Specific Non-Reducing Ends in Heparins from Different Animal Origins: Building Blocks Analysis Using Reductive Amination Tagging by Sulfanilic Acid
Abstract
1. Introduction
2. Results
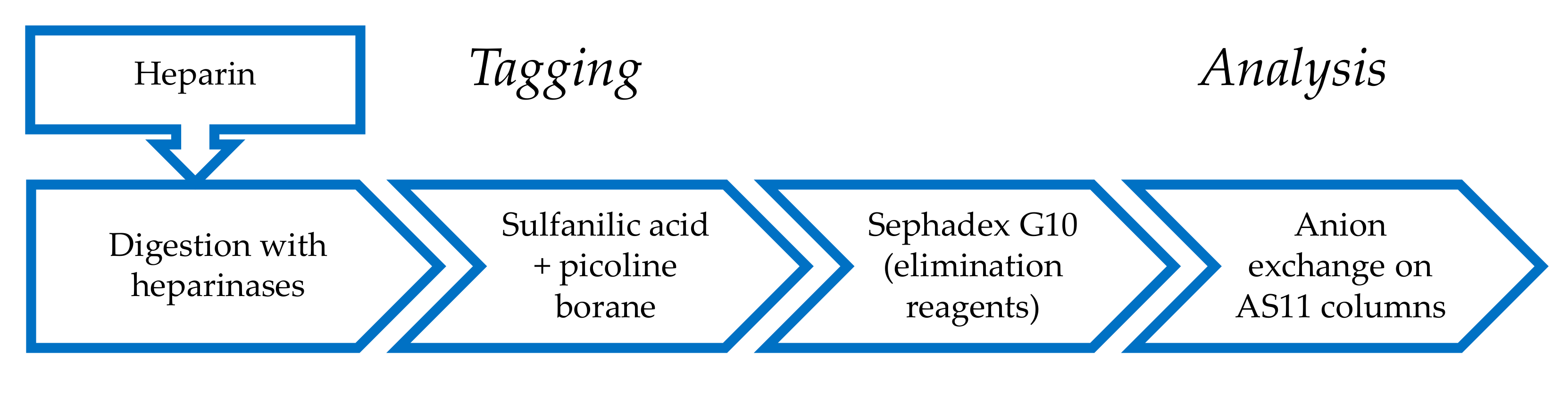
2.1. Derivatization of Enzymatically Digested Heparin with Sulfanilic Acid
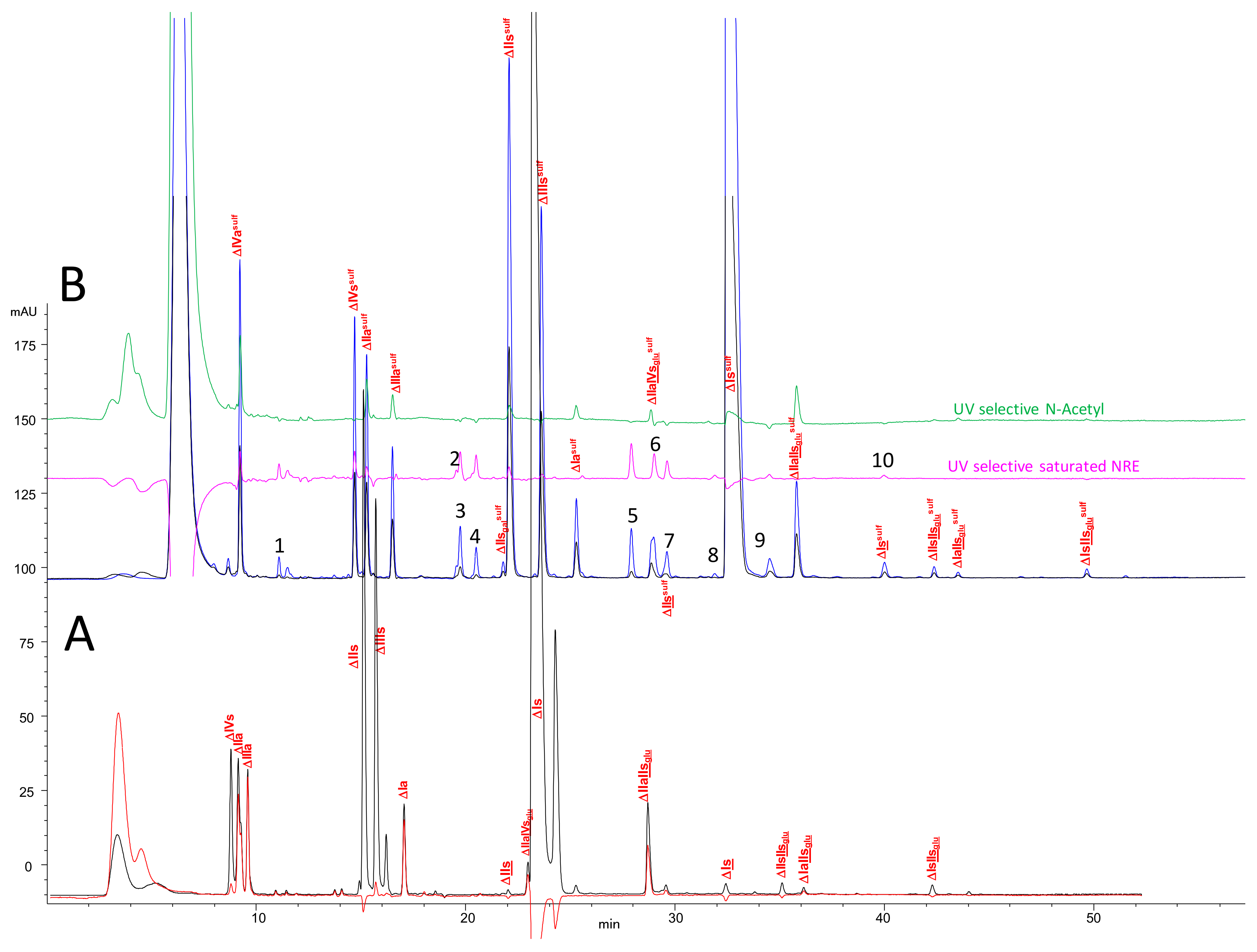
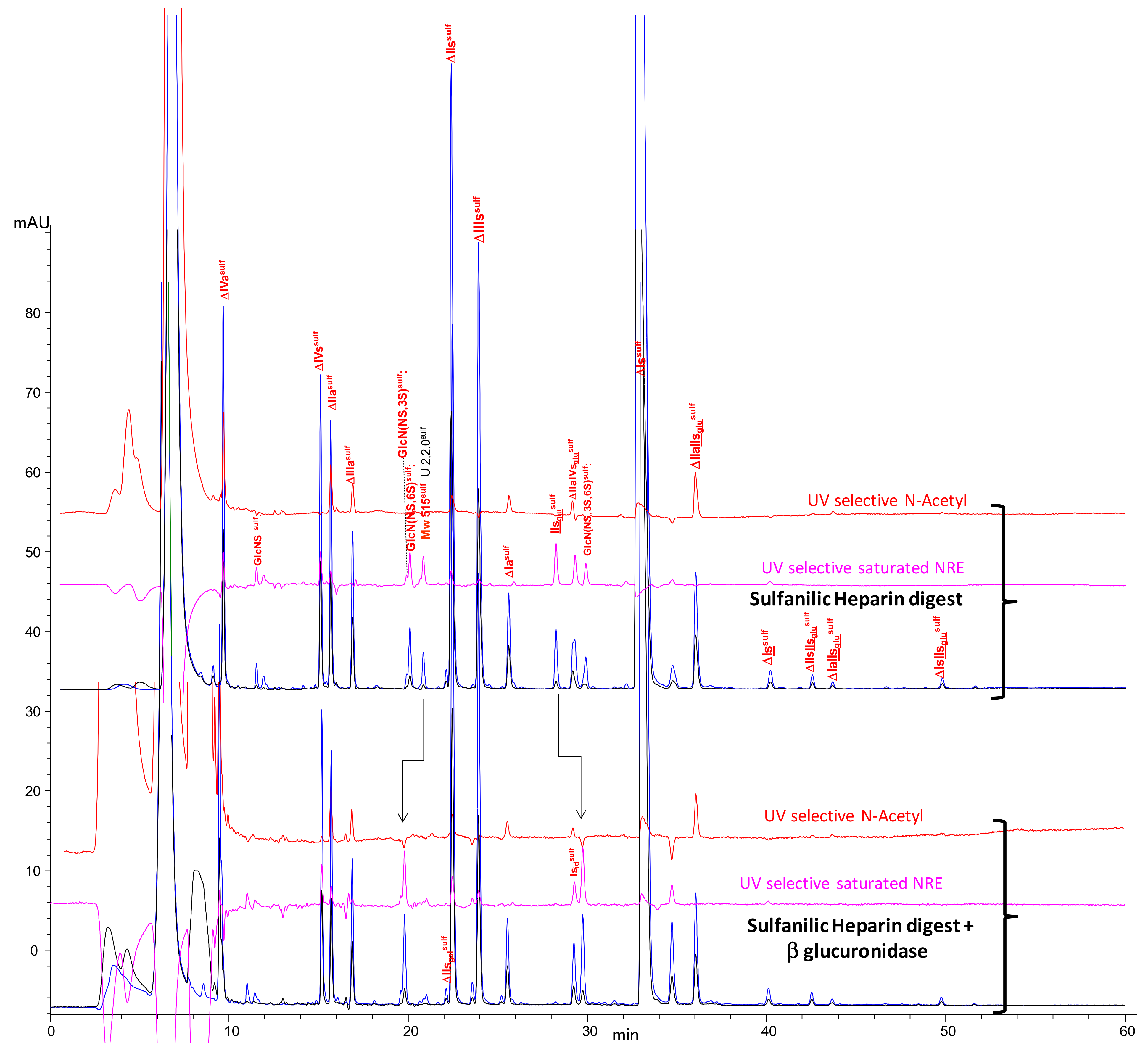
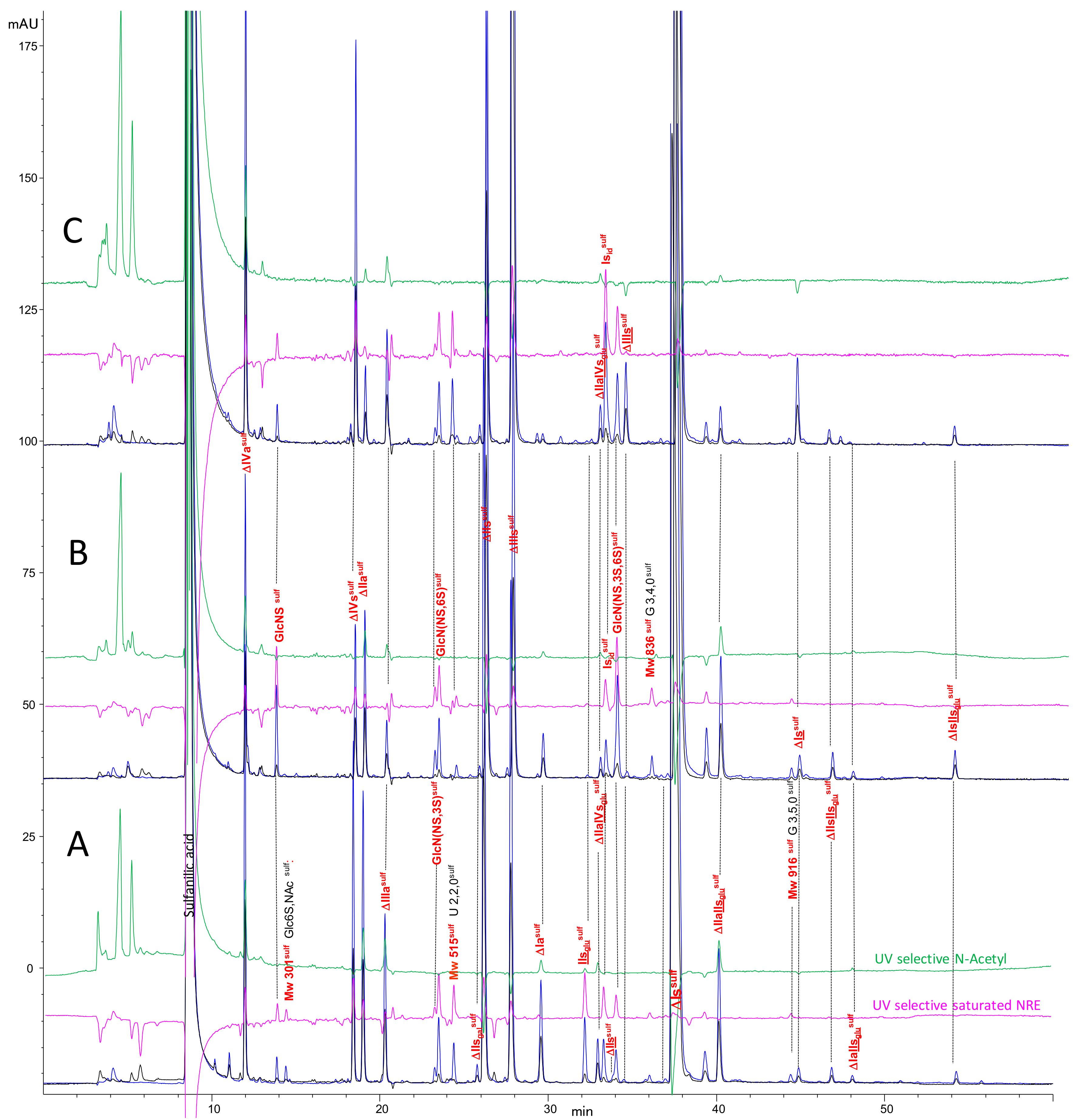
2.2. Chromatography of the Heparin Digests
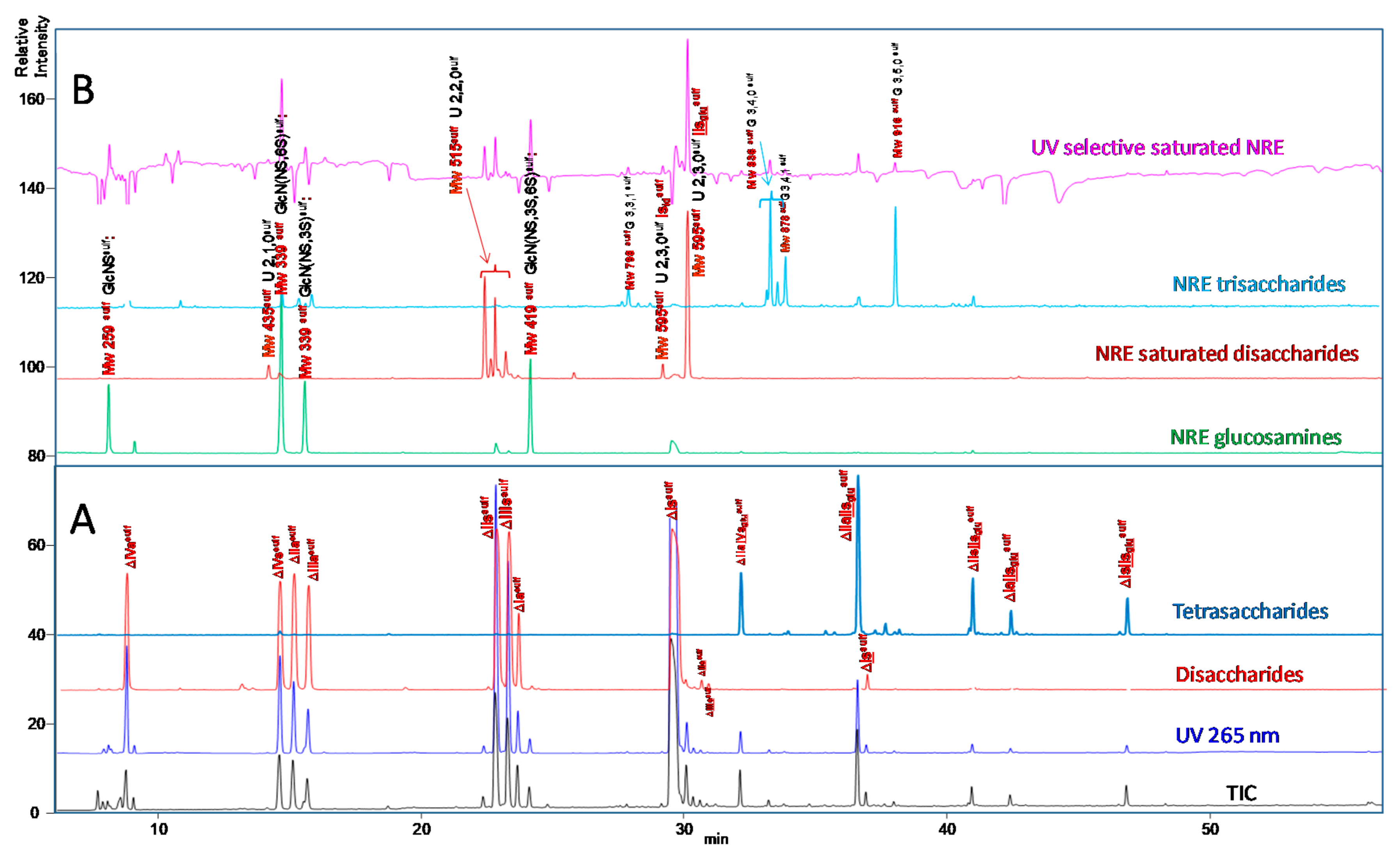
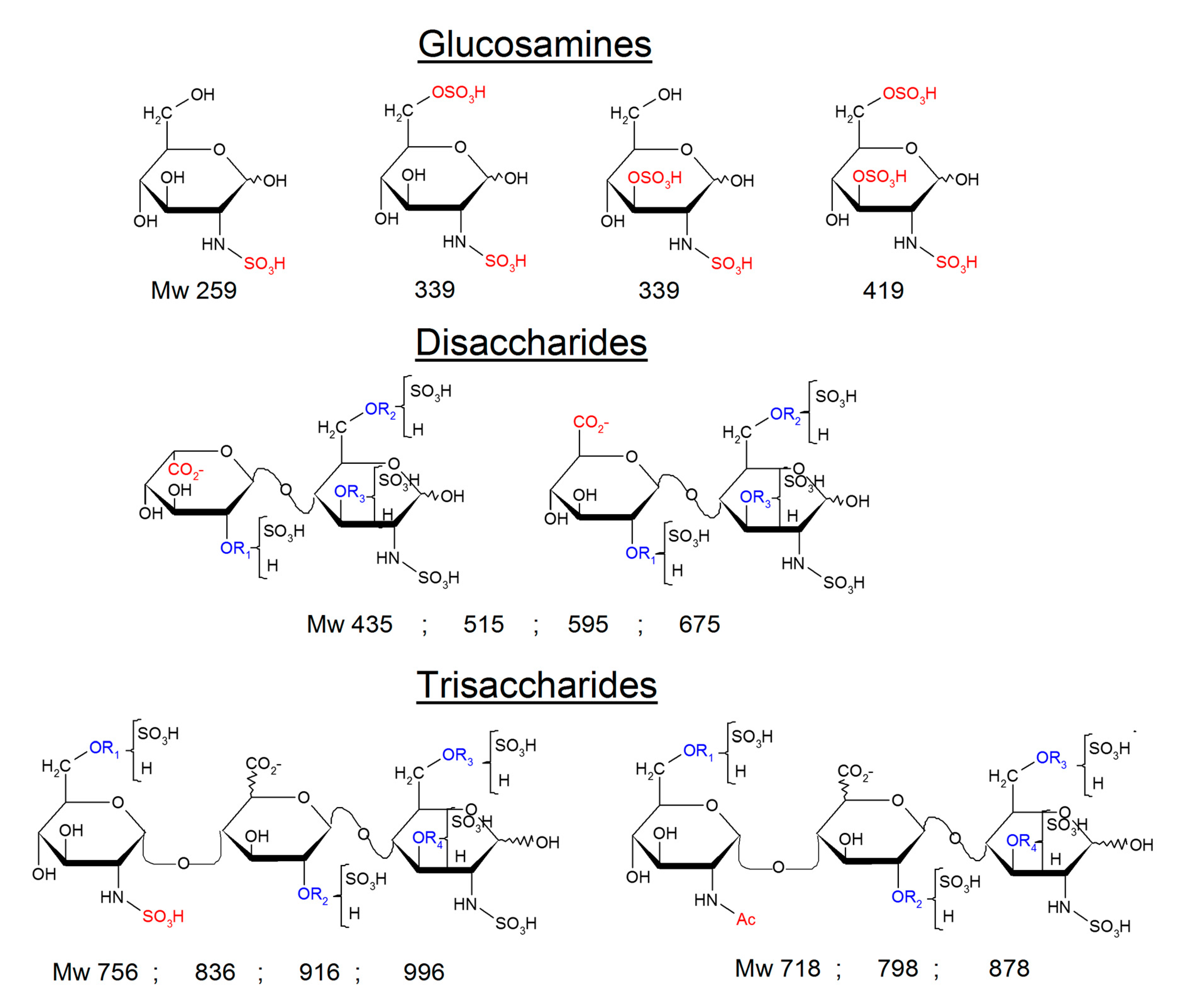
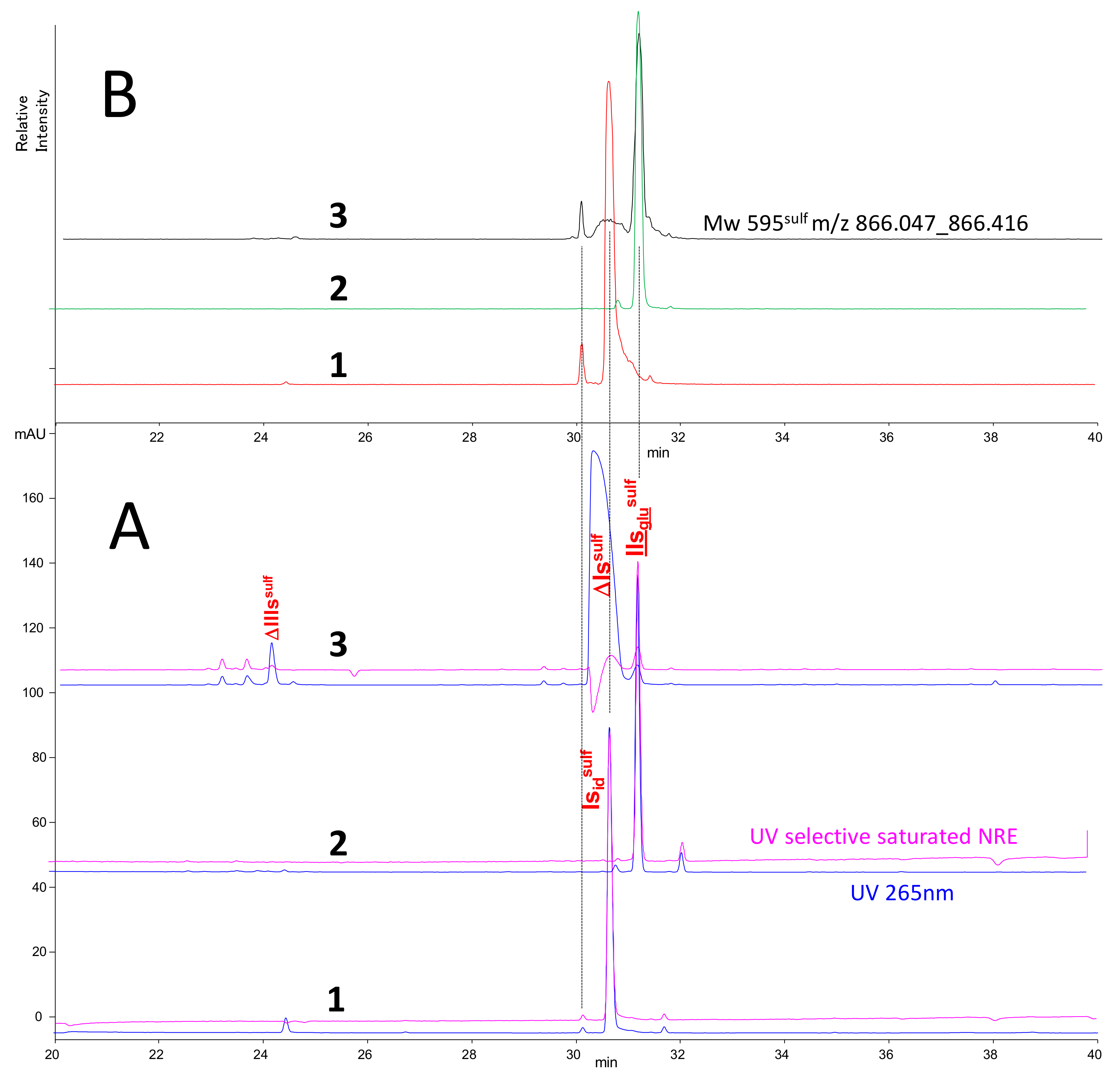
2.3. Identification of NRE Building Blocks
2.4. Treatment by β-Glucuronidase
2.5. Treatment by Δ4-5-Glycuronidase
2.6. Optimization of Chromatographic Conditions for the AS11 Method
2.7. Quantification of Building Blocks
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Heparin Lyase Digestions
4.3. Reductive Amination by Sulfanilic Acid
4.4. Analysis by SAX Chromatography on AS11 Columns
4.5. Ion Pair LC/MS
4.6. Identification of NRE Tetra and Disaccharides
4.7. Reaction with Exoglycosidases: β-d-Glucuronidase
4.8. Reaction with Δ4-5 Glycuronidase
5. Conclusions
Supplementary Materials
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AT | Antithrombin |
| BSA | Bovine serum albumin |
| EMA | European Medicines Agency |
| FDA | Food and Drug Administration |
| HFIP | 1,1,1,3,3,3-hexafluoro-2-propanol |
| HPTA | Heptylamine |
