Proteomic Profiling of the Human Fetal Multipotent Mesenchymal Stromal Cells Secretome
Abstract
1. Introduction
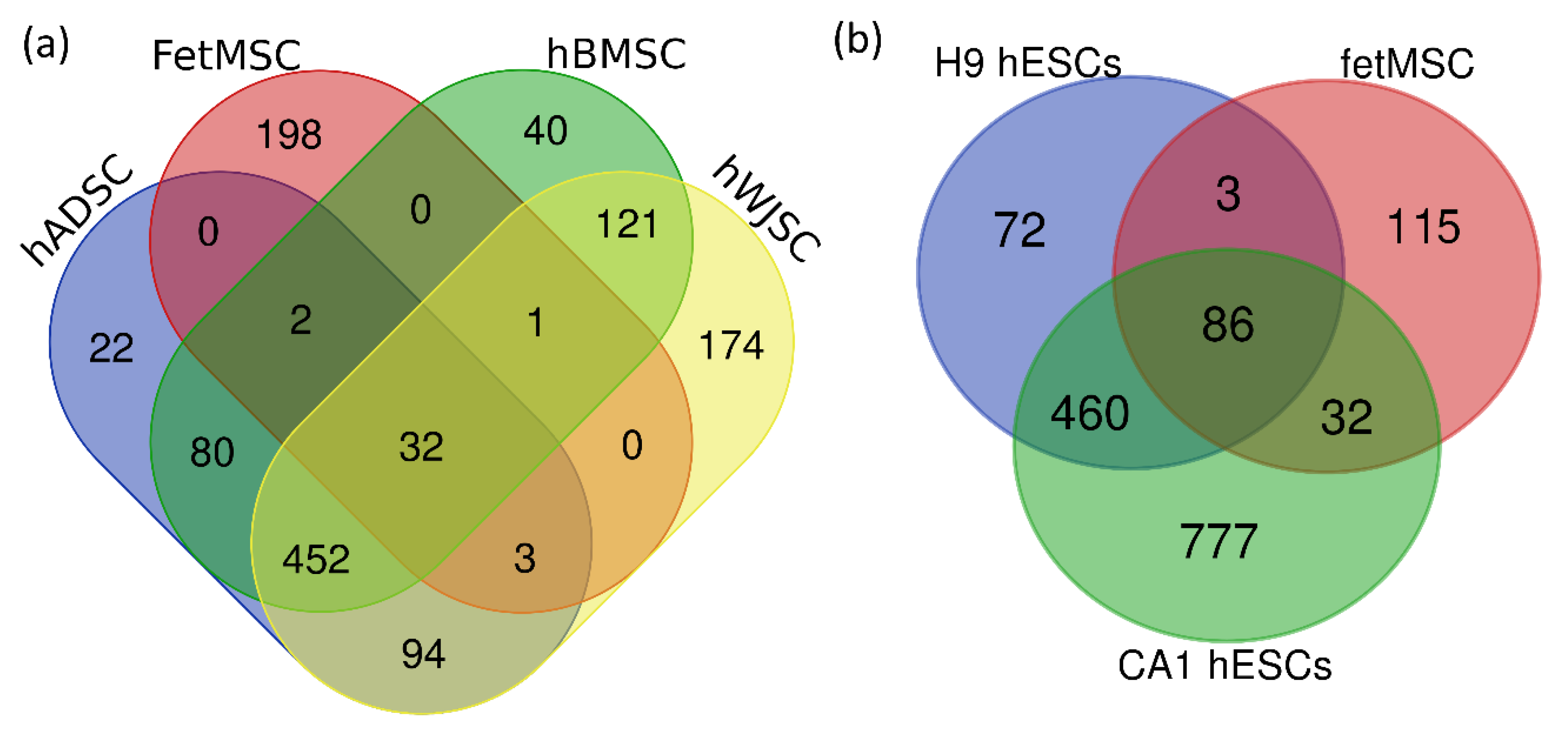
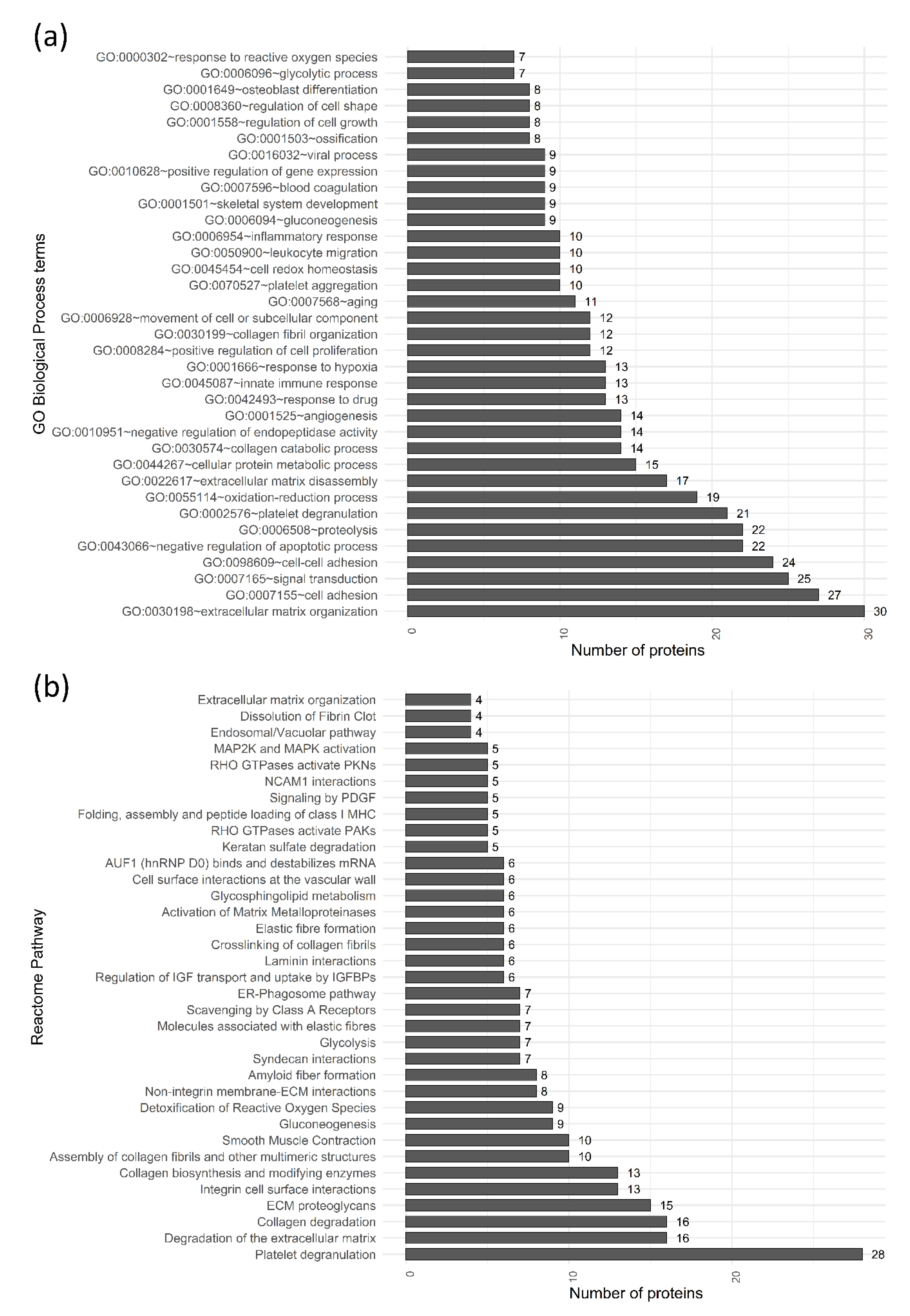
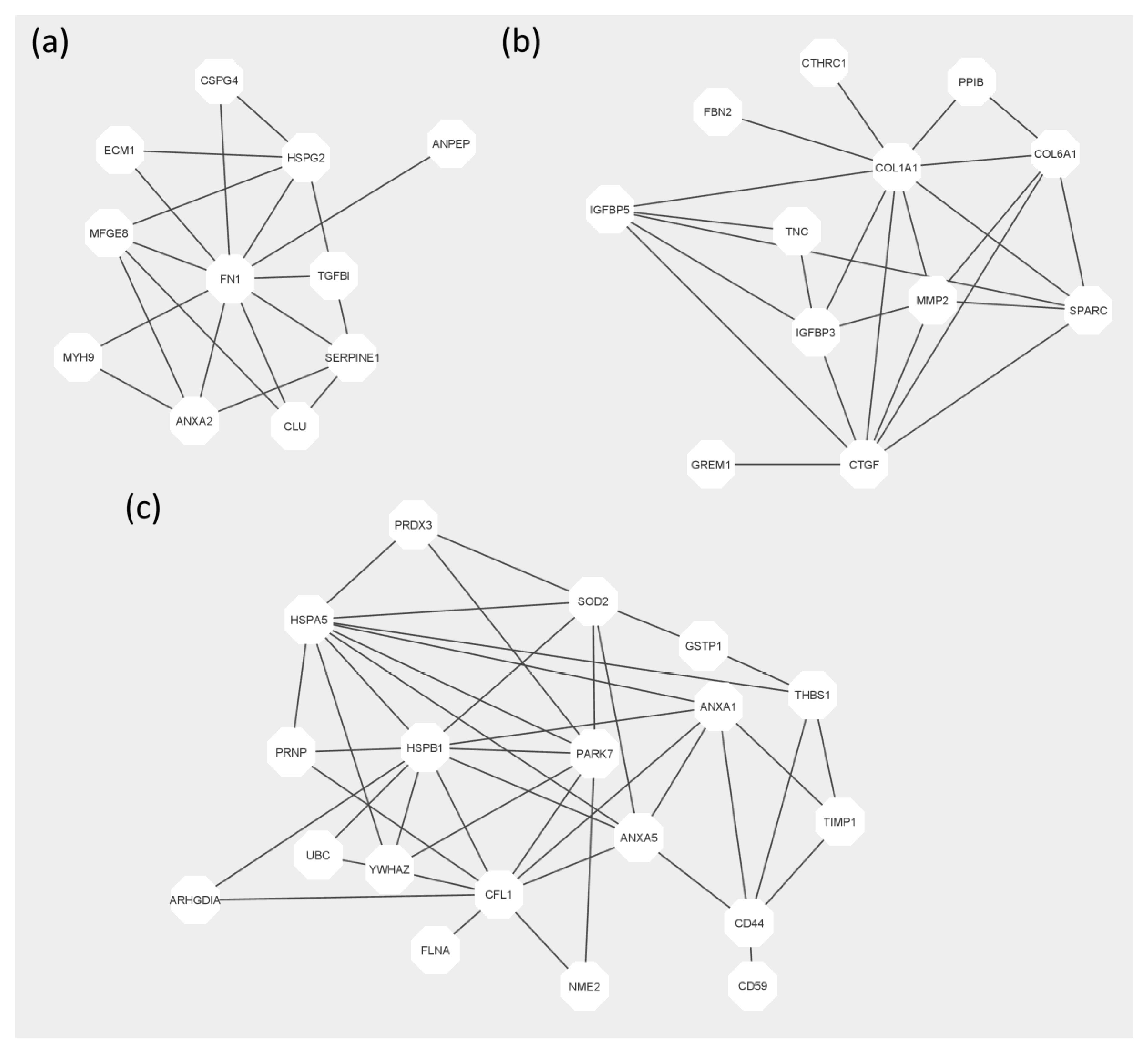
2. Results
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. FetMSCs Secretome Harvest
4.3. Protein Isolation
4.4. LC-MALDI MS/MS Analysis
4.5. Protein Identification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Webster, A. Conclusion: Regenerative Medicine—A New Paradigm? In The Global Dynamics of Regenerative Medicine: A Social Science Critique; Webster, A., Ed.; Palgrave Macmillan: London, UK, 2013; pp. 217–227. [Google Scholar] [CrossRef]
- Gomzikova, M.O.; Rizvanov, A.A. Current trends in regenerative medicine: From cell to cell-free therapy. BioNanoScience 2017, 7, 240–245. [Google Scholar] [CrossRef]
- Mahla, R.S. Stem cells applications in regenerative medicine and disease therapeutics. Int. J. Cell Biol. 2016, 2016, 6940283. [Google Scholar] [CrossRef] [PubMed]
- Volarevic, V.; Markovic, B.S.; Gazdic, M.; Volarevic, A.; Jovicic, N.; Arsenijevic, N.; Armstrong, L.; Djonov, V.; Lako, M.; Stojkovic, M. Ethical and safety issues of stem cell-based therapy. Int. J. Med. Sci. 2018, 15, 36. [Google Scholar] [CrossRef] [PubMed]
- Rikhtegar, R.; Pezeshkian, M.; Dolati, S.; Safaie, N.; Rad, A.A.; Mahdipour, M.; Nouri, M.; Jodati, A.R.; Yousefi, M. Stem cells as therapy for heart disease: iPSCs, ESCs, CSCs, and skeletal myoblasts. Biomed. Pharmacother. 2019, 109, 304–313. [Google Scholar] [CrossRef]
- Han, Y.; Li, X.; Zhang, Y.; Han, Y.; Chang, F.; Ding, J. Mesenchymal stem cells for regenerative medicine. Cells 2019, 8, 886. [Google Scholar] [CrossRef]
- Gnecchi, M.; He, H.; Liang, O.D.; Melo, L.G.; Morello, F.; Mu, H.; Noiseux, N.; Zhang, L.; Pratt, R.E.; Ingwall, J.S.; et al. Paracrine action accounts for marked protection of ischemic heart by Akt-modified mesenchymal stem cells. Nat. Med. 2005, 11, 367–368. [Google Scholar] [CrossRef]
