Insights into a Protein-Nanoparticle System by Paramagnetic Perturbation NMR Spectroscopy
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Sample Preparation
4.3. Spectroscopy
4.4. Spectroscopic Data Treatment
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kopp, M.; Kollenda, S.; Epple, M. Nanoparticle-Protein Interactions: Therapeutic Approaches and Supramolecular Chemistry. Acc. Chem. Res. 2017, 50, 1383–1390. [Google Scholar] [CrossRef]
- Brancolini, G.; Corazza, A.; Vuano, M.; Fogolari, F.; Mimmi, M.C.; Bellotti, V.; Stoppini, M.; Corni, S.; Esposito, G. Probing the Influence of Citrate-Capped Gold Nanoparticles on an Amyloidogenic Protein. ACS Nano 2015, 9, 2600–2613. [Google Scholar] [CrossRef]
- Brancolini, G.; Maschio, M.C.; Cantarutti, C.; Corazza, A.; Fogolari, F.; Corni, S.; Esposito, G. Citrate stabilized Gold Nanoparticles interfere with Amyloid Fibril formation: D76N and ∆N6 β2-microglobulin Variants. Nanoscale 2018, 10, 4793–4806. [Google Scholar] [CrossRef]
- Cantarutti, C.; Raimondi, S.; Brancolini, G.; Corazza, A.; Giorgetti, S.; Ballico, M.; Zanini, S.; Palmisano, G.; Bertoncin, P.; Marchese, L.; et al. Citrate-stabilized Gold Nanoparticles hinder fibrillogenesis of a pathologic variant of β2-microglobulin. Nanoscale 2017, 9, 3941–3951. [Google Scholar] [CrossRef]
- Cantarutti, C.; Bertoncin, P.; Corazza, A.; Giorgetti, S.; Mangione, P.P.; Bellotti, V.; Fogolari, F.; Esposito, G. Short-chain Alkanethiol Coating for Small-Size Gold Nanoparticles Supporting Protein Stability. Magnetochemistry 2017, 3, 40. [Google Scholar] [CrossRef]
- Cantarutti, C.; Raj, G.; Fogolari, F.; Giorgetti, S.; Corazza, A.; Bellotti, V.; Naumov, P.; Esposito, G. Interference of citrate-stabilized gold nanoparticles on β2-microglobulin oligomeric association. Chem. Commun. 2018, 54, 5422–5425. [Google Scholar] [CrossRef]
