditions for MP1 detection, (b) MP1 spiked in mouse plasma at LQC 0.6 ng/mL, (c) mouse plasma from pre-clinical study sample at 0.5 h time point showing MP1, (d) blank plasma using the conditions for PL3 (IS) detection, (e) PL3 (IS) spiked in plasma 0.5 µg/mL, (f) plasma from pre-clinical study sample at 0.5 h time point spiked with IS showing PL3.
Figure 2.
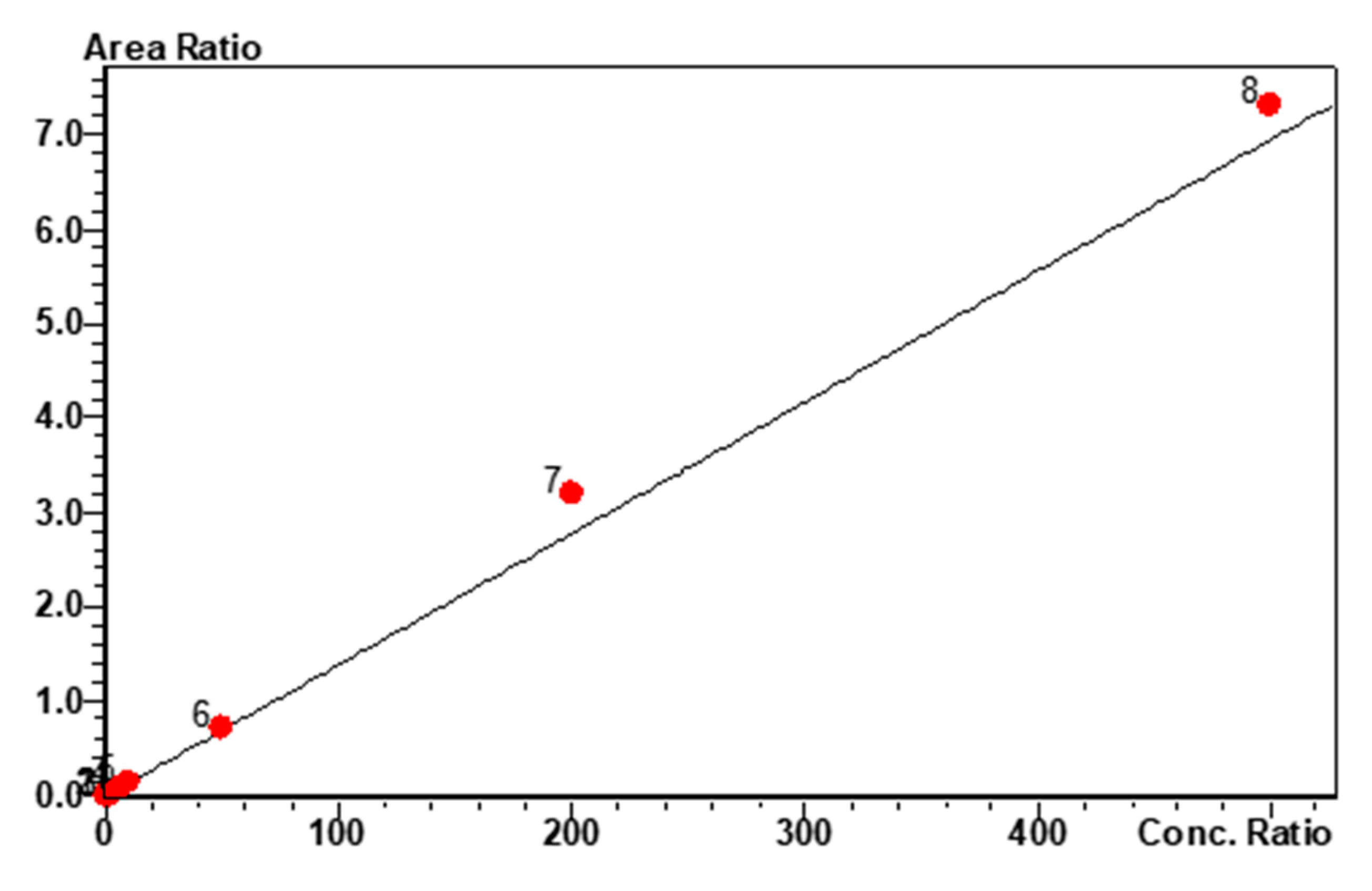
MP1 calibration curve and linearity. Linear calibration fit for MP1 over the concentration range of 0.2 to 500 ng/mL, r2 = 0.988 and %RSD = 9.97.
Figure 2.