- Teixeira, F.G.; Salgado, A.J. Mesenchymal stem cells secretome: Current trends and future challenges. Neural Regen. Res. 2020, 15, 75. [Google Scholar] [CrossRef]
- Hass, R.; Kasper, C.; Böhm, S.; Jacobs, R. Different populations and sources of human mesenchymal stem cells (MSC): A comparison of adult and neonatal tissue-derived MSC. Cell Commun. Signal. 2011, 9, 12. [Google Scholar] [CrossRef]
- Fekete, N.; Rojewski, M.T.; Fürst, D.; Kreja, L.; Ignatius, A.; Dausend, J.; Schrezenmeier, H. GMP-compliant isolation and large-scale expansion of bone marrow-derived MSC. PLoS ONE 2012, 7, e43255. [Google Scholar] [CrossRef]
- Teixeira, F.G.; Carvalho, M.M.; Sousa, N.; Salgado, A.J. Mesenchymal stem cells secretome: A new paradigm for central nervous system regeneration? Cell. Mol. Life Sci. 2013, 70, 3871–3882. [Google Scholar] [CrossRef]
- Sousa, B.R.; Parreira, R.C.; Fonseca, E.A.; Amaya, M.J.; Tonelli, F.M.; Lacerda, S.M.; Lalwani, P.; Santos, A.K.; Gomes, K.N.; Ulrich, H.; et al. Human adult stem cells from diverse origins: An overview from multiparametric immunophenotyping to clinical applications. Cytom. A 2014, 85, 43–77. [Google Scholar] [CrossRef] [PubMed]
- Bara, J.J.; Richards, R.G.; Alini, M.; Stoddart, M.J. Concise review: Bone marrow-derived mesenchymal stem cells change phenotype following in vitro culture: Implications for basic research and the clinic. Stem Cells 2014, 32, 1713–1723. [Google Scholar] [CrossRef] [PubMed]
- Assoni, A.; Coatti, G.; Valadares, M.C.; Beccari, M.; Gomes, J.; Pelatti, M.; Mitne-Neto, M.; Carvalho, V.M.; Zatz, M. Different donors mesenchymal stromal cells secretomes reveal heterogeneous profile of relevance for therapeutic use. Stem Cells Dev. 2017, 26, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Pennings, I.; van Dijk, L.A.; van Huuksloot, J.; Fledderus, J.O.; Schepers, K.; Braat, A.K.; Hsiao, E.C.; Barruet, E.; Morales, B.M.; Verhaar, M.C.; et al. Effect of donor variation on osteogenesis and vasculogenesis in hydrogel cocultures. J. Tissue Eng. 2019, 13, 433–445. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Y.; Teoh, S.H.; Chong, M.S.; Schantz, J.T.; Fisk, N.M.; Choolani, M.A.; Chan, J. Superior osteogenic capacity for bone tissue engineering of fetal compared with perinatal and adult mesenchymal stem cells. Stem Cells 2009, 27, 126–137. [Google Scholar] [CrossRef]
