Design, Synthesis, and Biological Evaluation of Pyridazinones Containing the (2-Fluorophenyl) Piperazine Moiety as Selective MAO-B Inhibitors
Abstract
1. Introduction
2. Results and Discussion
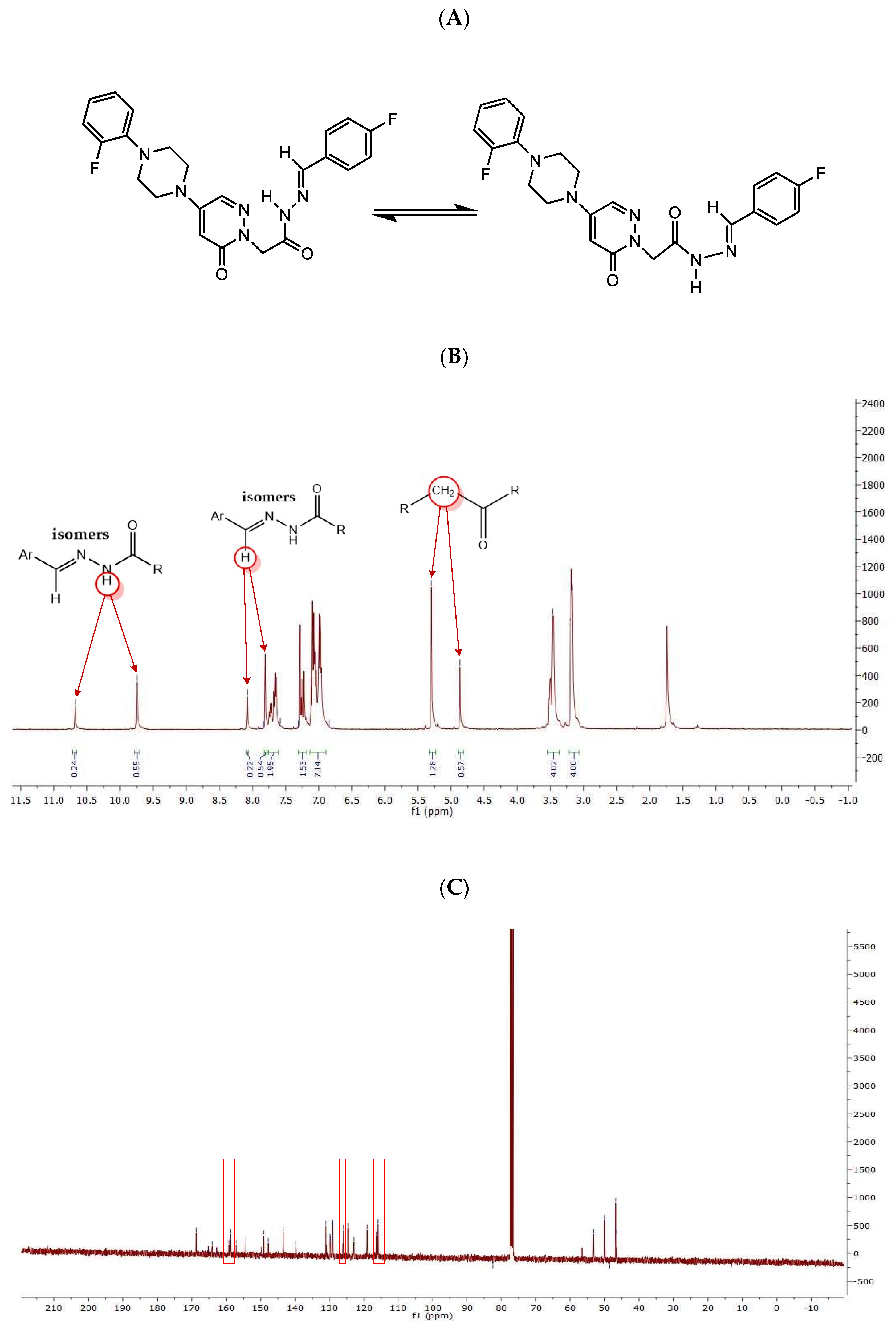
2.1. Chemistry
2.2. MAO Inhibitory Activities
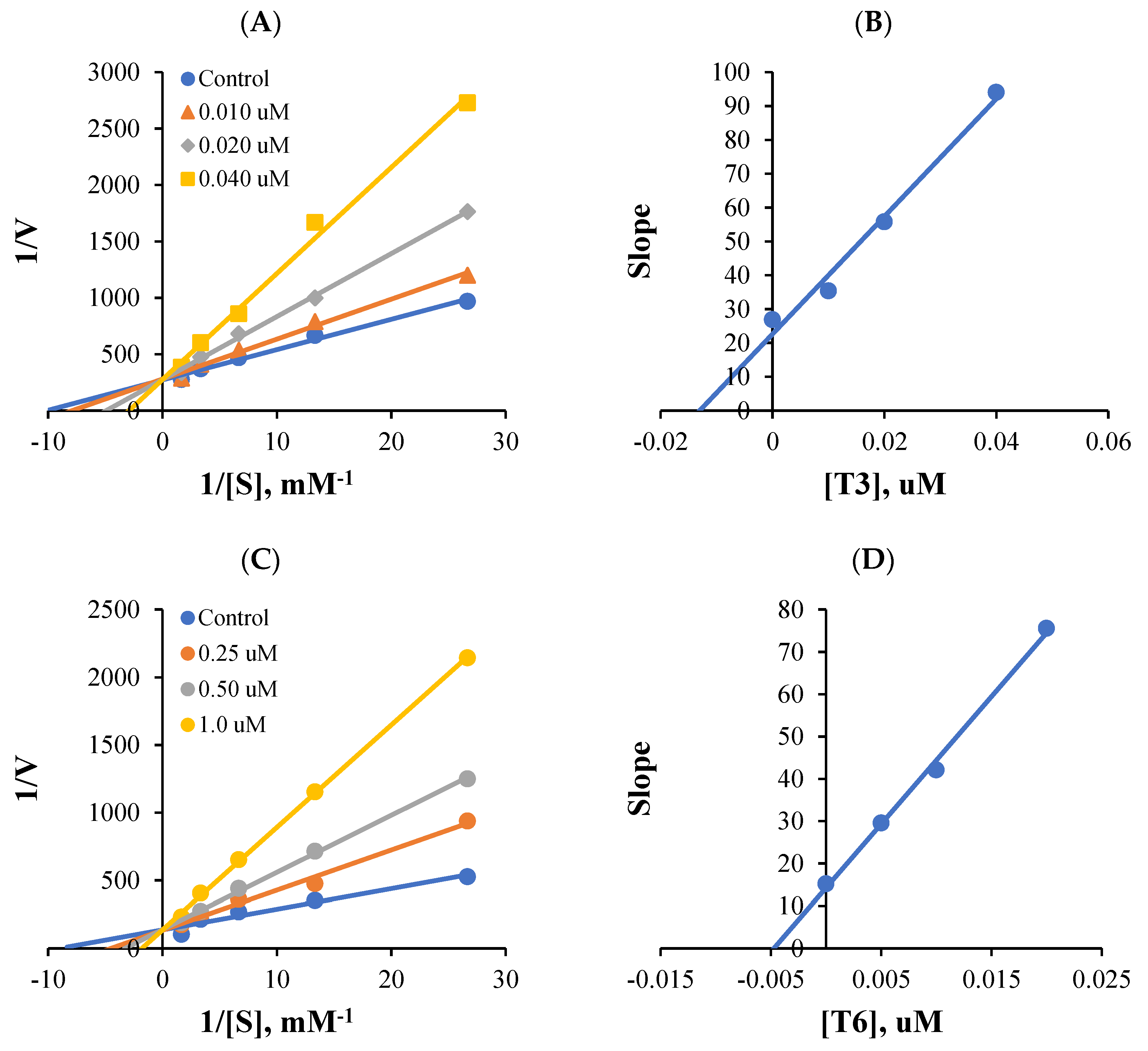
2.3. Kinetics of MAO-B Inhibitions
2.4. Reversibility Studies
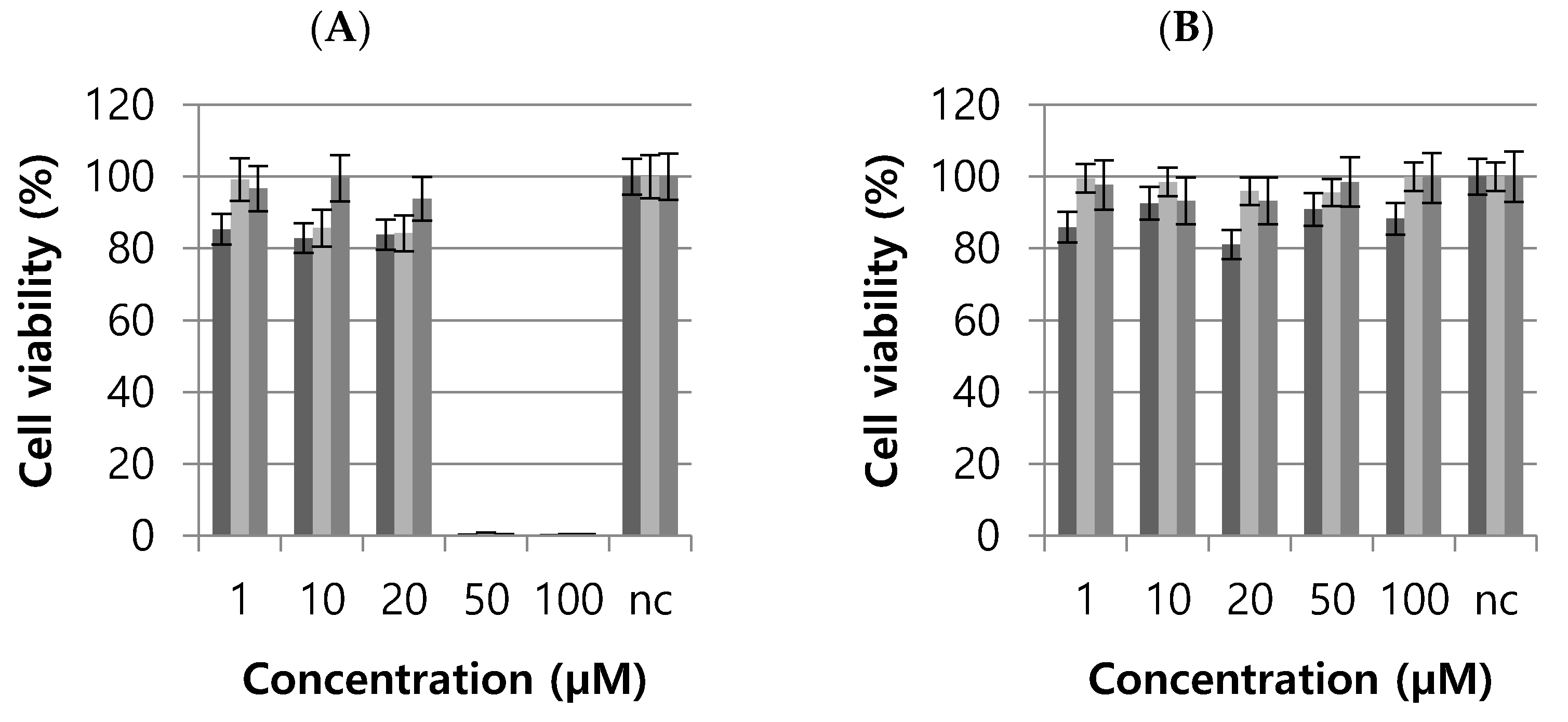
2.5. Cytotoxic Effects of the Active Pyridazinone Derivatives
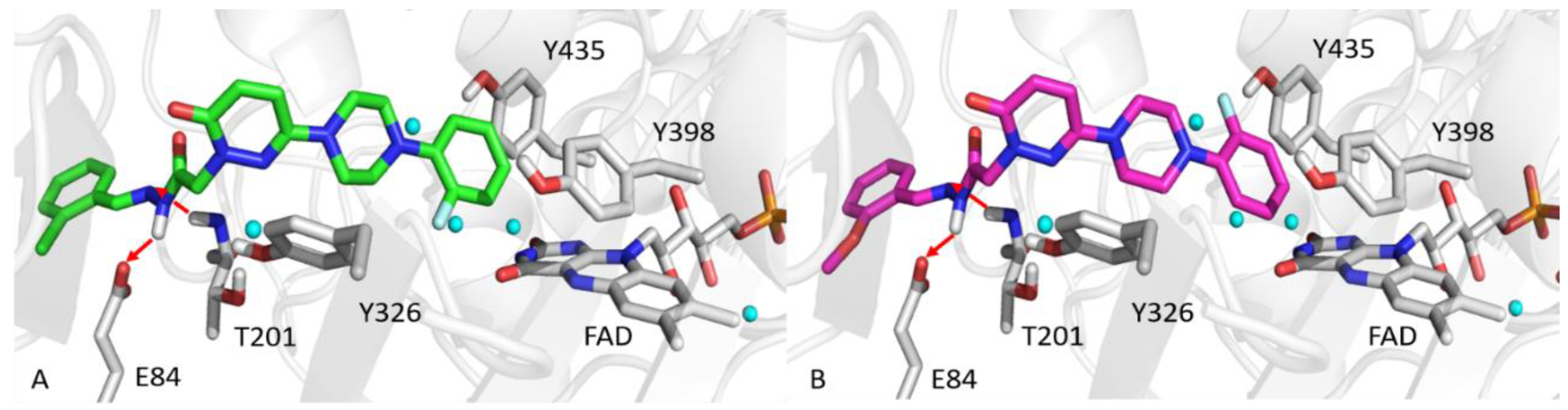
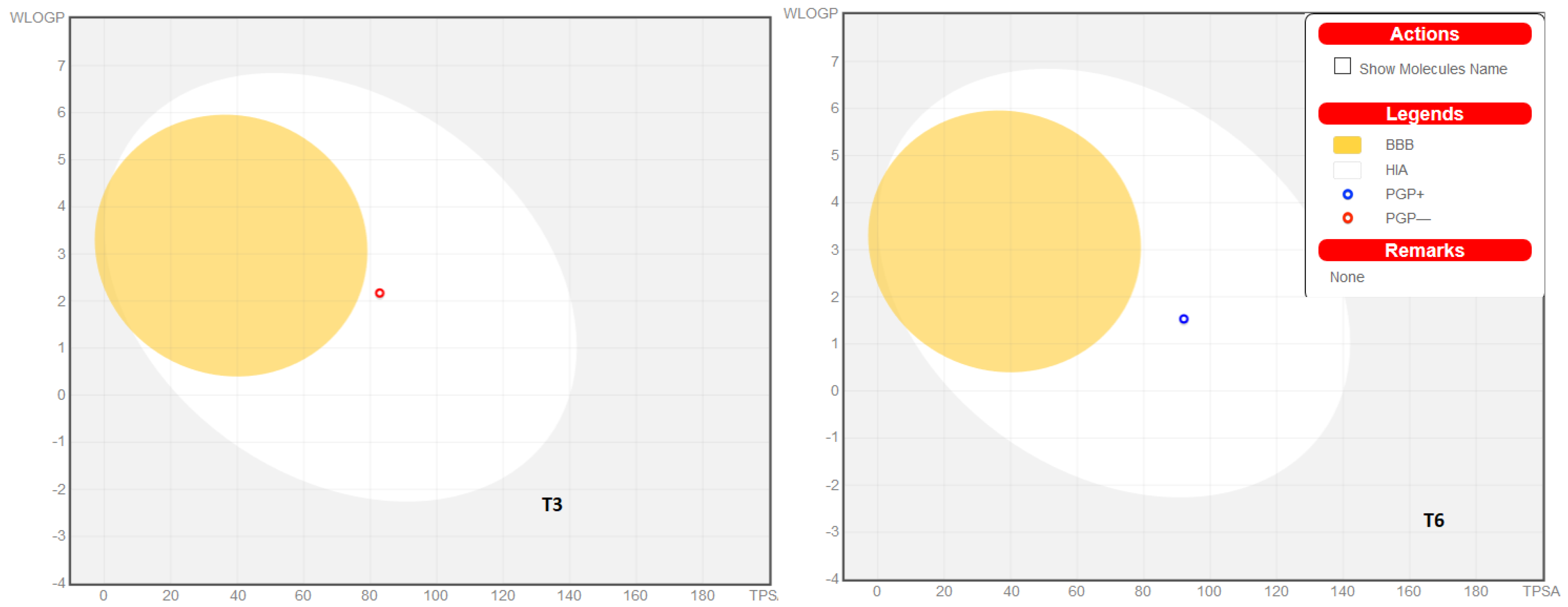
2.6. Molecular Docking Studies
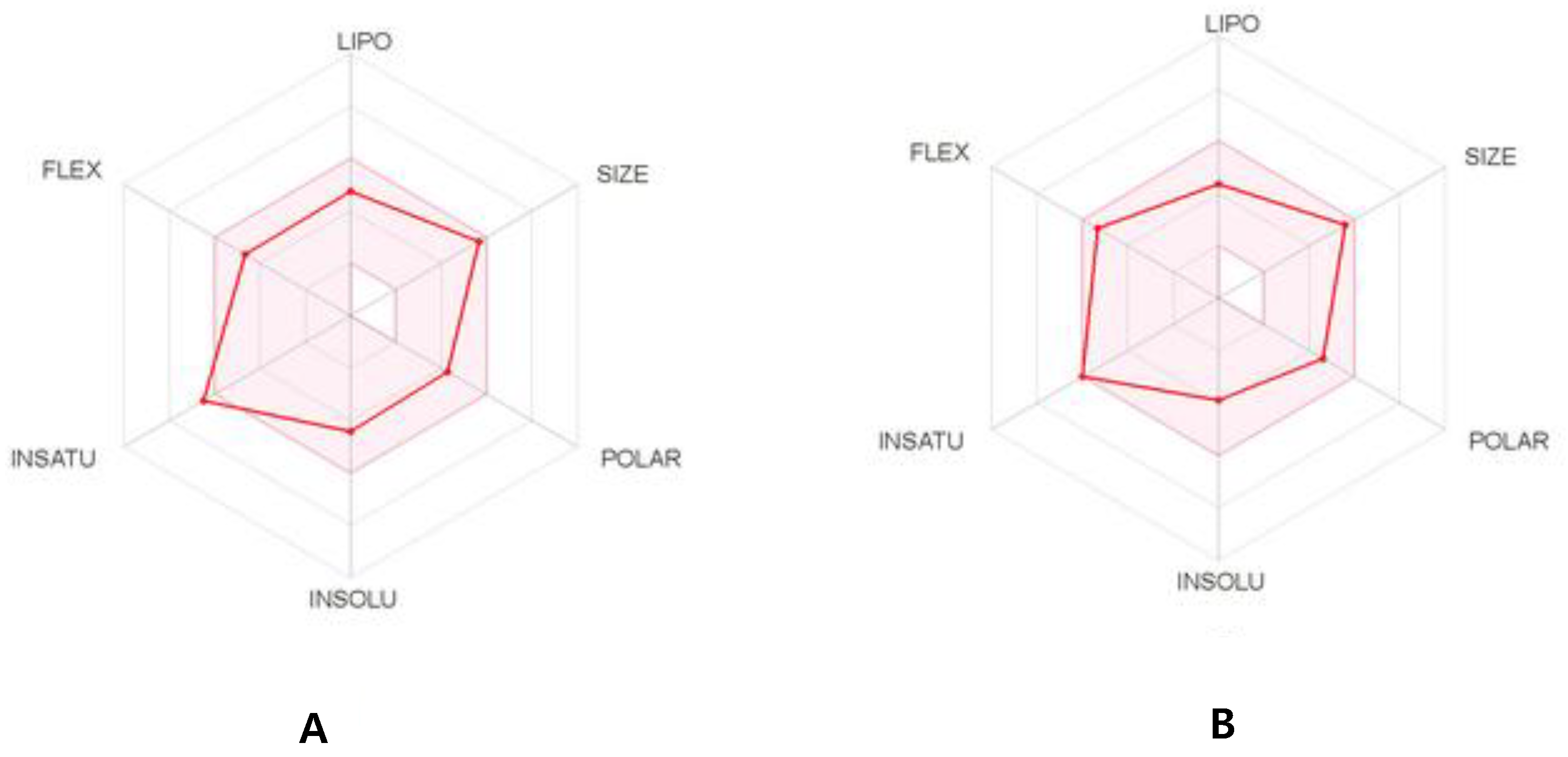
2.7. ADME Prediction
3. Materials and Methods
3.1. Chemical Studies
3.2. Synthesis of Intermediates
3.2.1. Synthesis of 3-chloro-6-[4-(2-fluorophenyl)piperazine-1-yl]pyridazine (1)
3.2.2. Synthesis of 6-(4-(2-fluorophenyl)piperazine-1-yl)-3(2H)-pyridazinone (2)
3.2.3. Synthesis of ethyl 6-(4-(2-fluorophenyl)piperazine-1-yl)-3(2H)-pyridazinone-2-yl-acetate (3)
3.2.4. Synthesis of 6-(4-(2-fluorophenyl)piperazine-1-yl)-3(2H)-pyridazinone-2-yl-acetohydrazide (4)
3.2.5. General Procedure for the Synthesis of T1–T12
3.2.6. 6-[4-(2-Fluorophenyl)piperazine-1-yl]-3(2H)-pyridazinone-2-acetyl-2-(3-bromobenzal)hydrazon (T8)
3.2.7. 6-[4-(2-Fluorophenyl)piperazine-1-yl]-3(2H)-pyridazinone-2-acetyl-2-(2-fluorobenzal)hydrazon (T9)
3.2.8. 6-[4-(2-Fluorophenyl)piperazine-1-yl]-3(2H)-pyridazinone-2-acetyl-2-(4-florobenzal)hydrazon (T10)
3.2.9. 6-[4-(2-Fluorophenyl)piperazine-1-yl]-3(2H)-pyridazinone-2-acetyl-2-(4-methoxybenzal)hydrazon (T11)
3.2.10. 6-[4-(2-Fluorophenyl)piperazine-1-yl]-3(2H)-pyridazinone-2-acetyl-2-(2-methylbenzal)hydrazon (T12)
3.3. Enzyme Assays
3.4. Enzyme Inhibitions and Kinetics
3.5. Analysis of Inhibitor Reversibilities
3.6. Cytotoxicity
3.7. Molecular Docking Studies
3.8. ADME Prediction