| LC/MS | Liquid chromatography-mass spectrometry |
| LMWH | Low-molecular-weight heparin |
| Mw | Mass average molecular weight |
| Ph. Eur. | European Pharmacopoeia |
| SAX | Strong anion exchange |
| CTA-SAX | Cetyl trimethyl ammonium dynamic strong anion exchange |
| USP | US Pharmacopeia |
| NRE | Non-reducing end |
| TIC | Total ionic current |
| RIC | Reconstructed ionic current |
| GPC | Gel permeation chromatography |
References
- European Pharmacopoiea Heparin Sodium Monograph. Available online: https://pheur.edqm.eu/app/10-0/content/10-0/0333E.htm?highlight=on&terms=heparin%20sodium&terms=sodium&terms=heparin&terms=0333%20heparin%20sodium&terms=0333 (accessed on 1 October 2020).
- USP Heparin Sodium Monograph. U.S. Pharmacopeial Convention Online. 2012. Available online: https://online.uspnf.com/uspnf/document/1_GUID-B46DC142-C09C-4B60-A166-7ED954703D9B_3_en-US?source=Search%20Results&highlight=heparin%20sodium (accessed on 1 October 2020).
- Guerrini, M.; Beccati, D.; Shriver, Z.; Naggi, A.; Viswanathan, K.; Bisio, A.; Capila, I.; Lansing, J.C.; Guglieri, S.; Fraser, B.; et al. Oversulfated chondroitin sulfate is a contaminant in heparin associated with adverse clinical events. Nat. Biotechnol. 2008, 26, 669–675. [Google Scholar] [CrossRef] [PubMed]
