Euodia pasteuriana Methanol Extract Exerts Anti-Inflammatory Effects by Targeting TAK1 in the AP-1 Signaling Pathway
Abstract
1. Introduction
2. Results
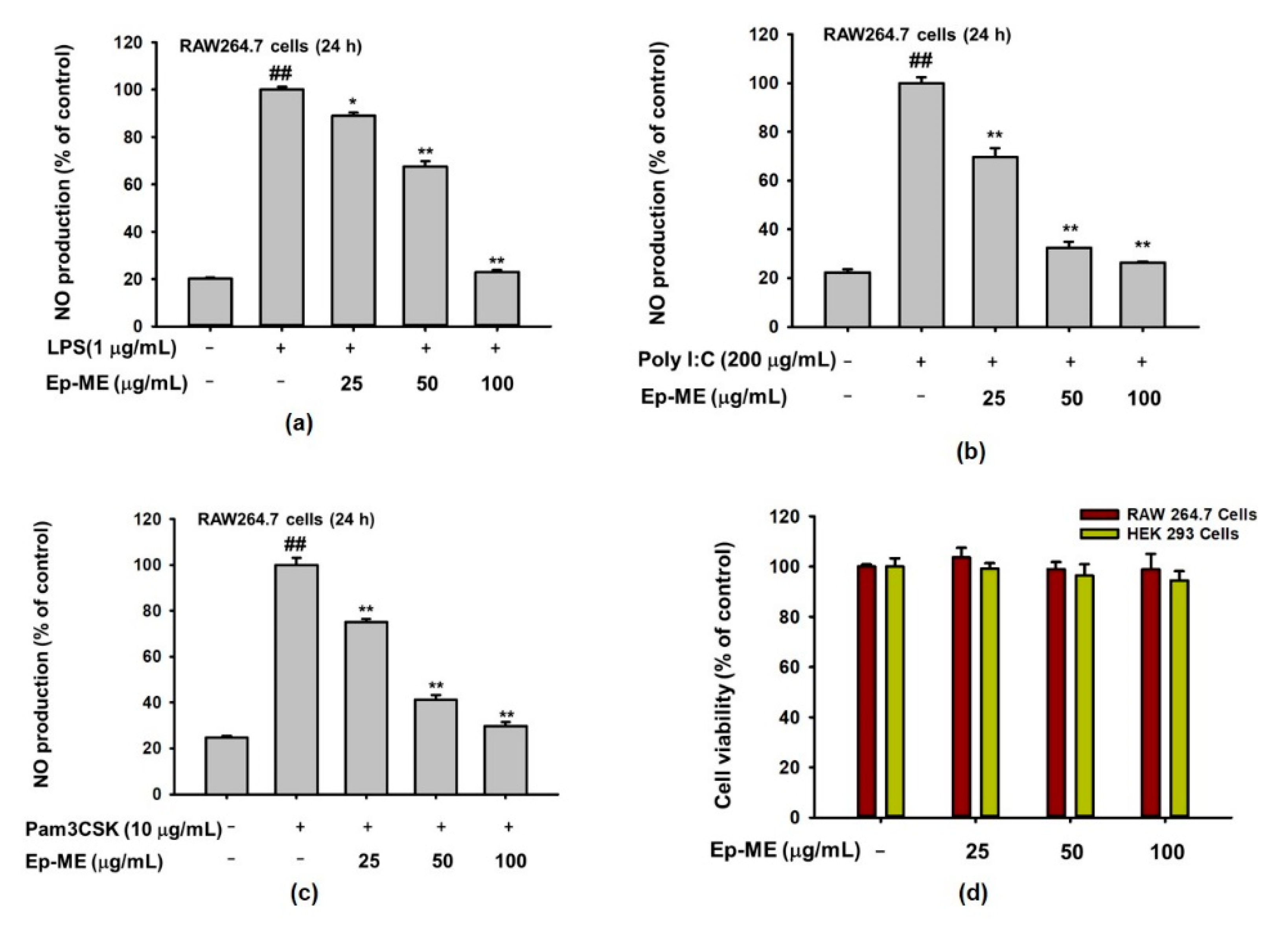
2.1. Effects of Ep-ME on Production of NO and Cytotoxicity
2.2. Effect of Ep-ME on Expression of Inflammatory Genes in LPS-Stimulated RAW264.7 Cells
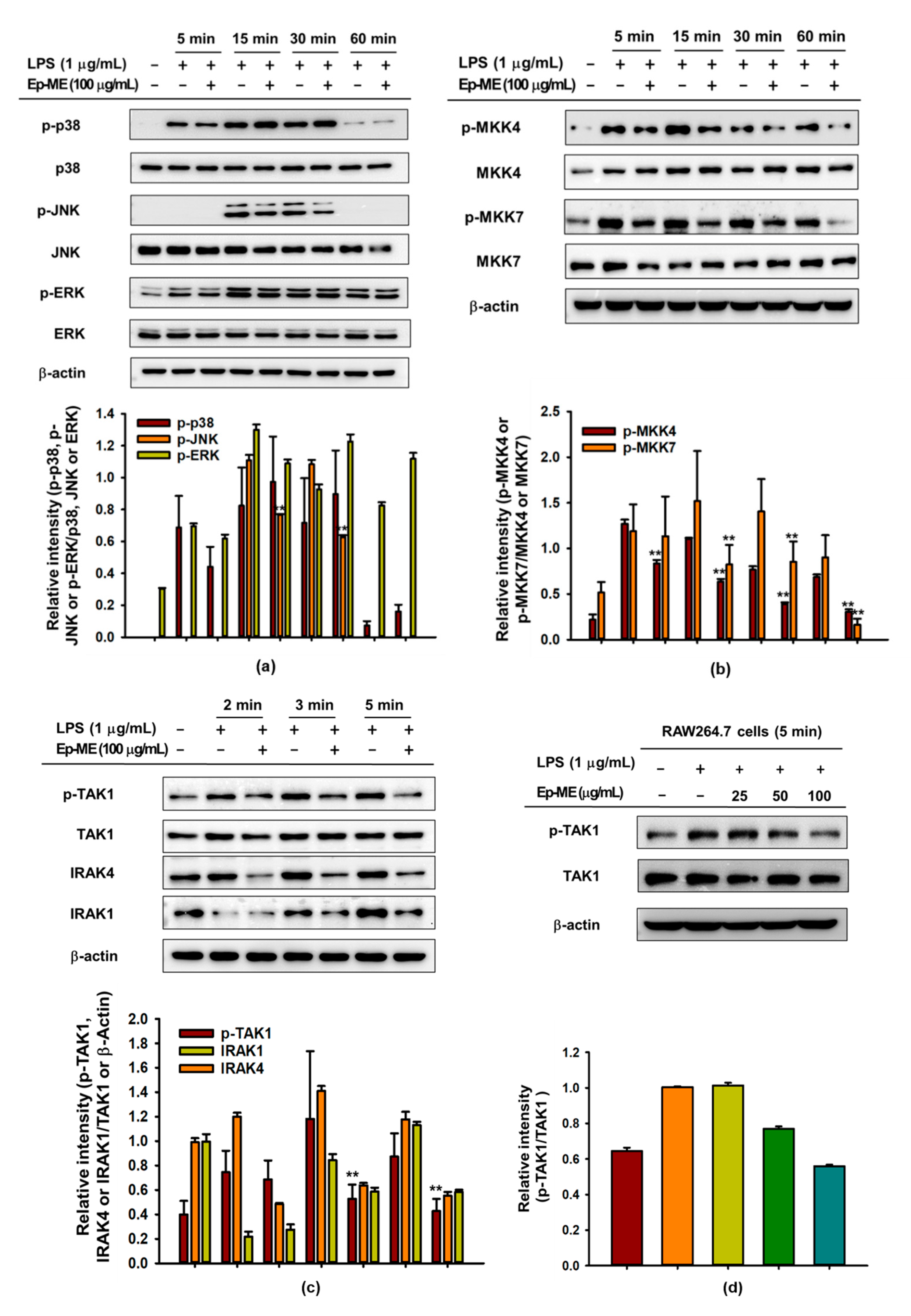
2.3. Effect of Ep-ME on Activation of the AP-1 Upstream Signaling Pathway
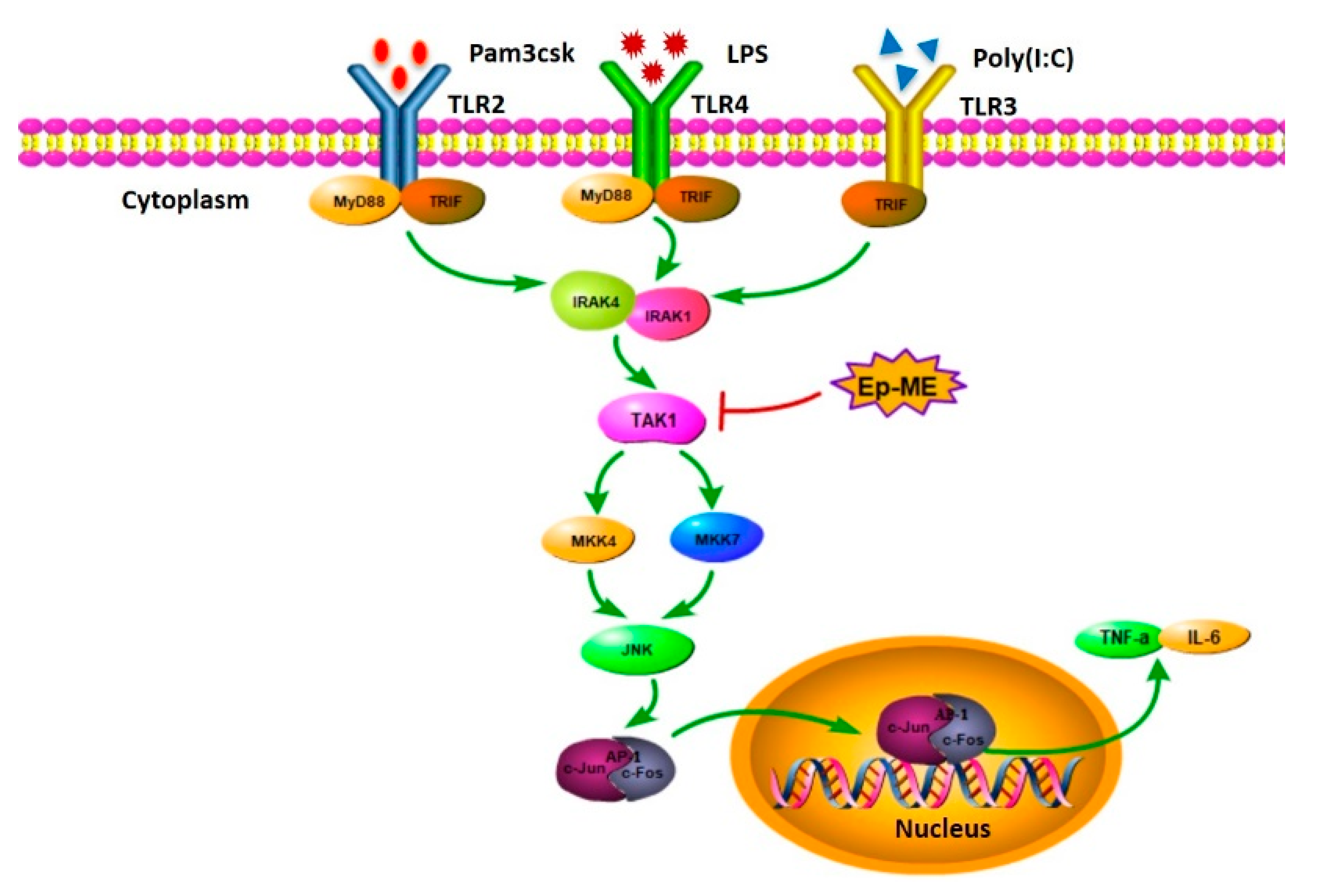
2.4. Anti-Inflammatory Effects of Ep-ME by Targeting TAK1 Kinase
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Cell Line and Cell Culture
4.3. Determination of NO Production
4.4. In Vitro Cell Viability Assay
4.5. mRNA Expression Analysis Using Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
4.6. Luciferase Reporter Gene Activity Assay
4.7. Preparation of Nuclear Extracts and Whole-Cell Lysates
4.8. Western Blot Analysis
4.9. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Takeuchi, O.; Akira, S. Pattern recognition receptors and inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef] [PubMed]
- Bagaitkar, J. Cellular dynamics of resolving inflammation. Blood J. Am. Soc. Hematol. 2014, 124, 1701–1703. [Google Scholar] [CrossRef] [PubMed]
- Helming, L. Inflammation: Cell recruitment versus local proliferation. Curr. Biol. 2011, 21, 548–550. [Google Scholar] [CrossRef] [PubMed]
- Lund, M.E.; To, J.; O’Brien, B.A.; Donnelly, S. The choice of phorbol 12-myristate 13-acetate differentiation protocol influences the response of THP-1 macrophages to a pro-inflammatory stimulus. J. Immunol. Methods 2016, 430, 64–70. [Google Scholar] [CrossRef]
- Kobayashi, E.H.; Suzuki, T.; Funayama, R.; Nagashima, T.; Hayashi, M.; Sekine, H.; Tanaka, N.; Moriguchi, T.; Motohashi, H.; Nakayama, K. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat. Commun. 2016, 7, 1–14. [Google Scholar] [CrossRef]