- Cantarutti, C.; Bertoncin, P.; Posocco, P.; Hunashal, Y.; Giorgetti, S.; Bellotti, V.; Fogolari, F.; Esposito, G. The interaction of β2-microglobulin and gold nanoparticles: Impact of coating, charge and size. J. Mater. Chem. B 2018, 6, 5964–5974. [Google Scholar] [CrossRef]
- Gejyo, F.; Yamada, T.; Odani, S.; Nakagawa, Y.; Arakawa, M.; Kunitomo, T.; Kataoka, H.; Suzuki, M.; Hirasawa, Y.; Shirahama, T.; et al. A new form of amyloid protein associated with chronic hemodialysis was identified as β2-microglobulin. Biochem. Biophys. Res. Commun. 1985, 129, 701–706. [Google Scholar] [CrossRef]
- Valleix, S.; Gillmore, J.D.; Bridoux, F.; Mangione, P.P.; Dogan, A.; Nedelec, B.; Boimard, M.; Touchard, G.; Goujon, J.M.; Lacombe, C.; et al. Hereditary Systemic Amyloidosis Dur to Asp76Asn Variant of β2-Microglobulin. N. Engl. J. Med. 2012, 366, 2276–2283. [Google Scholar] [CrossRef]
- Verdone, G.; Corazza, A.; Viglino, P.; Pettirossi, F.; Giorgetti, S.; Mangione, P.; Andreola, A.; Stoppini, M.; Bellotti, V.; Esposito, G. The solution structure of human β2-microglobulin reveals the prodromes of its amyloid transition. Protein Sci. 2002, 11, 487–499. [Google Scholar] [CrossRef]
- Esposito, G.; Corazza, A.; Viglino, P.; Verdone, G.; Pettirossi, F.; Fogolari, F.; Makek, A.; Giorgetti, S.; Mangione, P.; Stoppini, M.; et al. Solution structure of β2-microglobulin and insights into fibrillogenesis. BBA Proteins Proteom. 2005, 1753, 76–84. [Google Scholar] [CrossRef]
- Rennella, E.; Corazza, A.; Giorgetti, S.; Fogolari, F.; Viglino, P.; Porcari, R.; Verga, L.; Stoppini, M.; Bellotti, V.; Esposito, G. Folding and Fibrillogenesis: Clues from β2-microglobulin. J. Mol. Biol. 2010, 401, 286–297. [Google Scholar] [CrossRef]