- Fareed, J.; Jeske, W.; Ramacciotti, E. Porcine mucosal heparin shortage crisis! What are the options? Clin. Appl. Thromb. Hemost. 2019, 25, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Spelta, F.; Liverani, L.; Peluso, A.; Marinozzi, M.; Urso, E.; Guerrini, M.; Naggi, A. SAX-HPLC and HSQC NMR spectroscopy: Orthogonal methods for characterizing heparin batches composition. Front. Med. 2019, 6, 78. [Google Scholar] [CrossRef] [PubMed]
- Mauri, L.; Marinozzi, M.; Phatak, N.; Karfunkle, M.; St. Ange, K.; Guerrini, M.; Keire, D.A.; Linhardt, R.J. 1D and 2D-HSQC NMR: Two methods to distinguish and characterize heparin from, different animal and tissue sources. Front. Med. 2019, 6, 142. [Google Scholar] [CrossRef]
- Mourier, P.; Anger, P.; Martinez, C.; Herman, F.; Viskov, C. Quantitative compositional analysis of heparin using exhaustive heparinase digestion and strong anion exchange chromatography. Anal. Chem. Res. 2015, 3, 46–53. [Google Scholar] [CrossRef]
- Sun, X.; Sheng, A.; Liu, X.; Shi, F.; Jin, L.; Xie, S.; Zhang, F.; Linhardt, R.J.; Chi, L. Comprehensive identification and quantitation of basic building blocks for Low-Molecular Weight Heparin. Anal. Chem. 2016, 88, 7738–7744. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, D.; Sun, X.; Bai, X.; Jin, L.; Chi, L. Liquid chromatography-diode array detection-mass spectrometry for compositional analysis of low molecular weight heparins. Anal. Biochem. 2014, 451, 35–41. [Google Scholar] [CrossRef]