- Krylova, T.A.; Kol’tsova, A.M.; Zenin, V.V.; Musorina, A.S.; Iakovleva, T.K.; Polianskaia, G.G. Comparative characteristics of new mesenchymal stem cell lines derived from human embryonic stem cells, bone marrow and foreskin. Tsitologiia 2012, 54, 5–16. [Google Scholar] [CrossRef]
- Krylova, T.A.; Musorina, A.S.; Zenin, V.V.; Iakovleva, T.K.; Polianskaia, G.G. Comparative characteristics of mesenchymal stem cell lines derived from bone marrow and muscle of limb of early human embryo. Tsitologiia 2014, 56, 562. [Google Scholar]
- Aleksandrova, S.A.; Nashchekina, Y.A.; Nadezhdin, S.V.; Vasilyev, S.A.; Savchenko, R.R.; Pokrovskaya, L.A.; Blinova, M.I.; Mikhailova, N.A.; Khotin, M.G. Osteoinductive properties of human mesenchymal stem cells secretome obtained by automatic cell cultivation system. Tsitologiia 2020, 62, 238–249. [Google Scholar]
- Kalra, H.; Simpson, R.J.; Ji, H.; Aikawa, E.; Altevogt, P.; Askenase, P.; Buzas, E. Vesiclepedia: A compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012, 10, e1001450. [Google Scholar] [CrossRef]
- Jiao, X.; Sherman, B.T.; Huang, D.W.; Stephens, R.; Baseler, M.W.; Lane, H.C.; Lempicki, R.A. DAVID-WS: A stateful web service to facilitate gene/protein list analysis. Bioinformatics 2012, 28, 1805–1806. [Google Scholar] [CrossRef]
- Kehl, D.; Generali, M.; Mallone, A.; Heller, M.; Uldry, A.C.; Cheng, P.; Gantenbein, B.; Hoerstrup, S.P.; Weber, B. Proteomic analysis of human mesenchymal stromal cell secretomes: A systematic comparison of the angiogenic potential. NPJ Regen. Med. 2019, 4, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Hughes, C.; Radan, L.; Chang, W.Y.; Stanford, W.L.; Betts, D.H.; Postovit, L.M.; Lajoie, G.A. Mass spectrometry–based proteomic analysis of the matrix microenvironment in pluripotent stem cell culture. Mol. Cell Proteom. 2012, 11, 1924–1936. [Google Scholar] [CrossRef] [PubMed]