- Mogensen, T.H. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 2009, 22, 240–273. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int. Immunol. 2009, 21, 317–337. [Google Scholar] [CrossRef]
- Vidya, M.K.; Kumar, V.G.; Sejian, V.; Bagath, M.; Krishnan, G.; Bhatta, R. Toll-like receptors: Significance, ligands, signaling pathways, and functions in mammals. Int. Rev. Immunol. 2018, 37, 20–36. [Google Scholar] [CrossRef]
- Schmitz, F.; Mages, J.; Heit, A.; Lang, R.; Wagner, H. Transcriptional activation induced in macrophages by Toll-like receptor (TLR) ligands: From expression profiling to a model of TLR signaling. Eur. J. Immunol. 2004, 34, 2863–2873. [Google Scholar] [CrossRef]
- Thompson, M.R.; Kaminski, J.J.; Kurt-Jones, E.A.; Fitzgerald, K.A. Pattern recognition receptors and the innate immune response to viral infection. Viruses 2011, 3, 920–940. [Google Scholar] [CrossRef]
- Lu, Y.-C.; Yeh, W.-C.; Ohashi, P.S. LPS/TLR4 signal transduction pathway. Cytokine 2008, 42, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Ivashkiv, L.B. Inflammatory signaling in macrophages: Transitions from acute to tolerant and alternative activation states. Eur. J. Immunol. 2011, 41, 2477–2481. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.S.; Jeong, D.; Nam, G.; Yi, Y.-S.; Yoon, D.H.; Kim, T.W.; Park, Y.C.; Hwang, H.; Rhee, M.H.; Hong, S.; et al. AP-1 pathway-targeted inhibition of inflammatory responses in LPS-treated macrophages and EtOH/HCl-treated stomach by Archidendron clypearia methanol extract. J. Ethnopharmacol. 2013, 146, 637–644. [Google Scholar] [CrossRef] [PubMed]
- Newton, K.; Dixit, V.M. Signaling in innate immunity and inflammation. Cold Spring Harb. Perspect. Biol. 2012, 4, a006049. [Google Scholar] [CrossRef]