- Pecorini, S.; Camurri, G.; Torrini, L.; Ferraresi, R. Highly sensitive real-time PCR method to identify species origin in heparinoids. Anal. Bioanal. Chem. 2020, 412, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Rudd, T.R.; Mauri, L.; Marinozzi, M.; Stancanelli, E.; Yates, E.A.; Naggi, A.; Guerrini, M. Multivariate analysis applied to complex biological medicines. Faraday Discuss. 2019, 218, 303–316. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, Y.; Han, X.; Yu, Y.; Chen, J.; Fu, L.; Zhang, F.; Linhardt, R.J.; Fareed, J.; Hoppensteadt, D.; Jeske, W.; et al. Chemometric analysis of porcine, bovine and ovine heparins. J. Pharm. Biomed. Anal. 2019, 164, 345–352. [Google Scholar] [CrossRef]
- Fu, L.; Li, G.; Yang, B.; Onishi, A.; Li, L.; Sun, P.; Zhang, F.; Linhardt, R.J. Structural characterization of pharmaceutical heparins prepared from different animal tissues. J. Pharm. Sci. 2013, 102, 1447–1457. [Google Scholar] [CrossRef] [PubMed]
- Mourier, P.A.J.; Guichard, O.; Herman, F.; Viskov, C. Isolation of a pure octadecasaccharide with antithrombin activity from an ultra-low-molecular weight heparin. Anal. Biochem. 2014, 453, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Mourier, P.A.J.; Viskov, C. Chromatographic analysis and sequencing approach of heparin oligosaccharides using cetyl trimethyl ammonium dynamically coated stationary phases. Anal. Biochem. 2004, 332, 299–313. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, A.; Sugahara, K. Microanalysis of glycosaminoglycan-derived oligosaccharides labeled with a fluorophore 2-aminobenzamide by high performance liquid chromatography: Application to disaccharide compositionanalysis and exosequencing of oligosaccharides. Anal. Biochem. 