- Eleuteri, S.; Fierabracci, A. Insights into the secretome of mesenchymal stem cells and its potential applications. Int. J. Mol. Sci. 2019, 20, 4597. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Ni, J.; Smits, P.; Underhill, C.B.; Xie, B.; Chen, Y.; Ding, I. Extracellular matrix protein 1 (ECM1) has angiogenic properties and is expressed by breast tumor cells. FASEB J. 2001, 15, 988–994. [Google Scholar] [CrossRef]
- Liu, W.; Hajjar, K.A. The annexin A2 system and angiogenesis. Biol. Chem. 2016, 397, 1005–1016. [Google Scholar] [CrossRef]
- Hashida, H.; Takabayashi, A.; Kanai, M.; Adachi, M.; Kondo, K.; Kohno, N.; Miyake, M. Aminopeptidase N is involved in cell motility and angiogenesis: Its clinical significance in human colon cancer. Gastroenterology 2002, 122, 376–386. [Google Scholar] [CrossRef]
- Silvestre, J.S.; Théry, C.; Hamard, G.; Boddaert, J.; Aguilar, B.; Delcayre, A.; Clergue, M. Lactadherin promotes VEGF-dependent neovascularization. Nat. Med. 2005, 11, 499–506. [Google Scholar] [CrossRef]
- Shimo, T.; Nakanishi, T.; Nishida, T.; Asano, M.; Kanyama, M.; Kuboki, T.; Takigawa, M. Connective tissue growth factor induces the proliferation, migration, and tube formation of vascular endothelial cells in vitro, and angiogenesis in vivo. J. Biochem. 1999, 126, 137–145. [Google Scholar] [CrossRef]
- Son, H.N.; Nam, J.O.; Kim, S.; Kim, I.S. Multiple FAS1 domains and the RGD motif of TGFBI act cooperatively to bind αvβ3 integrin, leading to anti-angiogenic and anti-tumor effects. Biochim. Biophys. Acta Mol. Cell Res. 2013, 1833, 2378–2388. [Google Scholar] [CrossRef]
- Stefansson, S.; Petitclerc, E.; Wong, M.K.; McMahon, G.A.; Brooks, P.C.; Lawrence, D.A. Inhibition of angiogenesis in vivo by plasminogen activator inhibitor-1. J. Biol. Chem. 2001, 276, 8135–8141. [Google Scholar] [CrossRef]