- Hess, J.; Angel, P.; Schorpp-Kistner, M. AP-1 subunits: Quarrel and harmony among siblings. J. Cell Sci. 2004, 117, 5965–5973. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Kang, Y.-G.; Kim, Y.-J.; Lee, T.R.; Yoo, B.C.; Jo, M.; Kim, J.H.; Kim, J.-H.; Kim, D.; Cho, J.Y. Dehydroabietic acid suppresses inflammatory response via suppression of Src-, Syk-, and TAK1-mediated pathways. Int. J. Mol. Sci. 2019, 20, 1593. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Yi, Y.-S.; Son, Y.-J.; Han, S.Y.; Kim, D.H.; Nam, G.; Hossain, M.A.; Kim, J.-H.; Park, J.; Cho, J.Y. BIOGF1K, a compound K-rich fraction of ginseng, plays an antiinflammatory role by targeting an activator protein-1 signaling pathway in RAW264. 7 macrophage-like cells. J. Ginseng Res. 2018, 42, 233. [Google Scholar] [CrossRef]
- Yu, T.; Yang, Y.; Kwak, Y.-S.; Song, G.G.; Kim, M.-Y.; Rhee, M.H.; Cho, J.Y. Ginsenoside Rc from Panax ginseng exerts anti-inflammatory activity by targeting TANK-binding kinase 1/interferon regulatory factor-3 and p38/ATF-2. J. Ginseng Res. 2017, 41, 127–133. [Google Scholar] [CrossRef]
- Franks, A.L.; Slansky, J.E. Multiple associations between a broad spectrum of autoimmune diseases, chronic inflammatory diseases and cancer. Anticancer Res. 2012, 32, 1119–1136. [Google Scholar]
- Barnes, P.J.; Karin, M. Nuclear factor-κB—A pivotal transcription factor in chronic inflammatory diseases. N. Engl. J. Med. 1997, 336, 1066–1071. [Google Scholar] [CrossRef]
- Yi, Y.-S.; Cho, J.Y.; Kim, D. Cerbera manghas methanol extract exerts anti-inflammatory activity by targeting c-Jun N-terminal kinase in the AP-1 pathway. J. Ethnopharmacol. 2016, 193, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Zhao, Q.-L.; Jawaid, P.; Rehman, M.U.; Sakurai, H.; Kondo, T. Enhancement of hyperthermia-induced apoptosis by 5Z-7-oxozeaenol, a TAK1 inhibitor, in A549 cells. Cell Stress Chaperones 2016, 21, 873–881. [Google Scholar] [CrossRef] [PubMed]