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Harilal, S.; Jose, J.; Parambi, D.G.T.; Kumar, R.; Mathew, G.E.; Uddin, M.S.; Kim, H.; Mathew, B. Advancements in nanotherapeutics for Alzheimer’s disease: Current perspectives. J. Pharm. Pharmacol. 2019, 71, 1370–1383. [Google Scholar] [CrossRef] [PubMed]
- Geldenhuys, W.J.; Darvesh, A.S. Pharmacotherapy of Alzheimer’s disease: Current and future trends. Expert. Rev. Neurother. 2015, 5, 3–5. [Google Scholar] [CrossRef] [PubMed]
- Chiu, P.Y.; Wei, C.Y. Donepezil in the one-year treatment of dementia with Lewy bodies and Alzheimer’s disease. J. Neurol. Sci. 2017, 81, 22. [Google Scholar] [CrossRef]
- Mathew, B.; Parambi, D.G.T.; Mathew, G.E.; Uddin, M.S.; Inasu, S.T.; Kim, H.; Marathakam, A.; Unnikrishnan, M.K.; Carradori, S. Emerging therapeutic potentials of dual-acting MAO and AChE inhibitors in Alzheimer’s and Parkinson’s diseases. Arch. Pharm. 2019, 352, e1900177. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Baik, S.H.; Kang, S.; Cho, S.W.; Bae, J.; Cha, M.Y.; Sailor, M.J.; Jung, I.M.; Ahn, K.H. Close correlation of monoamine oxidase activity with progress of Alzheimer’s disease in mice, observed by in vivo two-photon imaging. ACS Cent. Sci. 2016, 2, 967–975. [Google Scholar] [CrossRef] [PubMed]
- Kumar, B.; Prakash, V.; Kumar, V. A perspective on monoamine oxidase enzyme as drug target: Challenges and opportunities. Curr. Drug Targets. 2017, 18, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Finberg, J.P.M.; Rabey, J.M. Inhibitors of MAO-A and MAO-B in psychiatry and neurology. Front. Pharmacol. 2016, 7, 340. [Google Scholar] [CrossRef]
- Guglielmi, P.; Mathew, B.; Secci, D.; Carradori, S. Chalcones: Unearthing their therapeutic possibility as monoamine oxidase B inhibitors. Eur. J. Med. Chem. 2020, 112650. [Google Scholar] [CrossRef]
- Carradori, S.; Silvestri, R. New frontiers in selective human MAO-B inhibitors. J. Med. Chem. 2015, 58, 6717–6732. [Google Scholar] [CrossRef]