- Wang, P.; Teng, Z.; Liu, X.; Liu, X.; Kong, C.; Lu, S. The COL6A1 rs201153092 single nucleotide polymorphism, associates with thoracic ossification of the posterior longitudinal ligament. Mol. Med. Rep. 2020, 21, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Barthelemi, S.; Robinet, J.; Garnotel, R.; Antonicelli, F.; Schittly, E.; Hornebeck, W.; Lorimier, S. Mechanical forces-induced human osteoblasts differentiation involves MMP-2/MMP-13/MT1-MMP proteolytic cascade. J. Cell. Biochem. 2012, 113, 760–772. [Google Scholar] [CrossRef] [PubMed]
- Sever, M.; Mammadov, B.; Guler, M.O.; Tekinay, A.B. Tenascin-C mimetic peptide nanofibers direct stem cell differentiation to osteogenic lineage. Biomacromolecules 2014, 15, 4480–4487. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Ogura, N.; Kato, R.; Tomoki, R.; Eda, T.; Okada, H.; Kondoh, T. Gene expression of Semaphorin 7A in human dental follicle cells. Oral. Maxillofac. Surg. 2014, 72, e194–e195. [Google Scholar] [CrossRef]
- Yoshiko, Y.; Maeda, N.; Aubin, J.E. Stanniocalcin 1 stimulates osteoblast differentiation in rat calvaria cell cultures. Endocrinology 2003, 144, 4134–4143. [Google Scholar] [CrossRef] [PubMed]
- Teplyuk, N.M.; Haupt, L.M.; Ling, L.; Dombrowski, C.; Mun, F.K.; Nathan, S.S.; Van Wijnen, A.J. The osteogenic transcription factor Runx2 regulates components of the fibroblast growth factor/proteoglycan signaling axis in osteoblasts. J. Cell. Biochem. 2009, 107, 144–154. [Google Scholar] [CrossRef]
- Hu, K.; Sun, H.; Gui, B.; Sui, C. Gremlin-1 suppression increases BMP-2-induced osteogenesis of human mesenchymal stem cells. Mol. Med. Rep. 2017, 15, 2186–2194. [Google Scholar] [CrossRef]
- Eguchi, K.; Akiba, Y.; Akiba, N.; Nagasawa, M.; Cooper, L.F.; Uoshima, K. Insulin-like growth factor binding Protein-3 suppresses osteoblast differentiation via bone morphogenetic protein-2. Biochem. Biophys. Res. Commun. 2018, 507, 465–470. [Google Scholar] [CrossRef]
- Canalis, E.; Economides, A.N.; Gazzerro, E. Bone morphogenetic proteins, their antagonists, and the skeleton. Endocr. Rev. 2003, 24, 218–235. [Google Scholar] [CrossRef]