MP1 calibration curve and linearity. Linear calibration fit for MP1 over the concentration range of 0.2 to 500 ng/mL, r2 = 0.988 and %RSD = 9.97.
Figure 3.
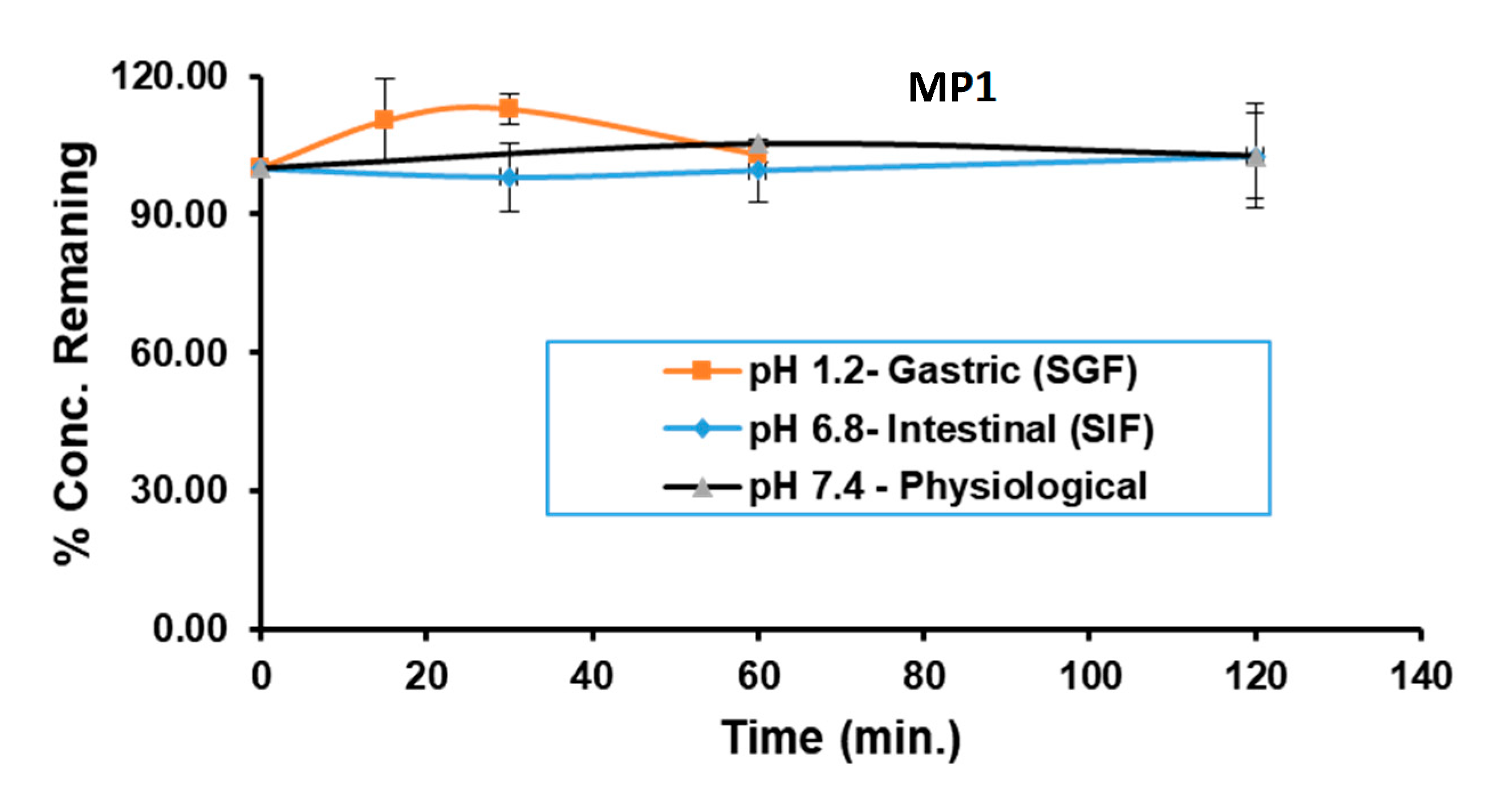
Gastrointestinal Stability of MP1. MP1 stability following a 1 h incubation in SGF (pH 1.2), a 2 h incubation in SIF (pH 6.8) and in buffer (pH 7.4).
Figure 3.
Gastrointestinal Stability of MP1. MP1 stability following a 1 h incubation in SGF (pH 1.2), a 2 h incubation in SIF (pH 6.8) and in buffer (pH 7.4).
Figure 4.