- Tripathi, R.K.P.; Ayyannan, S.R. Monoamine oxidase-B inhibitors as potential neurotherapeutic agents: An overview and update. Med. Res. Rev. 2019, 39, 1603–1706. [Google Scholar] [CrossRef]
- Guglielmi, P.; Carradori, S.; Ammazzalorso, A.; Secci, D. Novel approaches to the discovery of selective human monoamine oxidase-B inhibitors: Is there room for improvement? Expert Opin. Drug Discov. 2019, 14, 995–1035. [Google Scholar] [CrossRef] [PubMed]
- Kumar, B.; Sheetal, S.; Mantha, A.K.; Kumar, V. Recent developments on the structure-activity relationship studies of MAO inhibitors and their role in different neurological disorders. RSC Adv. 2016, 6, 42660–42683. [Google Scholar] [CrossRef]
- Mathew, B.; Haridas, A.; Suresh, J.; Mathew, G.E.; Ucar, G.; Jayaprakash, V. Monoamine oxidase inhibitory actions of chalcones. A mini review. Cent. Nerv. Syst. Agents Med. Chem. 2016, 16, 120–136. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.W.; Jang, B.K.; Cho, N.; Park, J.H.; Yeon, S.K.; Ju, E.J.; Lee, Y.S.; Han, G.; Pae, A.M.; Kim, D.J.; et al. Synthesis of a series of unsaturated ketone derivatives as selective and reversible monoamine oxidase inhibitors. Bioorg. Med. Chem. 2015, 23, 6486–6496. [Google Scholar] [CrossRef] [PubMed]
- Mathew, B.; Mathew, G.E.; Petzer, J.P.; Petzer, A. Structural exploration of synthetic chromones as selective MAO-B inhibitors. A Mini Review. Comb. Chem. High Throughput Screen. 2017, 20, 522–532. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, A.; Silva, T.; Yáñez, M.; Viña, D.; Orallo, F.; Ortuso, F.; Uriarte, E.; Alcaro, S.; Borges, F. Chromone, a privileged scaffold for the development of monoamine oxidase inhibition. J. Med. Chem. 2011, 54, 5165–5173. [Google Scholar] [CrossRef]