- Pinto, V.I.; Senini, V.W.; Wang, Y.; Kazembe, M.P.; McCulloch, C.A. Filamin A protects cells against force-induced apoptosis by stabilizing talin-and vinculin-containing cell adhesions. FASEB J. 2014, 28, 453–463. [Google Scholar] [CrossRef]
- Fujita, Y.; Kitagawa, M.; Nakamura, S.; Azuma, K.; Ishii, G.; Higashi, M.; Kishi, H.; Hiwasa, T.; Koda, K.; Nakajima, N.; et al. CD44 signaling through focal adhesion kinase and its anti-apoptotic effect. FEBS Lett. 2002, 528, 101–108. [Google Scholar] [CrossRef]
- Li, G.; Fridman, R.; Kim, H.R.C. Tissue inhibitor of metalloproteinase-1 inhibits apoptosis of human breast epithelial cells. Cancer Res. 1999, 59, 6267–6275. [Google Scholar] [CrossRef] [PubMed]
- Vizoso, F.J.; Eiro, N.; Cid, S.; Schneider, J.; Perez-Fernandez, R. Mesenchymal stem cell secretome: Toward cell-free therapeutic strategies in regenerative medicine. Int. J. Mol. Sci. 2017, 18, 1852. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Fan, X.; Hong, E.; Chen, J.J. Interaction of oncogenic papillomavirus E6 proteins with fibulin-1. Biochem. Biophys. Res. Commun. 2002, 296, 962–969. [Google Scholar] [CrossRef]
- Hilton, H.G.; Parham, P. Missing or altered self: Human NK cell receptors that recognize HLA-C. Immunogenetics 2017, 69, 567–579. [Google Scholar] [CrossRef]
- Kulpa, D.A.; Collins, K.L. The emerging role of HLA-C in HIV-1 infection. Immunology 2011, 134, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Stricher, F.; Macri, C.; Ruff, M.; Muller, S. HSPA8/HSC70 chaperone protein: Structure, function, and chemical targeting. Autophagy 2013, 9, 1937–1954. [Google Scholar] [CrossRef]
- Kaur, R.; Lal, S.K. The multifarious roles of heterogeneous ribonucleoprotein A1 in viral infections. Rev. Med. Virol. 2020, 30, e2097. [Google Scholar] [CrossRef]
- Newburg, D.S.; Peterson, J.A.; Ruiz-Palacios, G.M.; Matson, D.O.; Morrow, A.L.; Shults, J.; Guerrero, M.L.; Chaturvedi, P.; Newburga, S.O.; Scallan, C.D.; et al. Role of human-milk lactadherin in protectoin against symptomatic rotavirus infection. Lancet 1998, 351, 1160–1164. [Google Scholar] [CrossRef]