- Mangione, P.P.; Esposito, G.; Relini, A.; Raimondi, S.; Porcari, R.; Giorgetti, S.; Corazza, A.; Fogolari, F.; Penco, A.; Goto, Y.; et al. Amyloid fibrillogenesis of Asp76Asn β2-microglobulin and co-aggregation with the wild type: The crucial roles of shear flow, natural hydrophobic surfaces and chaperones. J. Biol. Chem. 2013, 288, 30917–30930. [Google Scholar] [CrossRef] [PubMed]
- Esposito, G.; Michelutti, R.; Verdone, G.; Viglino, P.; Hernandez, H.; Robinson, C.V.; Amoresano, A.; Dal Piaz, F.; Monti, M.; Pucci, P.; et al. Removal of the N-terminal hexapeptide from human β2-microglobulin facilitates protein aggregation and fibril formation. Protein Sci. 2000, 9, 831–845. [Google Scholar] [CrossRef]
- Linse, S.; Cabaleiro-Lago, C.; Xue, W.F.; Lynch, I.; Lindman, S.; Thulin, E.; Radford, S.E.; Dawson, K.A. Nucleation of Protein Fibrillation by Nanoparticles. Proc. Natl. Acad. Sci. USA 2007, 104, 8691–8696. [Google Scholar] [CrossRef]
- Hunashal, Y.; Cantarutti, C.; Giorgetti, S.; Marchese, L.; Molinari, H.; Niccolai, N.; Fogolari, F.; Esposito, G. Exploring exchange processes in proteins by paramagnetuc perturbation of NMR spectra. Phys. Chem. Chem Phys. 2020, 22, 6247–6259. [Google Scholar] [CrossRef]
- Esposito, G.; Lesk, A.M.; Molinari, H.; Motta, A.; Niccolai, N.; Pastore, A. Probing protein structure by solvent perturbation of nuclear magnetic resonance spectra. J. Mol. Biol. 1992, 224, 659–670. [Google Scholar] [CrossRef]
- Molinari, H.; Esposito, G.; Ragona, L.; Pegna, M.; Niccolai, N.; Brunne, R.M.; Lesk, A.M.; Zetta, L. Probing protein structure by solvent perturbation of NMR spectra: The surface accessibility of bovine pancreatic trypsin inhibitor. Biophys. J. 1997, 73, 382–396. [Google Scholar] [CrossRef][Green Version]
- Scarselli, M.; Bernini, A.; Segoni, C.; Molinari, H.; Esposito, G.; Lesk, A.M.; Laschi, F.; Temussi, P.A.; Niccolai, N. Tendamistat surface accessibility to the TEMPOL paramagnetic probe. J. Biomol. NMR 1999, 15, 125–133. [Google Scholar] [CrossRef]