1999, 269, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Drummond, K.J.; Yates, E.A.; Turnbull, J.E. Electrophoretic sequencing of heparin/heparan sulfate oligosaccharides using a highly sensitive fluorescent end label. Proteomics 2001, 1, 304–310. [Google Scholar] [CrossRef]
- Gupta, R.; Ponnusamy, M.P. Analysis of sulfates on low molecular weight heparin using mass spectrometry: Structural characterization of enoxaparin. Expert Rev. Proteom. 2018, 15, 503–513. [Google Scholar] [CrossRef]
- Viskov, C.; Just, M.; Laux, V.; Mourier, P.; Lorenz, M. Description of the chemical and pharmacological characteristics of a new hemisynthetic ultra-low-molecular-weight heparin, AVE5026. J. Thromb. Haemost. 2009, 7, 1143–1151. [Google Scholar] [CrossRef]
- Wu, Z.L.; Lech, M. Characterizing the non-reducing end structure of heparan sulfate. J. Biol. Chem. 2005, 280, 33749–33755. [Google Scholar] [CrossRef]
- Staples, G.O.; Shi, X.; Zaia, J. Extended N-sulfated domains reside at the nonreducing end of heparan sulfate chains. J. Biol. Chem. 2010, 285, 18336–18343. [Google Scholar] [CrossRef]
- Ogren, S.; Lindahl, U. Cleavage of macromolecular heparin by an enzyme from mouse mastocytoma. J. Biol. Chem. 1975, 250, 2690–2697. [Google Scholar]