- Van de Klundert, M.A.; Van den Biggelaar, M.; Kootstra, N.A.; Zaaijer, H.L. Hepatitis B virus protein X induces degradation of Talin-1. Viruses 2016, 8, 281. [Google Scholar] [CrossRef]
- Ramos, I.; Stamatakis, K.; Oeste, C.L.; Pérez-Sala, D. Vimentin as a Multifaceted Player and Potential Therapeutic Target in Viral Infections. Preprints 2020, 2020050041. [Google Scholar] [CrossRef]
- Dominici, M.L.B.K.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.C.; Krause, D.S.; Horwitz, E.M. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2012; Available online: https://www.R-project.Org (accessed on 11 November 2020).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar] [CrossRef]

Sample Availability: Samples of the compounds are not available from the authors. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lobov, A.A.; Yudintceva, N.M.; Mittenberg, A.G.; Shabelnikov, S.V.; Mikhailova, N.A.; Malashicheva, A.B.; Khotin, M.G. Proteomic Profiling of the Human Fetal Multipotent Mesenchymal Stromal Cells Secretome. Molecules 2020, 25, 5283. https://doi.org/10.3390/molecules25225283
Lobov AA, Yudintceva NM, Mittenberg AG, Shabelnikov SV, Mikhailova NA, Malashicheva AB, Khotin MG. Proteomic Profiling of the Human Fetal Multipotent Mesenchymal Stromal Cells Secretome. Molecules. 2020; 25(22):5283. https://doi.org/10.3390/molecules25225283
Chicago/Turabian StyleLobov, Arseniy A., Natalia M. Yudintceva, Alexey G. Mittenberg, Sergey V. Shabelnikov, Natalia A. Mikhailova, Anna B. Malashicheva, and Mikhail G. Khotin. 2020. "Proteomic Profiling of the Human Fetal Multipotent Mesenchymal Stromal Cells Secretome" Molecules 25, no. 22: 5283. https://doi.org/10.3390/molecules25225283
APA StyleLobov, A. A., Yudintceva, N. M., Mittenberg, A. G., Shabelnikov, S. V., Mikhailova, N. A., Malashicheva, A. B., & Khotin, M. G. (2020). Proteomic Profiling of the Human Fetal Multipotent Mesenchymal Stromal Cells Secretome. Molecules, 25(22), 5283. https://doi.org/10.3390/molecules25225283