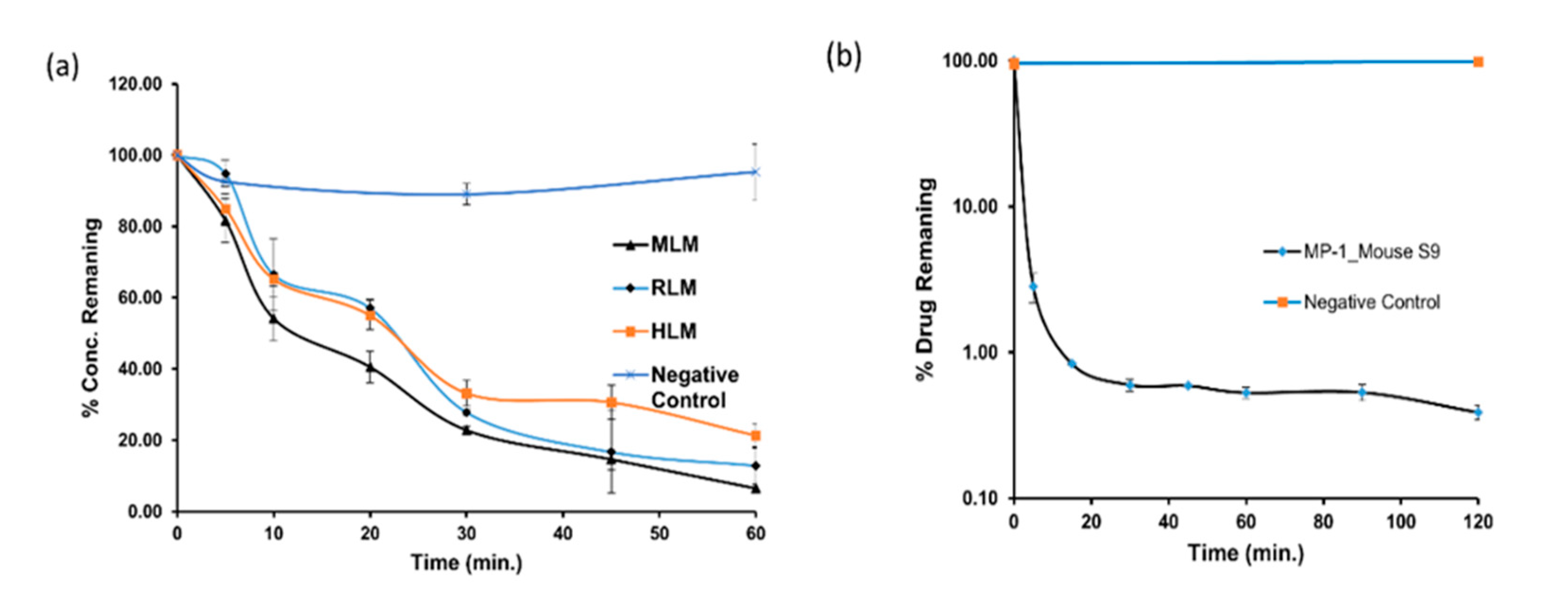
MP1 In-vitro metabolic stability in microsomes. Time-dependent metabolic depletion (% turnover or amount of drug remaining vs. incubation time) of MP1 in microsomes and mouse S9 fraction (a) interspecies microsomal metabolic stability of MP1 in presence of NADPH and absence of NADPH as a negative control (b) mouse S9 metabolic stability of MP1 in presence of NADPH and absence of NADPH as a negative control. Data shown as mean ± SD (n = 3).
Figure 4.
MP1 In-vitro metabolic stability in microsomes. Time-dependent metabolic depletion (% turnover or amount of drug remaining vs. incubation time) of MP1 in microsomes and mouse S9 fraction (a) interspecies microsomal metabolic stability of MP1 in presence of NADPH and absence of NADPH as a negative control (b) mouse S9 metabolic stability of MP1 in presence of NADPH and absence of NADPH as a negative control. Data shown as mean ± SD (n = 3).