- Niccolai, N.; Ciutti, A.; Spiga, O.; Scarselli, M.; Bernini, A.; Bracci, L.; Di Maro, D.; Dalvit, C.; Molinari, H.; Esposito, G.; et al. NMR Studies of Protein Surface Accessibility. J. Biol. Chem. 2001, 276, 42455–42461. [Google Scholar] [CrossRef]
- Benial, A.M.F.; Dhas, M.K.; Jawahar, A. Rotational Correlation Time Studies on Nitroxyl radicals Using 300 MHz ESR Spectrometer in High Viscous Liquid. Appl. Magn. Reson. 2011, 40, 311–319. [Google Scholar] [CrossRef]
- Niccolai, N.; Bonci, A.; Rustici, M.; Scarselli, M.; Neri, P.; Esposito, G.; Mascagni, P.; Motta, A.; Molinari, H. NMR delineation of inner and outer protons from paramagnetic relaxation perturbations in 1D and 2D spectra of peptides. J. Chem. Soc. Perkin Trans. 1991, 1453–1457. [Google Scholar] [CrossRef]
- Giorgetti, S.; Raimondi, S.; Pagano, K.; Relini, A.; Bucciantini, M.; Corazza, A.; Fogolari, F.; Codutti, L.; Salmona, M.; Mangione, P.; et al. Effect of tetracyclines on the dynamics of formation and destructuration of β2-microglobulin amyloid fibrils. J. Biol. Chem. 2011, 286, 2121–2131. [Google Scholar] [CrossRef] [PubMed]
- Esposito, G.; Garvey, M.; Alverdi, V.; Pettirossi, F.; Corazza, A.; Fogolari, F.; Polano, M.; Mangione, P.P.; Giorgetti, S.; Stoppini, M.; et al. Monitoring the interaction between β2-microglobulin and the molecular chaperone αB-crystallin by NMR and mass spectrometry. αB-Crystallin dissociates β2-microglobulin oligomers. J. Biol. Chem. 2013, 288, 17844–17858. [Google Scholar] [CrossRef] [PubMed]
- Williamson, M.P. Using chemical shift perturbation to characterise ligand binding. Prog. Nucl. Magn. Reson. Spectrosc. 2013, 73, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, V.; Katou, H.; Hagihara, Y.; Hasegawa, K.; Naiki, H.; Goto, Y. Mapping the core of of β2-microglobulin by H/D exchange. Nat. Struct. Biol. 2002, 9, 332–336. [Google Scholar] [CrossRef]