- Reis, J.; Cagide, F.; Chavarria, D.; Silva, T.B.; Fernandes, C.; Gaspar, A.; Uriarte, E.; Remiao, F.; Alcaro, S.; Ortuso, F.; et al. Discovery of new chemical entities of old targets. Insight on the lead optimization of chromones based monoamine oxidase B inhibitors. J. Med. Chem. 2016, 59, 5879–5893. [Google Scholar] [CrossRef]
- Badavath, V.N.; Baysal, I.; Ucar, G.; Sinha, B.N.; Jayaprakash, V. Monoamine oxidase inhibitory activity of novel pyrazoline analogues: Curcumin based design and synthesis. ACS Med. Chem. Lett. 2016, 7, 56–61. [Google Scholar] [CrossRef]
- Secci, D.; Bolasco, A.; Chimenti, P.; Carradori, S. The state of the art of pyrazole derivatives as monoamine oxidase inhibitors and antidepressant/anticonvulsant agents. Curr. Med. Chem. 2011, 18, 5114–5144. [Google Scholar] [CrossRef]
- Mathew, B.; Suresh, J.; Anbazhagan, S.; Mathew, G.E. Pyrazoline. A promising scaffold for the inhibition of monoamine oxidase. Cent. Nerv. Syst. Agents Med. Chem. 2013, 13, 195–206. [Google Scholar] [CrossRef]
- Matos, M.J.; Vina, D.; Vazquez-Rodriquez, S.; Uriarte, E.; Santana, L. Focusing on new monoamine oxidase inhibitors. Differently substituted coumarins as an interesting scaffold. Curr. Top. Med. Chem. 2012, 12, 2210–2233. [Google Scholar] [CrossRef] [PubMed]
- Guglielmi, P.; Secci, D.; Petzer, A.; Bagetta, D.; Chimenti, P.; Rotondi, G.; Ferrante, C.; Recinella, L.; Leone, S.; Alcaro, S.; et al. Benzo[b]tiophen-3-ol derivatives as effective inhibitors of human monoamine oxidase: Design, synthesis, and biological activity. J. Enzym. Inhib. Med. Chem. 2019, 34, 1511–1525. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.M.; Rangarajan, T.M.; Chaudhary, R.; Singh, R.P.; Singh, M.; Singh, R.P.; Tondo, A.R.; Gambacorta, N.; Nicolotti, O.; Mathew, B.; et al. Novel class of chalcone oxime ethers as potent monoamine oxidase-B and acetylcholinesterase inhibitors. Molecules 2020, 25, 2356. [Google Scholar] [CrossRef] [PubMed]