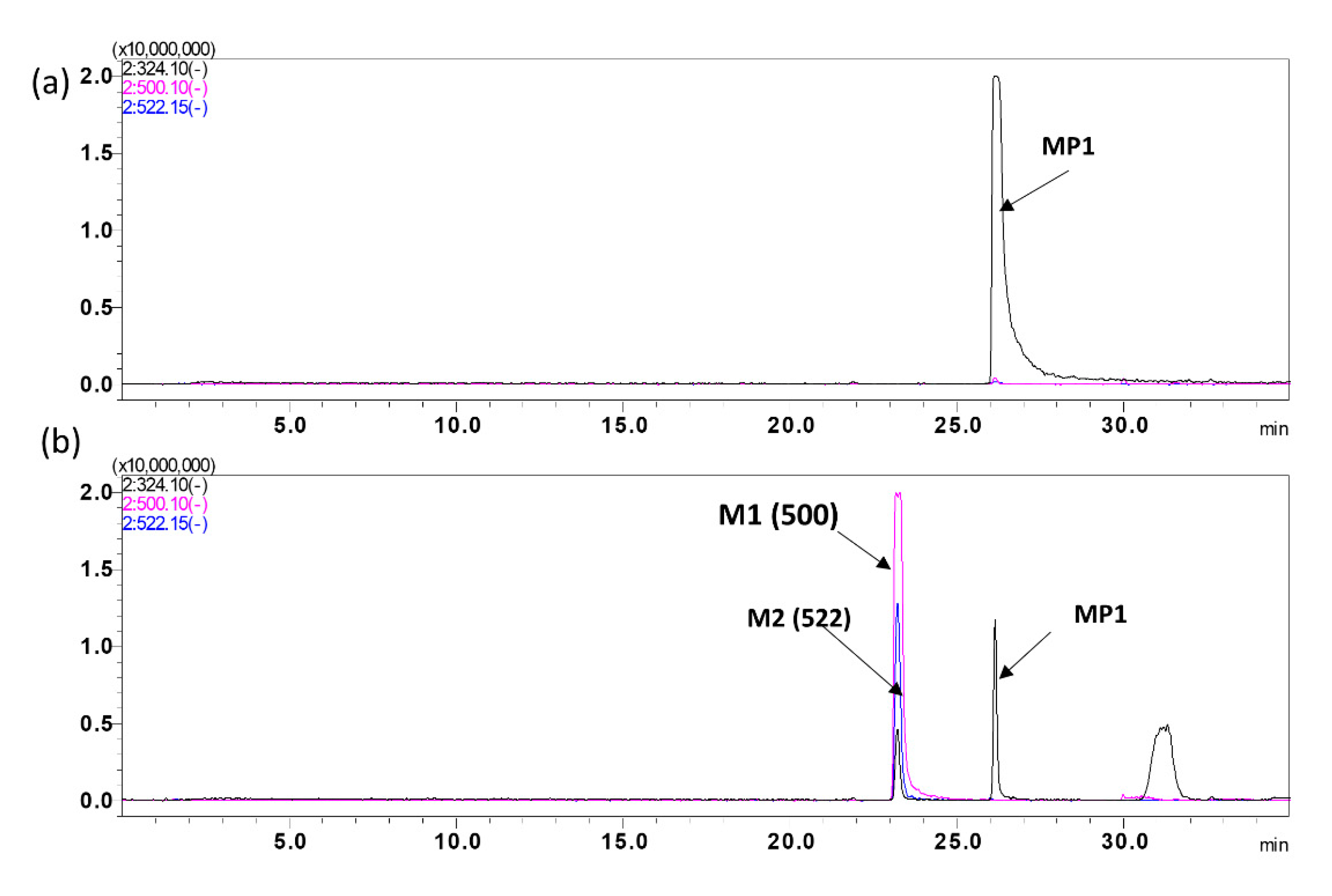
Figure 5.
MP1 Metabolite identification. Representative overly chromatogram of MP1 and its metabolites, utilizing a 35 min gradient for separation after, (a) 0 min incubation, and (b) 90 min incubation.
Figure 5.
MP1 Metabolite identification. Representative overly chromatogram of MP1 and its metabolites, utilizing a 35 min gradient for separation after, (a) 0 min incubation, and (b) 90 min incubation.