- Bodenhausen, G.; Ruben, D.J. Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem. Phys. Lett. 1980, 69, 185–189. [Google Scholar] [CrossRef]
- Palmer, A.G.; Cavanagh, J.; Wright, P.E.; Rance, M. Sensitivity improvement in proton-detected two-dimensional heteronuclear correlation NMR spectroscopy. J. Magn. Reson. 1991, 93, 151–170. [Google Scholar] [CrossRef]
- Schleucher, J.; Schwendinger, M.; Sattler, M.; Schmidt, P.; Schedletzky, O.; Glaser, S.J.; Sørensen, O.W.; Griesinger, C. A general enhancement scheme in heteronuclear multidimensional NMR employing pulsed field gradients. J. Biomol. NMR 1994, 4, 301–306. [Google Scholar] [CrossRef]
- Grzesiek, S.; Bax, A. The Importance of Not Saturating H2O in Protein NMR. Application to Sensitivity Enhancement and NOE Measurements. J. Am. Chem. Soc. 1993, 115, 12593–12594. [Google Scholar] [CrossRef]
- Kay, L.E.; Torchia, D.E.; Bax, A. Backbone Dynamics of Proteins Studied by 15N Inverse Detected Heteronuclear NMR Spectroscopy: Application to Staphylococcal Nuclease. Biochemistry 1989, 28, 8972–8979. [Google Scholar] [CrossRef] [PubMed]
- Knowles, P.F.; Marsh, D.; Rattle, H.W.E. Magnetic Resonance of Bio-Molecules: An Introduction to the Theory and Practice of NMR and ESR of Biological Systems; Wiley: London, UK, 1976. [Google Scholar]
- Kivelson, D. Theory of ESR Linewidths of Free Radicals. J. Phys. Chem. 1960, 33, 1094–1106. [Google Scholar] [CrossRef]
- Ceccon, A.; Tugarinov, V.; Bax, A.; Clore, G.M. Global Dynamics and Exchange Kinetics of a Protein on the Surface of Nanoparticles Revealed by Relaxation-Based Solution NMR Spectroscopy. J. Am. Chem. Soc. 2016, 138, 5789–5792. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Yadav, I.; Aswal, V.K.; Kohlbrecher, J. Structure and Interaction of Nanoparticle-Protein Complexes. Langmuir 2018, 34, 5679–5695. [Google Scholar] [CrossRef]
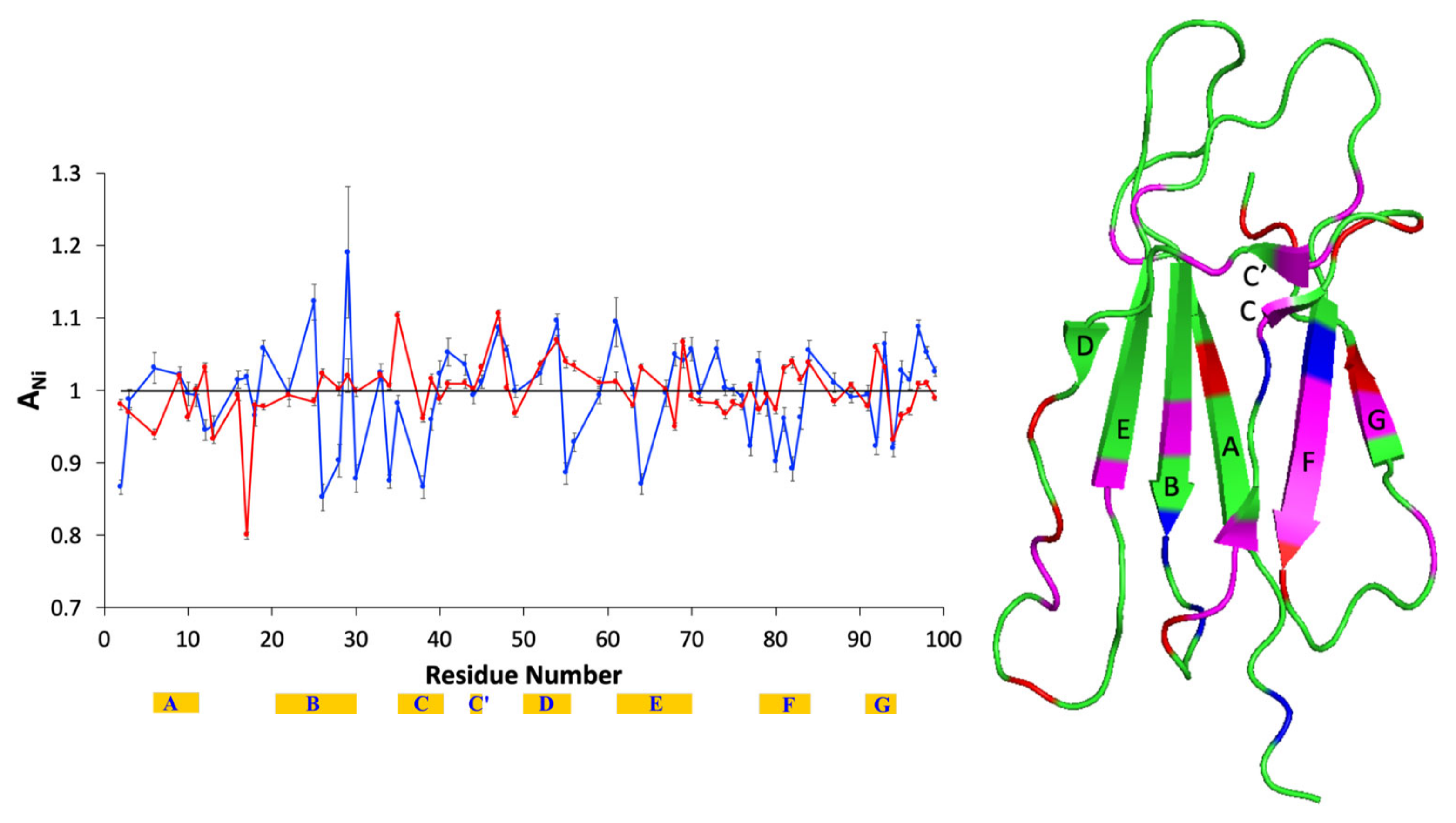
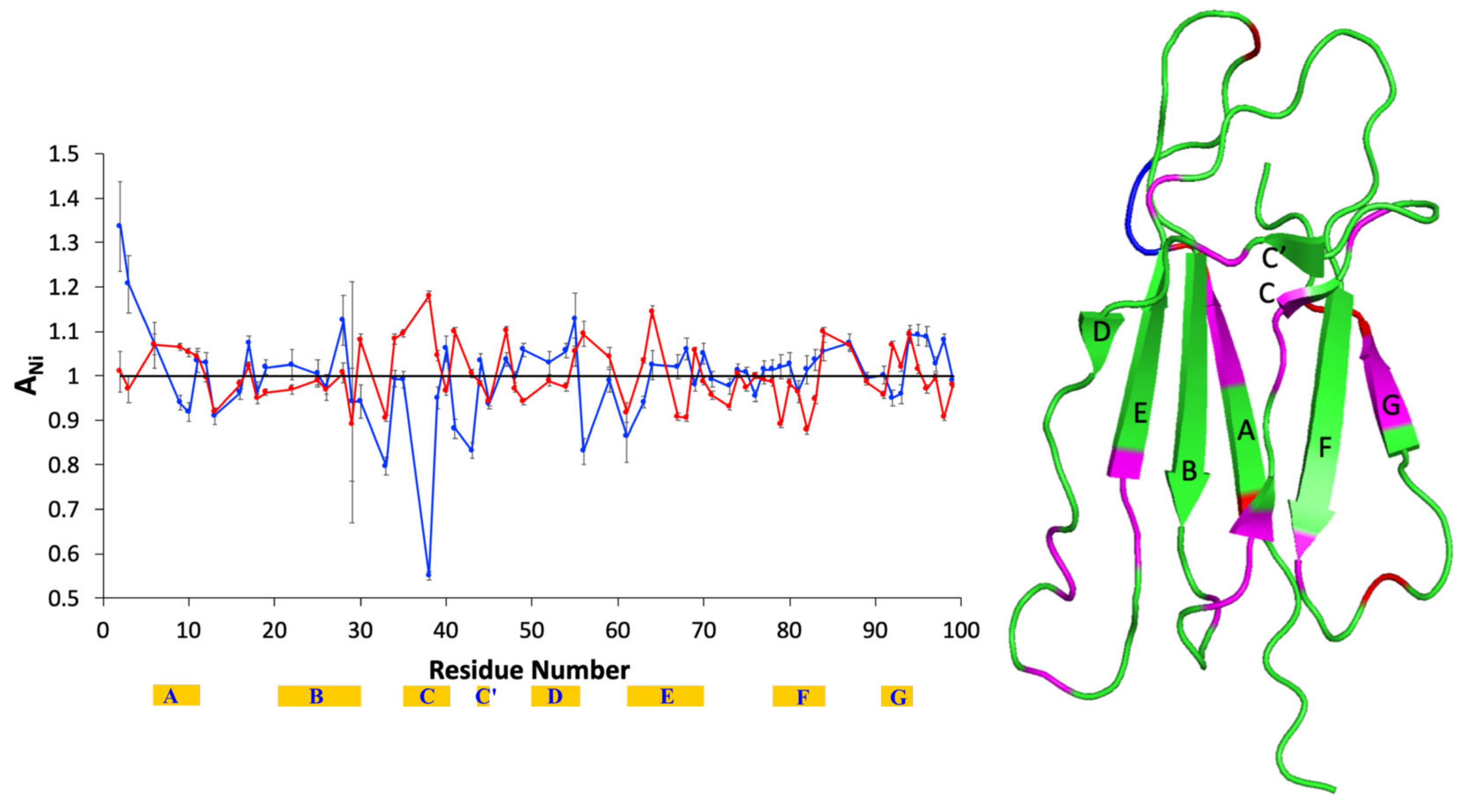