- Sharma, J.; Al-Omran, A.; Parvathy, S.J.I. Role of nitric oxide in inflammatory diseases. Inflammopharmacology 2007, 15, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-M.; An, J. Cytokines, inflammation, and pain. Int. Anesthesiol. Clin. 2007, 45, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Fingleton, B. Matrix metalloproteinases as regulators of inflammatory processes. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 2036–2042. [Google Scholar] [CrossRef]
- Van Lint, P.; Libert, C. Chemokine and cytokine processing by matrix metalloproteinases and its effect on leukocyte migration and inflammation. J. Leukoc. Biol. 2007, 82, 1375–1381. [Google Scholar] [CrossRef]
- Parks, W.C.; Wilson, C.L.; López-Boado, Y.S. Matrix metalloproteinases as modulators of inflammation and innate immunity. Nat. Rev. Immunol. 2004, 4, 617–629. [Google Scholar] [CrossRef]
- Schönbeck, U.; Mach, F.; Libby, P. Generation of Biologically Active IL-1β by Matrix Metalloproteinases: A Novel Caspase-1-Independent Pathway of IL-1β Processing. J. Immunol. 1998, 161, 3340–3346. [Google Scholar]
- Itoh, T.; Matsuda, H.; Tanioka, M.; Kuwabara, K.; Itohara, S.; Suzuki, R. The Role of Matrix Metalloproteinase-2 and Matrix Metalloproteinase-9 in Antibody-Induced Arthritis. J. Immunol. 2002, 169, 2643–2647. [Google Scholar] [CrossRef]
- Liu, J.; Cao, X. Cellular and molecular regulation of innate inflammatory responses. Cell. Mol. Immunol. 2016, 13, 711–721. [Google Scholar] [CrossRef]
- Garces de los Fayos Alonso, I.; Liang, H.-C.; Turner, S.D.; Lagger, S.; Merkel, O.; Kenner, L. The Role of Activator Protein-1 (AP-1) Family Members in CD30-Positive Lymphomas. Cancers 2018, 10, 93. [Google Scholar] [CrossRef] [PubMed]