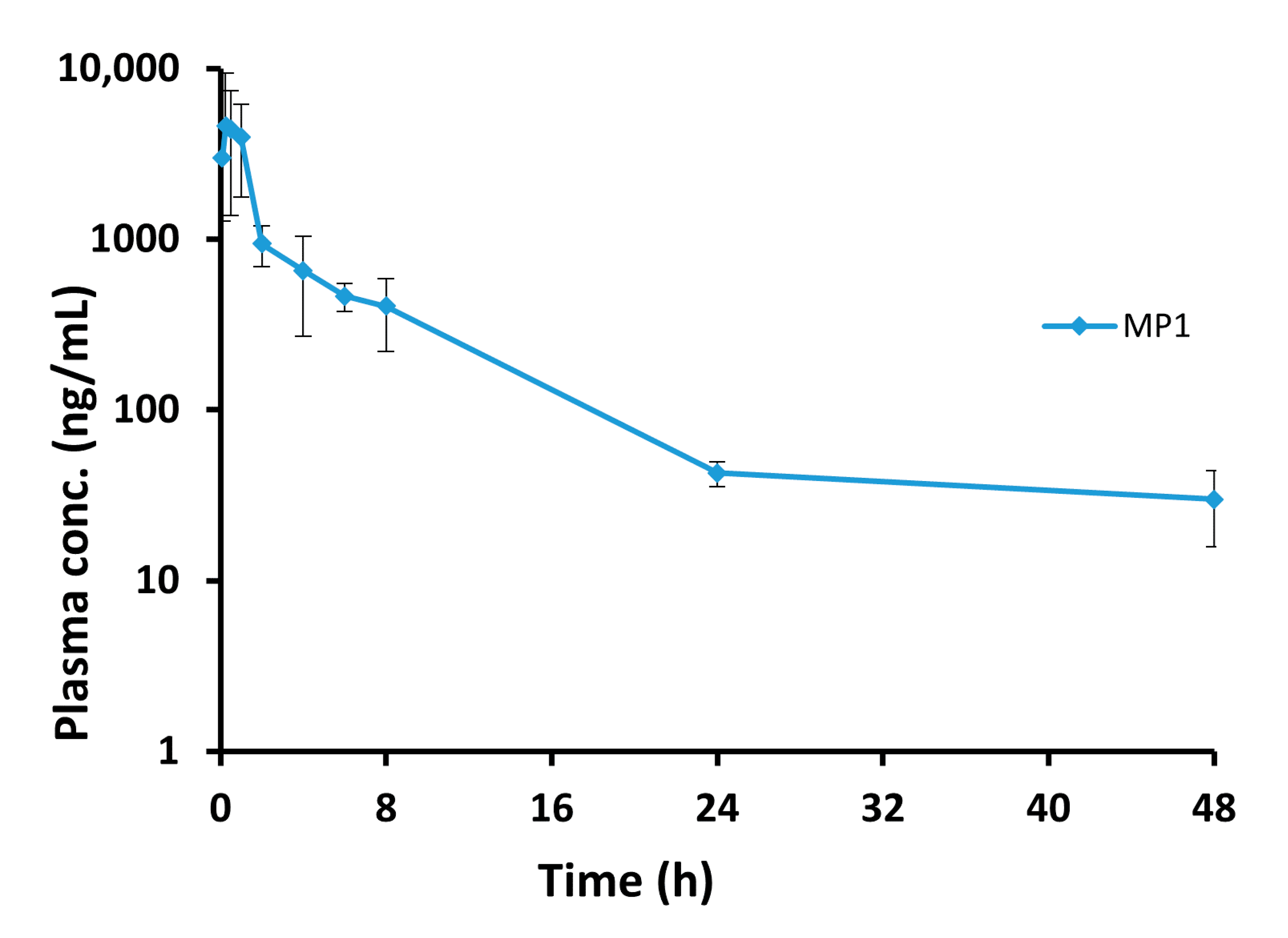
Figure 6.
The blood concentration vs. time profile for the MP1. Plasma concentration-time profile after 15 mg/kg oral administration of MP1 (Mean ± SD, n = 4 at each time).
Figure 6.
The blood concentration vs. time profile for the MP1. Plasma concentration-time profile after 15 mg/kg oral administration of MP1 (Mean ± SD, n = 4 at each time).