- Maliyakkal, N.; Eom, B.H.; Heo, J.H.; Almoyad, M.A.A.; Parambi, D.G.T.; Gambacorta, N.; Nicolotti, O.; Beeran, A.A.; Kim, H.; Mathew, B. A new potent and selective monoamine oxidase-B inhibitor with extended conjugation in chalcone framework. 1-[4-(morpholin-4-yl)phenyl]-5-phenylpenta-2,4-dien-1-one. Chem. Med. Chem. 2020, 15, 1629–1633. [Google Scholar] [CrossRef] [PubMed]
- Hagenow, J.; Hagenow, S.; Grau, K.; Khanfar, M.; Hefke, L.; Proschak, E.; Stark, H. Reversible small molecule inhibitors of MAO A and MAO B with anilide motifs. Drug Des. Dev. Ther. 2020, 14, 371–393. [Google Scholar] [CrossRef] [PubMed]
- Kavully, F.S.; Oh, J.M.; Dev, S.; Kaipakasseri, S.; Palakkathondi, A.; Vengamthodi, A.; Azeez, R.F.A.; Tondo, A.R.; Nicolotti, O.; Kim, H.; et al. Design of enamides as new selective monoamine oxidase-B inhibitors. J. Pharm. Pharmacol. 2020, 72, 916–926. [Google Scholar] [CrossRef]
- Turan-Zitouni, G.; Hussein, W.; Saglık, B.N.; Tabbi, A.; Korkut, B. Design, synthesis and biological evaluation of novel N-pyridyl-hydrazone derivatives as potential monoamine oxidase (MAO) inhibitors. Molecules 2018, 23, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Dubey, S.; Bhosle, P.A. Pyridazinone: An important element of pharmacophore possessing broad spectrum of activity. Med. Chem. Res. 2015, 24, 3579–3598. [Google Scholar] [CrossRef]
- Özdemir, Z.; Alagöz, M.A.; Uslu, H.; Karakurt, A.; Erikci, A.; Ucar, G.; Uysa, M. Synthesis, molecular modelling and biological activity of some pyridazinone derivatives as selective human monoamine oxidase-B inhibitors. Pharmacol. Rep. 2020, 72, 692–704. [Google Scholar] [CrossRef]
- Özdemir, Z.; Başak-Türkmen, N.; Ayhan, İ.; Çiftçi, O.; Uysal, M. Synthesis of new 6-[4-(2-fluorophenylpiperazine-1-yl)]-3(2h)-pyridazinone-2-acethyl-2-(substitutedbenzal)hydrazine derivatives and evaluation of their cytotoxic effects in liver and colon cancer cell lines. Pharm. Chem. J. 2019, 52, 923–929. [Google Scholar] [CrossRef]