- Zenz, R.; Eferl, R.; Scheinecker, C.; Redlich, K.; Smolen, J.; Schonthaler, H.B.; Kenner, L.; Tschachler, E.; Wagner, E.F. Activator protein 1 (Fos/Jun) functions in inflammatory bone and skin disease. Arthritis Res. Ther. 2008, 10, 201. [Google Scholar] [CrossRef] [PubMed]
- Wagner, E.F. Bone development and inflammatory disease is regulated by AP-1 (Fos/Jun). Ann. Rheum. Dis. 2010, 69 (Suppl. 1), i86–i88. [Google Scholar] [CrossRef] [PubMed]
- Shin, K.K.; Park, J.G.; Hong, Y.H.; Aziz, N.; Park, S.H.; Kim, S.; Kim, E.; Cho, J.Y. Anti-Inflammatory Effects of Licania macrocarpa Cuatrec Methanol Extract Target Src-and TAK1-Mediated Pathways. Evid. Based Complementary Altern. Med. 2019, 2019, 4873870. [Google Scholar] [CrossRef] [PubMed]
- Aashaq, S.; Batool, A.; Andrabi, K.I. TAK1 mediates convergence of cellular signals for death and survival. Apoptosis 2019, 24, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ou, G.; He, Y.; Ren, L.; Yang, X.; Zeng, M. Resveratrol attenuates the MSU crystal-induced inflammatory response through the inhibition of TAK1 activity. Int. Immunopharmacol. 2019, 67, 62–68. [Google Scholar] [CrossRef]
- Kim, H.G.; Kim, M.-Y.; Cho, J.Y. Alisma canaliculatum ethanol extract suppresses inflammatory responses in LPS-stimulated macrophages, HCl/EtOH-induced gastritis, and DSS-triggered colitis by targeting Src/Syk and TAK1 activities. J. Ethnopharmacol. 2018, 219, 202–212. [Google Scholar] [CrossRef]
- Kim, E.; Kang, Y.-G.; Kim, J.H.; Kim, Y.-J.; Lee, T.R.; Lee, J.; Kim, D.; Cho, J.Y. The antioxidant and anti-inflammatory activities of 8-hydroxydaidzein (8-HD) in activated macrophage-like RAW264. 7 cells. Int. J. Mol. Sci. 2018, 19, 1828. [Google Scholar] [CrossRef]
- Wu, J.; Powell, F.; Larsen, N.A.; Lai, Z.; Byth, K.F.; Read, J.; Gu, R.-F.; Roth, M.; Toader, D.; Saeh, J.C. Mechanism and in vitro pharmacology of TAK1 inhibition by (5 Z)-7-oxozeaenol. ACS Chem. Biol. 2013, 8, 643–650. [Google Scholar] [CrossRef]