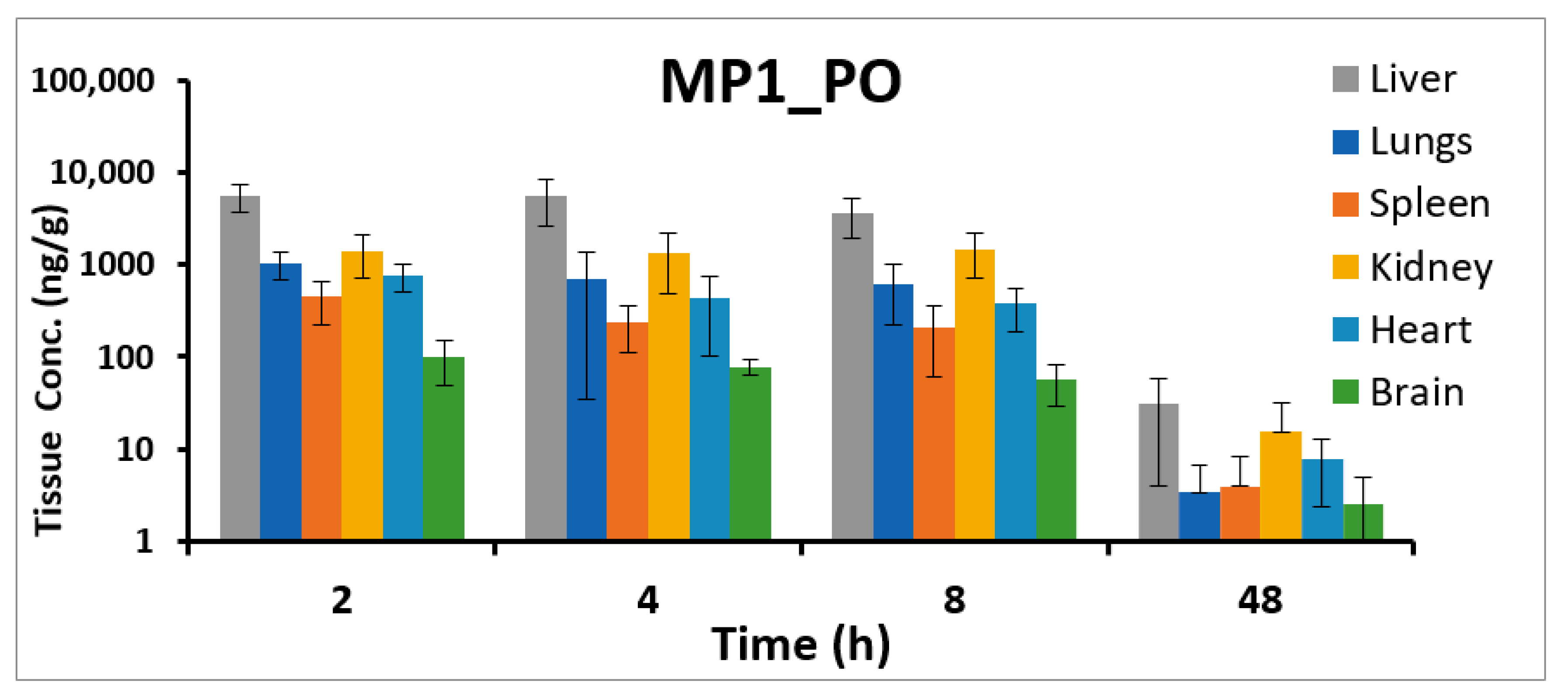
Figure 7.
MP1 tissue concentrations. Tissue concentrations (ng/g) at different time points in liver, lungs, heart, kidney, brain, and spleen after 15 mg/kg oral administration of MP1 in mice (Mean ± SD, n = 4).
Figure 7.
MP1 tissue concentrations. Tissue concentrations (ng/g) at different time points in liver, lungs, heart, kidney, brain, and spleen after 15 mg/kg oral administration of MP1 in mice (Mean ± SD, n = 4).
Table 1.
MP1 accuracy and precision. Intra–assay and inter–assay accuracy and precision of MP1 in mouse plasma (n = 6).
Table 1.
MP1 accuracy and precision. Intra–assay and inter–assay accuracy and precision of MP1 in mouse plasma (n = 6).
| Nominal Conc. (ng/mL) | Accuracy | Precision |
|---|
| %Bias Intra–Assay | %Bias Inter–Assay | %RSD Intra–Assay | %RSD Inter–Assay |
|---|
| LLOQ (0.2 ng/mL) | 7.3 | 4.8 | 10.8 | 8.3 |
| LQC (0.6 ng/mL) | 3.9 | −2.7 | 4.4 | 12.9 |
| MQC (100 ng/mL) | −4.7 | −14.0 | 1.4 | 13.6 |
| HQC (375 ng/mL) | −13.4 | −12.0 | 5.7 | 4.4 |
Table 2.
Recovery and matrix effect. Assessment of the recovery and matrix effect of MP1 in mouse plasma, (Mean ± SD, n = 3).
Table 2.
Recovery and matrix effect. Assessment of the recovery and matrix effect of MP1 in mouse plasma, (Mean ± SD, n = 3).
| Nominal Conc. (ng/mL) | % Extraction Recoveries
(Mean ± SD, n = 3) | % Matrix Effect
(Mean ± SD, n = 3) |
|---|
| LQC (0.6 ng/mL) | 95.8 ± 2.6 | 103.7 ± 8.6 |
| MQC (100 ng/mL) | 89.1 ± 10.6 | 90.5 ± 4.9 |
| HQC (375 ng/mL) | 90.8 ± 3.4 | 99.6 ± 7.6 |
| Internal standard (IS) (0.5 ng/mL) | 87.3 ± 3.8 | 105.1 ± 11 |
Table 3.