- Jacobsson, K.G.; Lindahl, U. Degradation of heparin proteoglycan in cultured mouse mastocytoma cells. Biochem. J. 1987, 246, 409–415. [Google Scholar] [CrossRef] [PubMed]
- Vlodavsky, I.; Ilan, N.; Nadir, Y.; Brenner, B.; Katz, B.Z.; Naggi, A.; Torri, G.; Casu, B.; Sasisekharan, R. Heparanase, heparin and the coagulation system in cancer progression. Thromb. Res. 2007, 120 (Suppl. 2), S112–S120. [Google Scholar] [CrossRef]
- Peterson, S.B.; Liu, J. Unraveling the specificity of heparanase utilizing synthetic substrates. J. Biol. Chem. 2010, 285, 14504–14513. [Google Scholar] [CrossRef] [PubMed]
- Peterson, S.; Liu, J. Deciphering mode of action of heparanase using structurally defined oligosaccharides. J. Biol. Chem. 2012, 287, 34836–34843. [Google Scholar] [CrossRef]
- Lawrence, R.; Brown, J.R.; Al-Mafraji, K.; Lamanna, W.C.; Beitel, J.R.; Boons, G.; Esko, J.D.; Crawfordb, B.E. Disease-specific non-reducing end carbohydrate biomarkers for mucopolysaccharidoses. Nat. Chem. Biol. 2012, 8, 197–204. [Google Scholar] [CrossRef]
- Ruhaak, L.R.; Steenvoorden, E.; Koeleman, C.A.; Deelder, A.M.; Wuhrer, M. 2-picoline-borane: A non-toxic reducing agent for oligosaccharide labeling by reductive amination. Proteomics 2010, 10, 2330–2336. [Google Scholar] [CrossRef]
- An Overview of the Principles of MSE, the Engine that Drives MS Performance. Waters White Papers. Available online: https://www.waters.com/webassets/cms/library/docs/720004036en.pdf (accessed on 1 October 2020).