- Ninomiya-Tsuji, J.; Kajino, T.; Ono, K.; Ohtomo, T.; Matsumoto, M.; Shiina, M.; Mihara, M.; Tsuchiya, M.; Matsumoto, K. A resorcylic acid lactone, 5Z-7-oxozeaenol, prevents inflammation by inhibiting the catalytic activity of TAK1 MAPK kinase kinase. J. Biol. Chem. 2003, 278, 18485–18490. [Google Scholar] [CrossRef]
- Zhou, H.; Li, S.; Wang, G. Euxanthone Ameliorates Sevoflurane-Induced Neurotoxicity in Neonatal Mice. J. Mol. Neurosci. 2019, 68, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Shi, Y.; Zhang, H.; Mine, Y.; Tsao, R. Anti-inflammatory and anti-oxidative activities of daidzein and its sulfonic acid ester derivatives. J. Funct. Foods 2017, 35, 635–640. [Google Scholar] [CrossRef]
- Choi, E.Y.; Jin, J.Y.; Lee, J.Y.; Choi, J.I.; Choi, I.S.; Kim, S.J. Anti-inflammatory effects and the underlying mechanisms of action of daidzein in murine macrophages stimulated with Prevotella intermedia lipopolysaccharide. J. Periodontal Res. 2012, 47, 204–211. [Google Scholar] [CrossRef]
- Han, S.Y.; Kim, J.; Kim, E.; Kim, S.H.; Seo, D.B.; Kim, J.H.; Shin, S.S.; Cho, J.Y. AKT-targeted anti-inflammatory activity of Panax ginseng calyx ethanolic extract. J. Ginseng Res. 2018, 42, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Kim, D.; Yoo, S.; Hong, Y.H.; Han, S.Y.; Jeong, S.; Jeong, D.; Kim, J.H.; Cho, J.Y.; Park, J. The skin protective effects of compound K, a metabolite of ginsenoside Rb1 from Panax ginseng. J. Ginseng Res. 2018, 42, 218–224. [Google Scholar] [CrossRef] [PubMed]


| Gene Name | Direction | Sequences (5′ to 3′) |
|---|---|---|
| MMP1 (Mouse) | Forward | ACAACGGAGACCGGCAAAAT |
| Reverse | GCTGGAAAGTGTGAGCAAGC | |
| MMP2 (Mouse) | Forward | GCCCCCATGAAGCCTTGTTT |
| Reverse | GTCAGTATCAGCATCGGGGG | |
| MMP3 (Mouse) | Forward | ACTCCCTGGGACTCTACCAC |