- Özdemir, Z.; Utku, S.; Mathew, B.; Carradori, S.; Orlando, G.; Simone, S.D.; Alagöz, M.A.; Özçelik, A.B.; Uysal, M.; Claudio Ferrante, F. Synthesis and biological evaluation of new 3(2H)-pyridazinone derivatives as non-toxic anti-cancer compounds against colon carcinoma cells. J. Enzym. Inhib. Med. Chem. 2020, 35, 1100–1109. [Google Scholar] [CrossRef] [PubMed]
- Vergara, S.; Carda, M.; Agut, R.; Yepes, L.M.; Velez, I.D.; Robledo, S.M.; Galeano, W.C. Synthesis, antiprotozoal activity and cytotoxicity in U-937 macrophages of triclosan–hydrazone hybrids. Med. Chem. Res. 2017, 26, 3262–3273. [Google Scholar] [CrossRef]
- Govindasami, T.; Pandey, A.; Palanivelu, N.; Pandey, A. Synthesis, Characterization and antibacterial activity of biologically important vanillin related hydrazone derivatives. Int. J. Org. Chem. 2011, 1, 71–77. [Google Scholar] [CrossRef]
- Mathew, B.; Mathew, G.E.; Ucar, G.; Joy, M.; Nafna, E.K.; Suresh, J. Monoamine oxidase inhibitory activity of methoxy-substituted chalcones. Int. J. Biol. Macromol. 2017, 104, 1321–1329. [Google Scholar] [CrossRef] [PubMed]
- Mathew, B.; Ucar, G.; Mathew, G.E.; Mathew, S.; Purapurath, P.K.; Moolayil, F.; Mohan, S.; Gupta, S.V. Monoamine oxidase inhibitory activity: Methyl- versus chloro-chalcone derivatives. Chem. Med. Chem. 2016, 11, 2649–2655. [Google Scholar] [CrossRef]
- Özçelik, A.B.; Özdemir, Z.; Sari, S.; Utku, S.; Uysal, M. A new series of pyridazinone derivatives as cholinesterases inhibitors: Synthesis, in vitro activity and molecular modeling studies. Pharmacol. Rep. 2019, 71, 1253–1263. [Google Scholar] [CrossRef]
- Mathew, B.; Baek, S.C.; Parambi, D.G.T.; Lee, J.P.; Joy, M.; Annie Rilda, P.R.; Randev, R.V.; Nithyamol, P.; Vijayan, V.; Inasu, S.T.; et al. Selected aryl thiosemicarbazones as a new class of multi-targeted monoamine oxidase inhibitors. Med. Chem. Comm. 2018, 9, 1871–1881. [Google Scholar] [CrossRef]