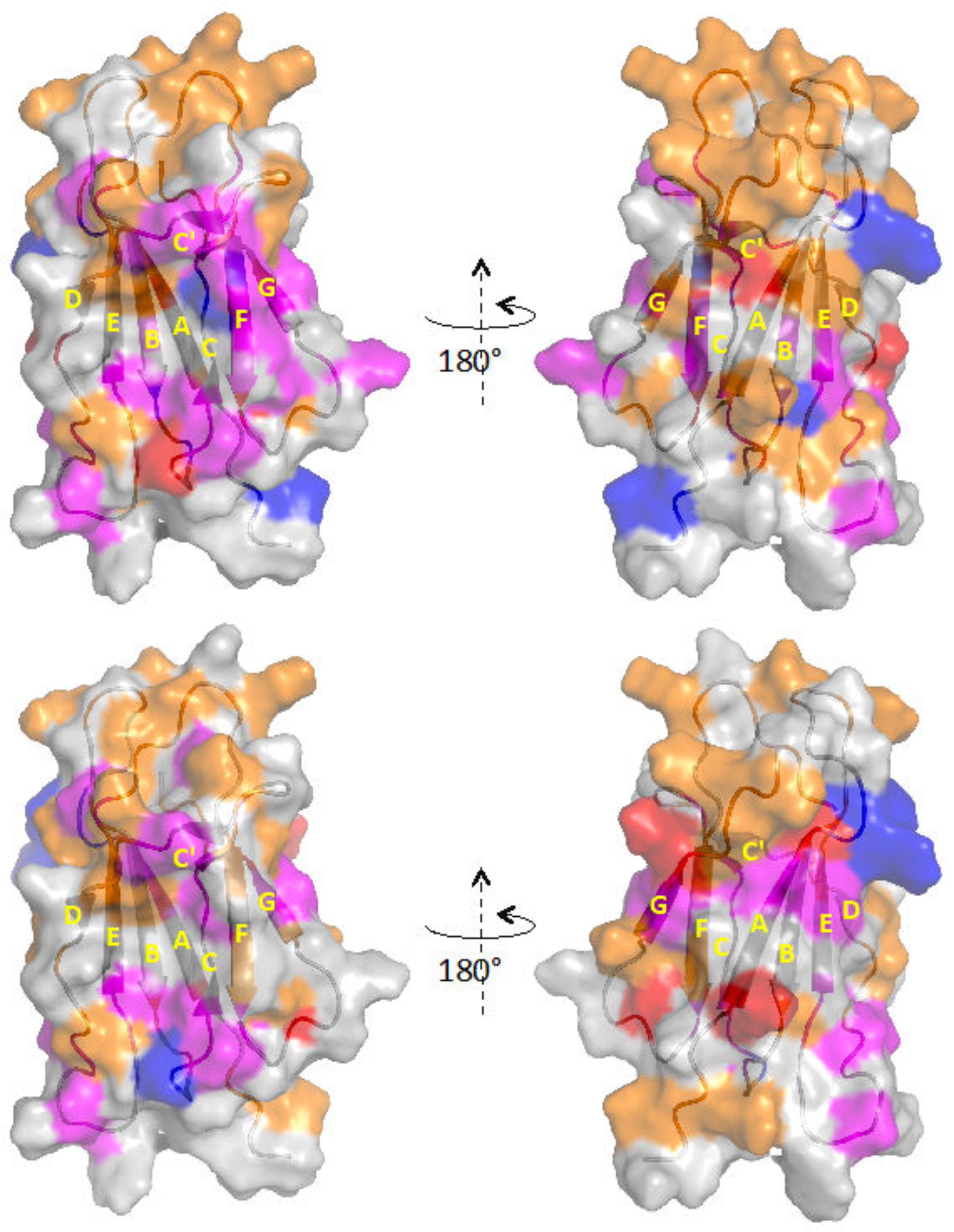
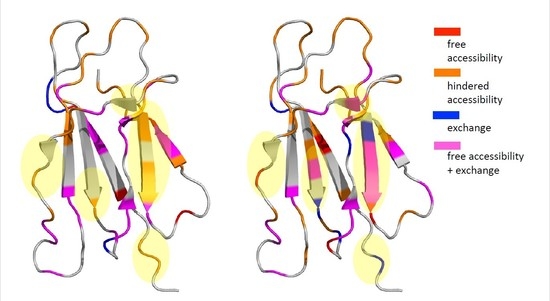
| Composition | Tempol | Tempol + AuNPs | Tempol + β2m | Tempol + AuNPs + β2m |
|---|---|---|---|---|
| [Tempol] | [AuNP] = 60 nM | [β2m] = 8 μM | [AuNP] = 60 nM; [β2m] = 8 μM | |
| 1.6 mM | 3.3 ± 0.2 | 3.1 ± 0.2 | 3.4 ± 0.2 | 3.3 ± 0.2 |
| 0.8 mM | 3.2 ± 0.2 | 3.0 ± 0.3 | 3.2 ± 0.3 | 2.9 ± 0.2 |
| 0.4 mM | 3.1 ± 0.3 | 3.2 ± 0.3 | 3.1 ± 0.3 | 3.1 ± 0.4 |
| Structure Region | AN > 1 | Type I AN [off-eq] > AN [eq] | Type II AN [off-eq] < AN [eq] |
|---|---|---|---|
| N-term, strand A | V9 Q2, I7 | K6, Y10 | Q2 Q2, I7 |
| loop AB | R12, K19 | H13, E16, N17, K19 N17 | R12 R12, H13 |
| strand B | Y26 | C25, G29 S28 | Y26, S28, F30 N21, F22, C25, F30 |
| loop BC | S33, D34 D34 | S33 | D34 D34 |
| strand C | I35, L39, K41 I35 | L40, K41 | I35, D38, L39 I35 |
| turn CC’, strand C’, loop C’D | G43, R45, E47 G43, E47, K48, V49 | G43, K48, V49 R45, K48 | R45, E47 N42 |
| strand D | S52, L54, S55, F56 F56 | L54 E50, L54 | S55, F56 S52, F56 |
| loop DE | D59 | S61 D59 | S61 |
| strand E | L64, E69 E69 | Y63, T68, F70 E69, F70 | L64, E69 L64 |
| loop EF | E77 K75 | T73, E74, K75, D76 T71, E74 | E77 |
| strand F | R81, V82, N83, H84 N83, H84 | T78 T78 | C80, R81, V82, N83 A79, H84 |
| loop FG | Q89 V85, L87 | L87 | Q89 V85 |
| strand G, C-term | I92, V93, R97, D98 K94 | V93, W95, D96, R97, D98, M99 V93, D96, D98, M99 | I92 I92, K94 |
| Structure Region | AN > 1 | Type I AN [off-eq] > AN [eq] | Type II AN [off-eq] < AN [eq] |
|---|---|---|---|
| N-term, strand A | K6, V9, Y10, S11 V9 | Q2, R3 K6, Y10 | V9, Y10 Q2 |
| loop AB | N17 | N17, K19 H13, E16, N17, K19 | R12, H13 R12 |
| strand B | F30 Y26 | F22, S28 C25, G29 | F30 Y26, S28, F30 |
| loop BC | D34 S33, D34 | S33, D34 D34 | |
| strand C | I35, D38, L39, K41 I35, L39, K41 | L40 L40, K41 | I35, D38, L39, K41 I35, D38, L39 |