Stability of MP1. Stability was tested in mouse plasma at different storage conditions, (Mean ± SD, n = 3).
Table 3.
Stability of MP1. Stability was tested in mouse plasma at different storage conditions, (Mean ± SD, n = 3).
| Nominal Conc. (ng/mL) | Measured Mean Conc. (ng/mL) | % Accuracy |
|---|
| Autosampler stability 4 °C, Mean ± SD, n = 3. |
| LQC (0.6 ng/mL) | 0.515 ± 0.107 | 85.8 ± 17.8 |
| MQC (100 ng/mL) | 102.8 ± 9.5 | 102.85 ± 9.45 |
| HQC (375 ng/mL) | 340.6 ± 12.8 | 90.8 ± 3.4 |
| Bench-top stability 21 °C, Mean ± SD, n = 3. |
| LQC (0.6 ng/mL) | 0.57 ± 0.03 | 94.2 ± 4.9 |
| MQC (100 ng/mL) | 114.8 ± 2.5 | 114.9 ± 2.6 |
| HQC (375 ng/mL) | 386.5 ± 15.2 | 103 ± 4 |
| Freeze-thaw stability −80 °C, up to 3 Cycle, Mean ± SD, n = 3. |
| LQC (0.6 ng/mL) | 0.67 ± 0.06 | 108.6 ± 8.9 |
| MQC (100 ng/mL) | 102.8 ± 11.95 | 101.4 ± 10 |
| HQC (375 ng/mL) | 366.3 ± 44.9 | 97.7 ± 11.96 |
| Long term stability −80 °C, 12 months, Mean ± SD, n = 3. |
| LQC (0.6 ng/mL) | 0.61± 0.02 | 100.3 ± 4.1 |
| MQC (100 ng/mL) | 92.40 ± 7.92 | 92.4 ± 7.9 |
| HQC (375 ng/mL) | 383.69± 79.94 | 101.8 ±13.5 |
Table 4.
The value MP1 the blood to plasma ratio (Kb/p). MP1 concentration in blood and plasma, (Mean ± SD, n = 3).
Table 4.
The value MP1 the blood to plasma ratio (Kb/p). MP1 concentration in blood and plasma, (Mean ± SD, n = 3).
| Time (min) | Blood Conc. | Plasma Conc. | |
|---|
| Mean (ng/mL) ± SD | Mean (ng/mL) ± SD | Kb/p |
|---|
| 0 | 294.1 ± 11.1 | 727.73 ± 49.16 | 0.40 |
| 30 | 490.7 ± 83.0 | 725.84 ± 157.67 | 0.68 |
| 60 | 490.8 ± 29.8 | 781.54 ± 24.53 | 0.63 |
Table 5.
MP1 Plasma Protein Binding. Plasma protein binding in mouse plasma (Mean ± SD values for MP1, n = 3).
Table 5.
MP1 Plasma Protein Binding. Plasma protein binding in mouse plasma (Mean ± SD values for MP1, n = 3).
| Nominal Conc. (µg/mL) | % Plasma Protein Bound ± SD |
|---|
| MP1 (1 µg/mL) | 99.96 ± 0.03 |
| MP1 (10 µg/mL) | 99.97 ± 0.02 |
Table 6.
In-vitro metabolic stability in mouse S9 fraction. MP1 estimates for CLint, t½ and CLint,H in mouse (MLM), rat (RLM) and human (HLM) liver microsomes, and mouse S9, (Mean ± SD, n = 3).
Table 6.
In-vitro metabolic stability in mouse S9 fraction. MP1 estimates for CLint, t½ and CLint,H in mouse (MLM), rat (RLM) and human (HLM) liver microsomes, and mouse S9, (Mean ± SD, n = 3).
| Parameters | MLM | RLM | HLM | Mouse S9 |
|---|
| t½ (min) | 15.64 ± 0.46 | 18.1± 3.73 | 27.47 ± 2.68 | 4.9 ± 0.2 |
| CLint (µL/min/mg protein) | 88.72 ± 2.66 | 80.01 ± 16.5 | 50.99 ± 5.25 | 142.3 ± 5.6 |
| CLint,H (mL/min/kg wgt) | 479.07 ± 14.36 | 288.02 ± 59.41 | 81.58 ± 8.39 | N/A |
Table 7.
MP1 metabolites chromatogram summary. MP1 metabolites biotransformation, mass shift, precursor ion and retention time.
Table 7.