- Lee, H.W.; Ryu, H.W.; Kang, M.G.; Park, D.; Oh, S.R.; Kim, H. Potent selective monoamine oxidase B inhibition by maackiain, a pterocarpan from the roots of Sophora flavescens. Bioorg. Med. Chem. Lett. 2016, 26, 4714–4719. [Google Scholar] [CrossRef]
- Baek, S.C.; Lee, H.W.; Ryu, H.W.; Kang, M.G.; Park, D.; Kim, S.H.; Cho, M.L.; Oh, S.R.; Kim, H. Selective inhibition of monoamine oxidase A by hispidol. Bioorg. Med. Chem. Lett. 2018, 15, 584–588. [Google Scholar] [CrossRef]
- Parambi, D.G.T.; Oh, J.M.; Baek, S.C.; Lee, J.P.; Tondo, A.R.; Nicolotti, O.; Kim, H.; Mathew, B. Design, synthesis and biological evaluation of oxygenated chalcones as potent and selective MAO-B inhibitors. Bioorg. Chem. 2019, 93, 103335. [Google Scholar] [CrossRef]
- Son, S.Y.; Ma, J.; Kondou, Y.; Yoshimura, M.; Yamashita, E.; Tsukihara, T. Structure of human monoamine oxidase A at 2.2-A resolution: The control of opening the entry for substrates/inhibitors. Proc. Natl. Acad. Sci. USA 2008, 105, 5739–5744. [Google Scholar] [CrossRef] [PubMed]
- Binda, C.; Wang, J.; Pisani, L.; Caccia, C.; Carotti, A.; Salvati, P.; Edmondson, D.E.; Mattevi, A. Structures of human monoamine oxidase B complexes with selective noncovalent inhibitors: Safinamide and coumarin analogs. J. Med. Chem. 2007, 50, 5848–5852. [Google Scholar] [CrossRef]
- Schrödinger Release 2018-2. Prime; Schrödinger, LLC: New York, NY, USA, 2018. [Google Scholar]
- Schrödinger Release 2018-2. Glide; Schrödinger, LLC: New York, NY, USA, 2018. [Google Scholar]
- Mangiatordi, G.F.; Alberga, D.; Pisani, L.; Gadaleta, D.; Trisciuzzi, D.; Farina, R.; Carotti, A.; Lattanzi, G.; Catto, M.; Nicolotti, O. A rational approach to elucidate human monoamine oxidase molecular selectivity. Eur. J. Pharm. Sci. 2017, 101, 90–99. [Google Scholar] [CrossRef] [PubMed]
- LigPrep; Schrödinger, LLC: New York, NY, USA, 2018.