- Sun, X.; Guo, Z.; Yu, M.; Lin, C.; Sheng, A.; Wang, Z.; Linhardt, R.J.; Chi, L. Hydrophilic interaction chromatography-multiple reaction monitoring mass spectrometry method for basic building block analysis of low molecular weight heparins prepared through nitrous acid depolymerization. J. Chromatogr. A 2017, 1479, 121–128. [Google Scholar] [CrossRef]
- Beecher, C.N.; Manighalam, M.S.; Nwachuku, A.F.; Larive, C.K. Screening enoxaparin tetrasaccharide SEC fractions for 3-O-sulfo-N-sulfoglucosamine residues using [1H,15N] HSQC NMR. Anal. Bioanal. Chem. 2016, 408, 1545–1555. [Google Scholar] [CrossRef]
- Yamada, S.; Sakamoto, K.; Tsuda, H.; Yoshida, K.; Sugahara, K.; Khoo, K.H.; Morris, H.R.; Dell, A. Structural studies on the tri- and tetrasaccharides isolated from porcine intestinal heparin and characterization of heparinase/heparitinases using them as substrates. Glycobiology 1994, 4, 69–78. [Google Scholar] [CrossRef]
- Wang, H.M.; Loganathan, D.; Linhardt, R.J. Determination of the pKa of glucuronic acid and the carboxy groups of heparin by 13C-nuclear-magnetic-resonance spectroscopy. Biochem. J. 1991, 278, 689–959. [Google Scholar] [CrossRef]
- Mochizuki, H.; Yoshida, K.; Gotoh, M.; Sugioka, S.; Kikuchi, N.; Kwon, Y.-D.; Tawada, A.; Maeyama, K.; Inaba, N.; Hiruma, T.; et al. Characterization of a Heparan Sulfate 3-O-Sulfotransferase-5, an Enzyme Synthesizing a Tetrasulfated Disaccharide. J. Biol. Chem. 2003, 278, 26780–26787. [Google Scholar] [CrossRef] [PubMed]
- Mourier, P.A.; Agut, C.; Souaifi-Amara, H.; Herman, F.; Viskov, C. Analytical and statistical comparability of generic enoxaparins from the US market withthe originator product. J. Pharm. Biomed. Anal. 2015, 115, 431–442. [Google Scholar] [CrossRef] [PubMed]
- Sadowski, R.; Gadzała-Kopciuch, R.; Kowalkowski, T.; Widomski, P.; Jujeczka, L.; Buszewski, B. Characterization of low-molecular-weight heparins by strong anion-exchange chromatography. J. AOAC Int. 2017, 100, 1706–1714. [Google Scholar] [CrossRef]
- Rudd, T.R.; Macchi, E.; Muzi, L.; Ferro, M.; Gaudesi, D.; Torri, G.; Casu, B.; Guerrini, M.; Yates, E.A. Unravelling structural information from complex mixtures utilizing correlation spectroscopy applied to HSQC Spectra. Anal. Chem. 2013, 85, 7487–7493. [Google Scholar] [CrossRef] [PubMed]
- Alekseeva, A.; Urso, E.; Mazzini, G.; Naggi, A. Heparanase as an additional tool for detecting structural peculiarities of heparin oligosaccharides. Molecules 2019, 24, 4403. [Google Scholar] [CrossRef] [PubMed]
- Naimy, H.; Buczek-Thomas, J.A.; Nugent, M.A.; Leymarie, N.; Zaia, J. Highly sulfated non-reducing end-derived heparan sulfate domains bind fibroblast growth factor-2 with high affinity and are enriched in biologically active fractions. J. Biol. Chem. 2011, 286, 19311–19319. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Huang, Y.; Buczek-Thomas, J.A.; Ethen, C.M.; Nugent, M.A.; Wu, Z.L.; Zaia, J. A liquid chromatography-mass spectrometry-based approach to characterize the substrate specificity of mammalian heparanase. J. Biol. Chem. 2014, 289, 34141–34151. [Google Scholar] [CrossRef]
- Guerrini, M.; Guglieri, S.; Casu, B.; Torri, G.; Mourier, P.; Boudier, C.; Viskov, C. Antithrombin-binding octasaccharides and role of extensions of the activepentasaccharide sequence in the specificity and strength of interaction: Evidence for very high affinity induced by an unusual glucuronic acid residue. J. Biol. Chem. 2008, 283, 26662–26675. [Google Scholar] [CrossRef]
- Mourier, P.A.J.; Guichard, O.Y.; Herman, F.; Viskov, C. Heparin sodium compliance to USP monograph: Structural elucidation of an atypical 2.18 ppm NMR signal. J. Pharm. Biomed. Anal. 2012, 67–68, 169–174. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |


| Nomenclature | |
| HexUA = Uronic acid | IdoA = l-iduronic acid |
| GlcA = d-glucuronic acid | ΔHexUA = 4,5-unsaturated uronic acid |
| GlcN = d-glucosamine | Man = d-mannosamine |
| NS = N-sulfate | NAc = N-acetyl |
| Mnt 6S2,5 anhydr = 6-O sulfated 2,5 anhydro Mannitol | 2S = 2-O-sulfate |
| 3S = 3-O-sulfate | 6S = 6-O-sulfate |
| GalA = d-galacturonic acid | Epoxy = Epoxised iduronic acid |
| SA = tagging with sulfanilic acid | w/w = weight/weight |