| turn CC’, strand C’, loop C’D | E47 G43, R45, E47 | E44, K48, V49 G43, K48, V49 | E47 R45, E47 |
| strand D | S55, F56 S52, L54, S55, F56 | S52, L54 L54 | S55, F56 S55, F56 |
| loop DE | D59 D59 | S61 | D59 |
| strand E | Y63, L64, E69 L64, E69 | Y67, T68, F70 Y63, T68, F70 | Y63, L64, E69 L64, E69 |
| loop EF | E77 | T71, T73, K75 T73, E74, K75, D76 | E77 |
| strand F | H84 R81, V82, N83, H84 | A79, C80, V82, N83 T78 | H84 C80, R81, V82, N83 |
| loop FG | L87 Q89 | L87 | Q89 |
| strand G, C-term | I92, V93, K94, W95 I92, V93, R97, D98 | K91, W95, D96, R97, D98 V93, W95, D96, R97, D98, M99 | I92, V93 I92 |
Sample Availability: Samples of Gold nanoparticles are commercially available from Sigma. Protein samples must be expressed and are currently not available from the authors. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hunashal, Y.; Cantarutti, C.; Giorgetti, S.; Marchese, L.; Fogolari, F.; Esposito, G. Insights into a Protein-Nanoparticle System by Paramagnetic Perturbation NMR Spectroscopy. Molecules 2020, 25, 5187. https://doi.org/10.3390/molecules25215187
Hunashal Y, Cantarutti C, Giorgetti S, Marchese L, Fogolari F, Esposito G. Insights into a Protein-Nanoparticle System by Paramagnetic Perturbation NMR Spectroscopy. Molecules. 2020; 25(21):5187. https://doi.org/10.3390/molecules25215187
Chicago/Turabian StyleHunashal, Yamanappa, Cristina Cantarutti, Sofia Giorgetti, Loredana Marchese, Federico Fogolari, and Gennaro Esposito. 2020. "Insights into a Protein-Nanoparticle System by Paramagnetic Perturbation NMR Spectroscopy" Molecules 25, no. 21: 5187. https://doi.org/10.3390/molecules25215187
APA StyleHunashal, Y., Cantarutti, C., Giorgetti, S., Marchese, L., Fogolari, F., & Esposito, G. (2020). Insights into a Protein-Nanoparticle System by Paramagnetic Perturbation NMR Spectroscopy. Molecules, 25(21), 5187. https://doi.org/10.3390/molecules25215187