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Reeta; Baek, S.C.; Lee, J.P.; Rangarajan, T.M.; Ayushee; Singh, R.P.; Singh, M.; Mangiatordi, G.F.; Nicolotti, O.; Kim, H.; et al. Ethyl acetohydroxamate incorporated chalcones: Unveiling a novel class of chalcones for multitarget monoamine oxidase-B inhibitors against Alzheimer’s disease. CNS Neurol. Disord. Drug Targets 2019, 8, 643–654. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are available from the authors. |



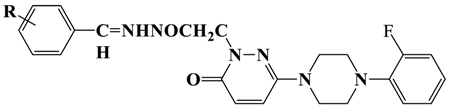 | |||||
|---|---|---|---|---|---|
| Compound No. | R | Yield (%) | P | MM | Molecular Formula |
| T1 | H | 79.90 | 226–228 | 434.48 | C23H23FN6O2 |
| T2 | 4-CH3 | 78.15 | 198–200 | 448.49 | C24H25FN6O2 |
| T3 | 2-Cl | 73.19 | 203–205 | 468.91 | C23H22ClFN6O2 |
| T4 | 4-Cl | 91.21 | 192–194 | 468.91 | C23H22ClFN6O2 |
| T5 | 4-Br | 85.69 | 209–211 | 513.36 | C23H22BrFN6O2 |
| T6 | 2-CH3O | 77.45 | 239–241 | 464.49 | C24H25FN6O3 |
| T7 | 4-N(CH3)2 | 68.77 | 237–239 | 477.53 | C25H28FN7O2 |
| T8 | 3-Br | 90.08 | 207–208 | 513.36 | C23H22BrFN6O2 |
| T9 | 2-F | 88.36 | 225–226 | 452.47 | C23H22 F2N6O2 |
| T10 | 4-F | 57.70 | 178–180 | 452.47 | C23H22 F2N6O2 |
| T11 | 4- OCH3 | 75.32 | 148–150 | 464.49 | C24H25FN6O3 |
| T12 | 2-CH3 | 72.21 | 244–245 | 448.49 | C24H25FN6O2 |
| Compounds | Residual Activity (%) | IC50 (µM) | SI b | ||
|---|---|---|---|---|---|
| MAO-A | MAO-B | MAO-A | MAO-B | ||
| (10 µM) | (1.0 µM) | ||||
| T1 | 96.6 ± 1.20 | 99.4 ± 0.85 | - | 7.68 ± 0.14 | - |
| T2 | 24.6 ± 2.86 | 4.22 ± 0.85 | 4.29 ± 0.12 | 0.10 ± 0.035 | 42.9 |
| T3 | 24.6 ± 0.97 | 3.01 ± 0.85 | 4.19 ± 0.27 | 0.039 ± 0.0028 | 107.4 |
| T4 | 75.2 ± 4.70 | 6.00 ± 0.94 | - | 0.099 ± 0.0069 | - |
| T5 | 84.0 ± 0.68 | 54.0 ± 2.83 | - | 1.68 ± 0.11 | - |
| T6 | 20.7 ± 0.62 | −15.3 ± 0.94 | 1.57 ± 0.80 | 0.013 ± 0.0016 | 120.8 |
| T7 | 25.4 ± 6.92 | 57.8 ± 9.58 | 4.43 ± 0.29 | 0.87 ± 0.056 | - |
| T8 | 73.3 ± 2.86 | 67.2 ± 9.58 | - | 2.67 ± 0.051 | - |
| T9 | 35.2 ± 0.56 | 2.60 ± 0.74 | 6.76 ± 0.46 | 0.20 ± 0.0017 | 33.8 |
| T10 | 33.7 ± 4.70 | 95.3 ± 6.70 | 7.35 ± 0.059 | 35.3 ± 0.39 | 0.21 |
| T11 | 70.4 ± 8.50 | 61.1 ± 2.98 | - | 6.86 ± 0.37 | - |
| T12 | 61.8 ± 1.65 | 17.9 ± 5.95 | - | 0.15 ± 0.021 | - |
| Toloxatone | 1.08 ± 0.025 | - | |||
| Lazabemide | - | 0.14 ± 0.011 | |||
| Clorgyline | 0.0070 ± 0.00070 | - | |||
| Pargyline | - | 0.030 ± 0.00083 | |||
| Compounds | Docking Score (kcal/mol) | |
|---|---|---|
| MAO-A | MAO-B | |
| T3 | −8.20 | −9.45 |
| T6 | −8.43 | −9.56 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Çeçen, M.; Oh, J.M.; Özdemir, Z.; Büyüktuncel, S.E.; Uysal, M.; Abdelgawad, M.A.; Musa, A.; Gambacorta, N.; Nicolotti, O.; Mathew, B.; et al. Design, Synthesis, and Biological Evaluation of Pyridazinones Containing the (2-Fluorophenyl) Piperazine Moiety as Selective MAO-B Inhibitors. Molecules 2020, 25, 5371. https://doi.org/10.3390/molecules25225371
Çeçen M, Oh JM, Özdemir Z, Büyüktuncel SE, Uysal M, Abdelgawad MA, Musa A, Gambacorta N, Nicolotti O, Mathew B, et al. Design, Synthesis, and Biological Evaluation of Pyridazinones Containing the (2-Fluorophenyl) Piperazine Moiety as Selective MAO-B Inhibitors. Molecules. 2020; 25(22):5371. https://doi.org/10.3390/molecules25225371
Chicago/Turabian StyleÇeçen, Muhammed, Jong Min Oh, Zeynep Özdemir, Saliha Ebru Büyüktuncel, Mehtap Uysal, Mohamed A. Abdelgawad, Arafa Musa, Nicola Gambacorta, Orazio Nicolotti, Bijo Mathew, and et al. 2020. "Design, Synthesis, and Biological Evaluation of Pyridazinones Containing the (2-Fluorophenyl) Piperazine Moiety as Selective MAO-B Inhibitors" Molecules 25, no. 22: 5371. https://doi.org/10.3390/molecules25225371
APA StyleÇeçen, M., Oh, J. M., Özdemir, Z., Büyüktuncel, S. E., Uysal, M., Abdelgawad, M. A., Musa, A., Gambacorta, N., Nicolotti, O., Mathew, B., & Kim, H. (2020). Design, Synthesis, and Biological Evaluation of Pyridazinones Containing the (2-Fluorophenyl) Piperazine Moiety as Selective MAO-B Inhibitors. Molecules, 25(22), 5371. https://doi.org/10.3390/molecules25225371