| PMH = Porcine mucosa heparin | BMH = Bovine mucosa heparin |
| OMH = Ovine mucosa heparin | |
| Structural Symbols | |
| ΔIVa = ΔHexUA − GlcNAc | ΔIVs = ΔHexUA − GlcNS |
| ΔIIa = ΔHexUA − GlcNAc(6S) | ΔIIIa = ΔHexUA(2S) − GlcNAc |
| ΔIIs = ΔHexUA − GlcN(NS,6S) | ΔIIIs = ΔHexUA(2S) − GlcN(NS) |
| ΔIa = ΔHexUA(2S) − GlcNAc(6S) | ΔIs = ΔHexUA(2S) − GlcN(NS,6S) |
| ΔIIs = ΔHexUA − GlcN(NS,3S,6S) | ΔIIIs = ΔHexUA(2S) − GlcN(NS,3S) |
| ΔIs = ΔHexUA(2S) − GlcN(NS,3S,6S) | IVsgal = GalA − GlcNS |
| IIsgal = GalA − GlcN(NS,6S) | IIsepoxy = GulA2,3epo − GlcN(NS,6S) |
| IIsglu = GlcA − GlcN(NS,6S) | IIIsid = IdoA(2S) − GlcNS |
| IVsglu = GlcA − GlcNS | Isid = IdoA(2S) − GlcN(NS,6S) |
| IIsglu = GlcA − GlcN(NS,3S,6S) | IVsglu = GlcA − GlcN(NS,3S) |
| Glyserox = Oxidized glycoserine (ΔGlcA-Gal-Gal-Xyl-COOH) | |
| ΔU(x,y,z) = Δ-unsaturated oligosaccharide, x saccharides units, y sulfates, z N-acetyl | |
| ΔU(x,y,z)sulf = ΔU(x,y,z) with sulfanilic acid reductive amination | |
| G(x,y,z) = Oligosaccharide with a glucosamine at its non-reducing end, x saccharides units, y sulfates, z N-acetyl | |
| G(x,y,z)sulf = G(x,y,z) with sulfanilic acid reductive amination | |
| Mw 595sulf = Oligosaccharide at Mw 595 Da with sulfanilic reductive amination (595 + 157 Da) | |
| The iduronic (id) or glucuronic (glu) structure of uronic acids is indicated for oligosaccharides, e.g., ΔIs-IIIid Underlined disaccharides have a 3-O sulfated glucosamine, e.g., IIsglu (GlcA-GlcNS,3S,6S) | |
| Heparin | PMH | OMH | BMH | |
|---|---|---|---|---|
| Unsaturated Building Blocks | ΔHexUA-GlcNAc | 2.3 | 1.6 | 3.0 |
| ΔHexGalA-GlcNS | 0.0 | 0.0 | 0.2 | |
| ΔHexUA-GlcNS | 2.3 | 1.1 | 2.9 | |
| ΔHexUA-GlcNAc(6S) | 2.3 | 1.4 | 0.7 | |
| ΔHexUA(2S)-GlcNAc | 1.4 | 0.5 | 1.0 | |
| ΔHexGalA-GlcN(NS,6S) | 0.2 | 0.2 | 0.2 | |
| ΔHexUA-GlcN(NS,6S) | 8.6 | 9.0 | 6.9 | |
| ΔHexUA(2S)-GlcN(NS) | 6.0 | 5.6 | 25.2 | |
| ΔHexUA(2S)-GlcNAc(6S) | 1.4 | 0.6 | 0.2 | |
| ΔHexUA-GlcNAc(6S)-GlcA-GlcN(NS,3S) | 0.9 | 0.5 | 0.9 | |
| ΔHexUA-GlcN(NS,3S,6S) | 0.1 | 0.3 | 0.2 | |
| ΔHexUA(2S)-GlcN(NS,3S) | 0.1 | 0.1 | 1.3 | |
| ΔHexUA(2S)-GlcN(NS,6S) | 61.8 | 65.6 | 43.9 | |
| ΔHexUA-GlcNAc(6S)-GlcA-GlcN(NS,3S,6S) | 3.7 | 3.4 | 1.1 | |
| ΔHexUA(2S)-GlcN(NS,3S,6S) | 0.3 | 0.5 | 1.6 | |
| ΔHexUA-GlcN(NS, 6S)-GlcA-GlcN(NS,3S,6S) | 0.2 | 0.8 | 0.4 | |
| ΔHexUA(2S)-GlcNAc(6S)-GlcA-GlcN(NS,3S,6S) | 0.2 | 0.2 | 0.1 | |
| ΔHexUA(2S)-GlcN(2S,6S)-GlcA-GlcN(NS,3S,6S) | 0.4 | 0.9 | 0.6 | |
| NRE Building Blocks | GlcNS | 0.1 | 0.4 | 0.2 |
| GlcNAc(6S) | 0.1 | 0.0 | 0.0 | |
| GlcN(NS,3S) | 0.1 | 0.2 | 0.1 | |
| GlcN(NS,6S) | 0.5 | 0.5 | 0.5 | |
| U(2,2,0) | 0.5 | 0.2 | 0.7 | |
| GlcA-GlcN(NS,3S,6S) | 0.9 | <0.1 | <0.1 | |
| IdoA(2S)-GlcN(NS,6S) | 0.7 | 0.6 | 2.1 | |
| GlcN(NS,3S,6S) | 0.3 | 1.1 | 0.8 | |
| G(3,4,0) | 0.2 | 0.5 | N.D. | |
| G(3,5,0) | 0.2 | 0.3 | 0.2 | |
| NRE | % NRE (Monosaccharides) | 2.0 | 2.3 | 2.5 |
| % Glucosamines | 51.2 | 81.6 | 48.7 | |
| % uronic acids | 48.8 | 18.4 | 51.3 | |
| Isid/IIsglu | 0.7 | 7.7 | 36.4 | |
| % 3-0 Sulfation | 36.4 | 37.7 | 23.7 | |
| Heparin | Sulfates/Carboxylates | 2.48 | 2.63 | 2.31 |
| NAc | 12.6 | 8.0 | 7.8 | |
| 6-OH | 16.3 | 12.6 | 38.7 | |
| 2-OH | 26.4 | 22.3 | 20.2 | |
| 3-OS | 5.0 | 5.7 | 5.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mourier, P.A.J. Specific Non-Reducing Ends in Heparins from Different Animal Origins: Building Blocks Analysis Using Reductive Amination Tagging by Sulfanilic Acid. Molecules 2020, 25, 5553. https://doi.org/10.3390/molecules25235553
Mourier PAJ. Specific Non-Reducing Ends in Heparins from Different Animal Origins: Building Blocks Analysis Using Reductive Amination Tagging by Sulfanilic Acid. Molecules. 2020; 25(23):5553. https://doi.org/10.3390/molecules25235553
Chicago/Turabian StyleMourier, Pierre A. J. 2020. "Specific Non-Reducing Ends in Heparins from Different Animal Origins: Building Blocks Analysis Using Reductive Amination Tagging by Sulfanilic Acid" Molecules 25, no. 23: 5553. https://doi.org/10.3390/molecules25235553
APA StyleMourier, P. A. J. (2020). Specific Non-Reducing Ends in Heparins from Different Animal Origins: Building Blocks Analysis Using Reductive Amination Tagging by Sulfanilic Acid. Molecules, 25(23), 5553. https://doi.org/10.3390/molecules25235553