MP1 metabolites chromatogram summary. MP1 metabolites biotransformation, mass shift, precursor ion and retention time.
| Peak ID | Biotransformation | Mass Shift | Precursor Ion (m/z) | Retention Time (Minutes) |
|---|
| | Parent (MP1) | 0 | 324.10 | 26.7 |
| M1 | Glucuronidation | 176 | 500.12 | 23.0 |
| M2 | Unknown | 198 | 522.15 | 23.0 |
Table 8.
MP1 plasma concentrations following oral administration. (15 mg/kg) to mice (Mean ± SD, n = 4).
Table 8.
MP1 plasma concentrations following oral administration. (15 mg/kg) to mice (Mean ± SD, n = 4).
| Time (h) | Mean Plasma Conc. (ng/mL) | SD |
|---|
| 0.08 | 2990.2 | 1705.6 |
| 0.25 | 2452.1 | 313.7 |
| 0.50 | 4412.7 | 3038.8 |
| 1 | 3985.7 | 2222.2 |
| 2 | 943.2 | 251.6 |
| 4 | 655.3 | 385.4 |
| 6 | 465.8 | 87.1 |
| 8 | 405.9 | 184.5 |
| 24 | 42.7 | 7.1 |
| 48 | 30.1 | 14.2 |
Table 9.
Summary of MP1 pharmacokinetic parameters. Pharmacokinetic parameters of MP1 after 15 mg/kg oral administration (Mean, SD, SE and CV%, n = 20).
Table 9.
Summary of MP1 pharmacokinetic parameters. Pharmacokinetic parameters of MP1 after 15 mg/kg oral administration (Mean, SD, SE and CV%, n = 20).
| PK Parameter | Mean | SD | SE | CV% |
|---|
| Cmax (ng/mL) | 4714.7 | ±2343.5 | 1171.8 | 49.7 |
| Tmax (h) | 0.6 | ±0.4 | 0.2 | 68.9 |
| t1/2 (h) | 9.2 | ±1.7 | 0.9 | 18.7 |
| AUC0–∞ (h × ng/mL) | 15367.8 | 5466.9 | 2733.4 | 35.6 |
| AUC0–last (h × ng/mL) | 14951.7 | ±5357.2 | 2678.6 | 35.8 |
| Vd (L/kg) | 13.9 | ±4.3 | 2.1 | 30.9 |
| CL (L/h/kg) | 1.1 | ±0.3 | 0.2 | 31.6 |
Table 10.
MP1 tissue concentrations. Tissue concentrations (ng/g) in liver, lungs, heart, kidney, brain, and spleen after 15 mg/kg oral administration of MP1 in mice (Mean ± SD, n = 4 at each time).
Table 10.
MP1 tissue concentrations. Tissue concentrations (ng/g) in liver, lungs, heart, kidney, brain, and spleen after 15 mg/kg oral administration of MP1 in mice (Mean ± SD, n = 4 at each time).
| Time (h) | Lung | Spleen | Kidney | Brain | Heart | Liver |
|---|
| Mean (ng/g) | SD | Mean (ng/g) | SD | Mean (ng/g) | SD | Mean (ng/g) | SD | Mean (ng/g) | SD | Mean (ng/g) | SD |
|---|
| 2 | 1036.2 | 338.0 | 445.4 | 219.9 | 1432.2 | 715.8 | 100.1 | 50.8 | 759.1 | 258.1 | 5613.8 | 1941.2 |
| 4 | 705.3 | 671.4 | 236.3 | 126.8 | 1351.6 | 874.8 | 77.7 | 14.9 | 433.0 | 331.3 | 5583.3 | 2912.9 |
| 8 | 619.2 | 392.5 | 206.6 | 145.2 | 1463.7 | 751.2 | 56.3 | 27.5 | 376.2 | 187.9 | 3615.3 | 1699.5 |
| 48 | 3.4 | 3.4 | 3.9 | 4.3 | 15.3 | 16.3 | 2.5 | 2.4 | 7.6 | 5.3 | 30.6 | 26.6 |
Table 11.
Summary of MS/MS parameters: precursor ion, fragment ions, voltage potential (Q1), collision energy (CE) and voltage potential (Q3) for MP1 and IS.
Table 11.
Summary of MS/MS parameters: precursor ion, fragment ions, voltage potential (Q1), collision energy (CE) and voltage potential (Q3) for MP1 and IS.
| Analytes | MRM Transition
m/z (Q1 → Q3) | Q1 (V) | Q3 (V) | CE (V) | Retention Time (min) |
|---|
| Target: MP1 | 324.10 > 168.30 | −11 | −10 | −22 | 1.9 |
| 324.10 > 304.30 | −11 | −20 | −18 | |
| Internal standard (IS): PL3 | 411.94 > 224.14 | −14 | −14 | −28 | 4.5 |