| Reverse | TTCTTCACGGTTGCAGGGAG | |
| MMP9 (Mouse) | Forward | TCTTCCCCAAAGACCTGAAA |
| Reverse | TGATGTTATGATGGTCCCAC | |
| TNF-α (Mouse) | Forward | TAGCCCACGTCGTAGCAAAC |
| Reverse | ACCCTGAGCCATAATCCCCT | |
| IL-6 (Mouse) | Forward | GCCTTCTTGGGACTGATGCT |
| Reverse | TGGAAATTGGGGTAGGAAGGAC | |
| GAPDH (Mouse) | Forward | ACCACAGTCCATGCCATCAC |
| Reverse | CCACCACCCTGTTGCTGTAG | |
| MMP1 (Human) | Forward | CACAGCTTCCCAGCGACTC |
| Reverse | GTCCCGATGATCTCCCCTGA | |
| MMP2 (Human) | Forward | CCCACTGAGGAGTCCAACAT |
| Reverse | CATTTACACGTCTGCGGATCT | |
| MMP3 (Human) | Forward | ATCCTACTGTTGCTGTGCGT |
| Reverse | CATCACCTCCAGAGTGTCGG | |
| GAPDH (Human) | Forward | GGTCACCAGGGCTGCTTTTA |
| Reverse | GATGGCATGGACTGTGGTCA |
Sample Availability: Samples of the compound, Euodia pasteuriana methanol extract is available from the authors. | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Kim, M.-Y.; Cho, J.Y. Euodia pasteuriana Methanol Extract Exerts Anti-Inflammatory Effects by Targeting TAK1 in the AP-1 Signaling Pathway. Molecules 2020, 25, 5760. https://doi.org/10.3390/molecules25235760
Zhang J, Kim M-Y, Cho JY. Euodia pasteuriana Methanol Extract Exerts Anti-Inflammatory Effects by Targeting TAK1 in the AP-1 Signaling Pathway. Molecules. 2020; 25(23):5760. https://doi.org/10.3390/molecules25235760
Chicago/Turabian StyleZhang, Jianmei, Mi-Yeon Kim, and Jae Youl Cho. 2020. "Euodia pasteuriana Methanol Extract Exerts Anti-Inflammatory Effects by Targeting TAK1 in the AP-1 Signaling Pathway" Molecules 25, no. 23: 5760. https://doi.org/10.3390/molecules25235760
APA StyleZhang, J., Kim, M.-Y., & Cho, J. Y. (2020). Euodia pasteuriana Methanol Extract Exerts Anti-Inflammatory Effects by Targeting TAK1 in the AP-1 Signaling Pathway. Molecules, 25(23), 5760. https://doi.org/10.3390/molecules25235760







