Ruthenium(IV) Complexes as Potential Inhibitors of Bacterial Biofilm Formation
Abstract
1. Introduction
2. Results and Discussion
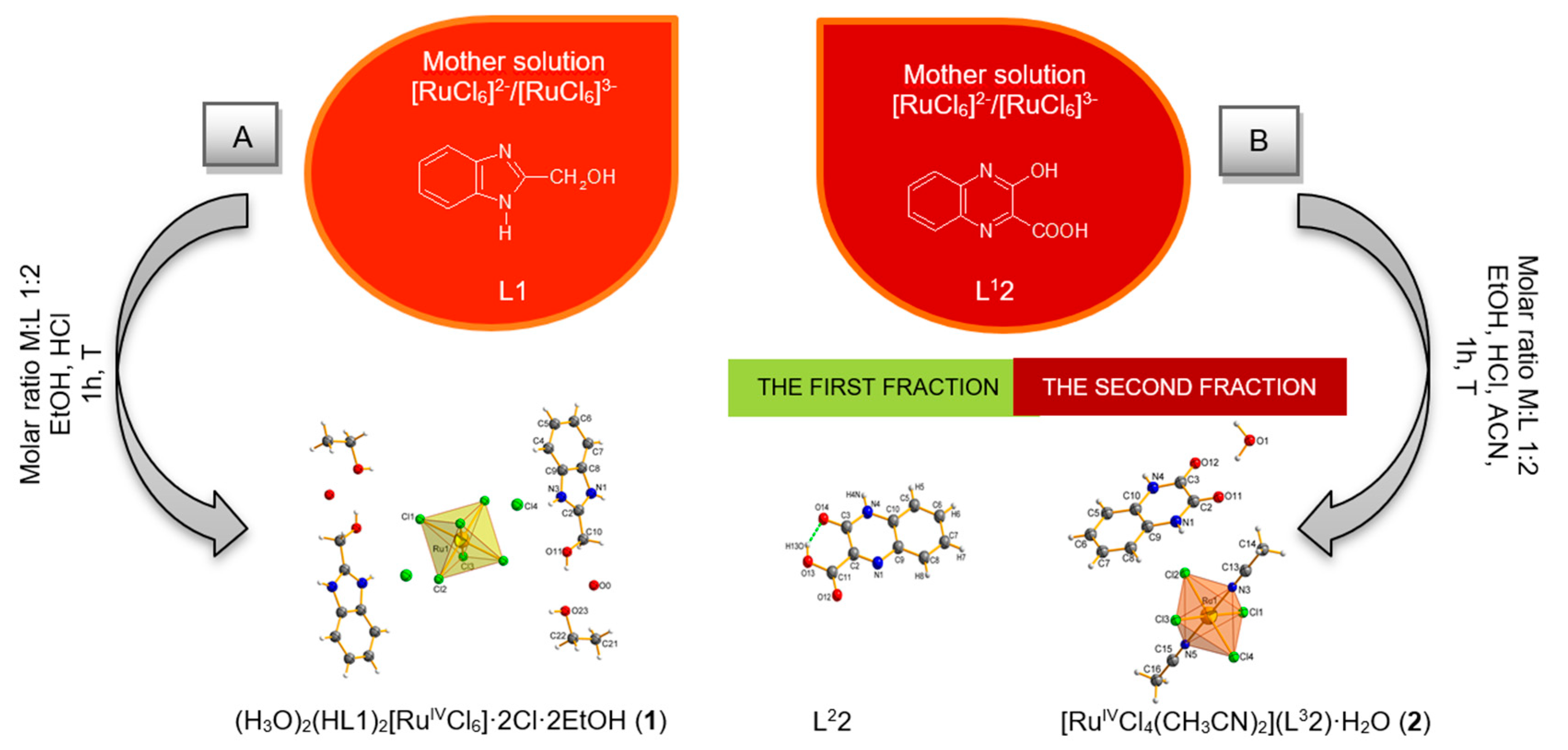
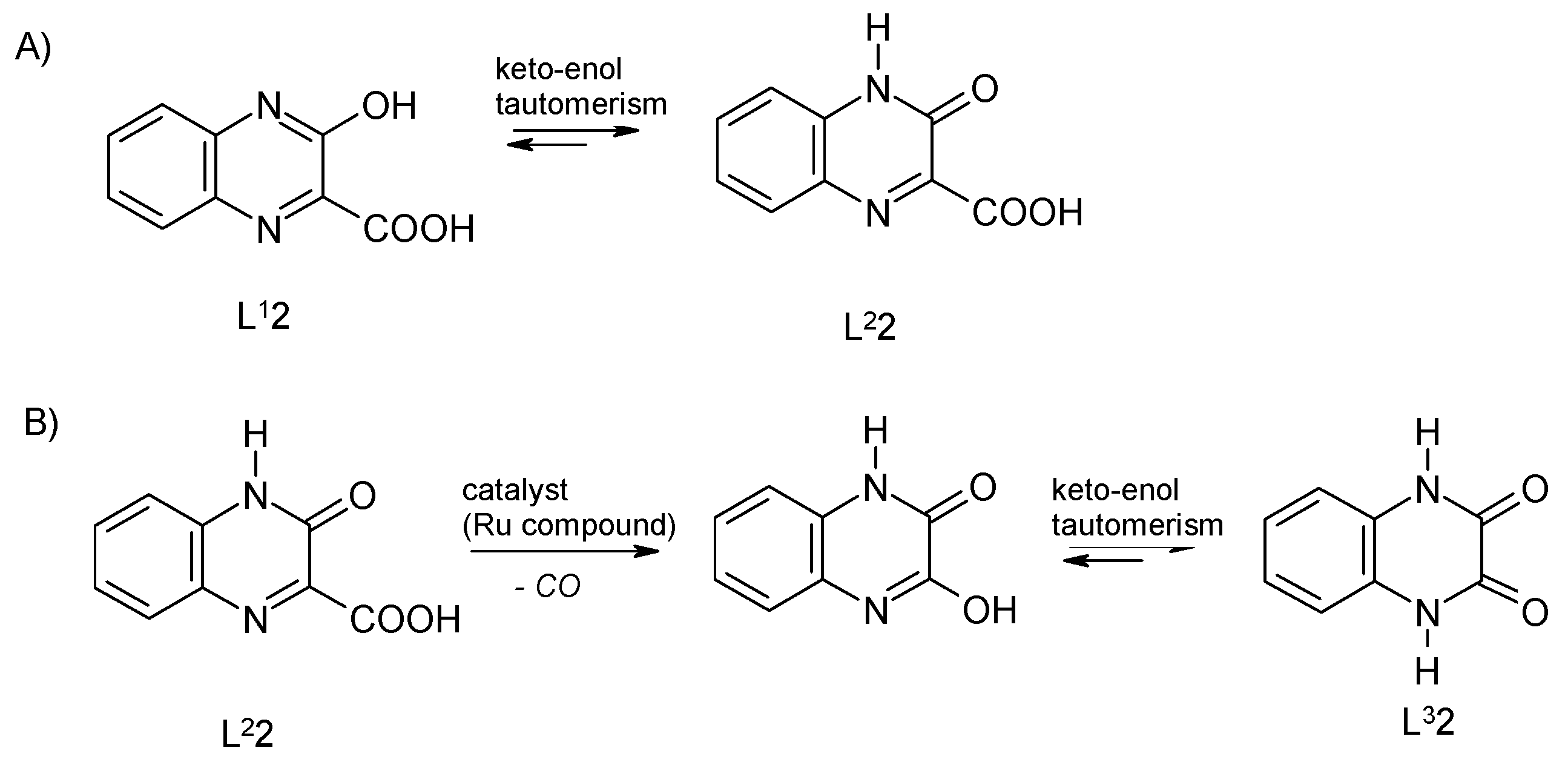
2.1. Syntheses and Characterization
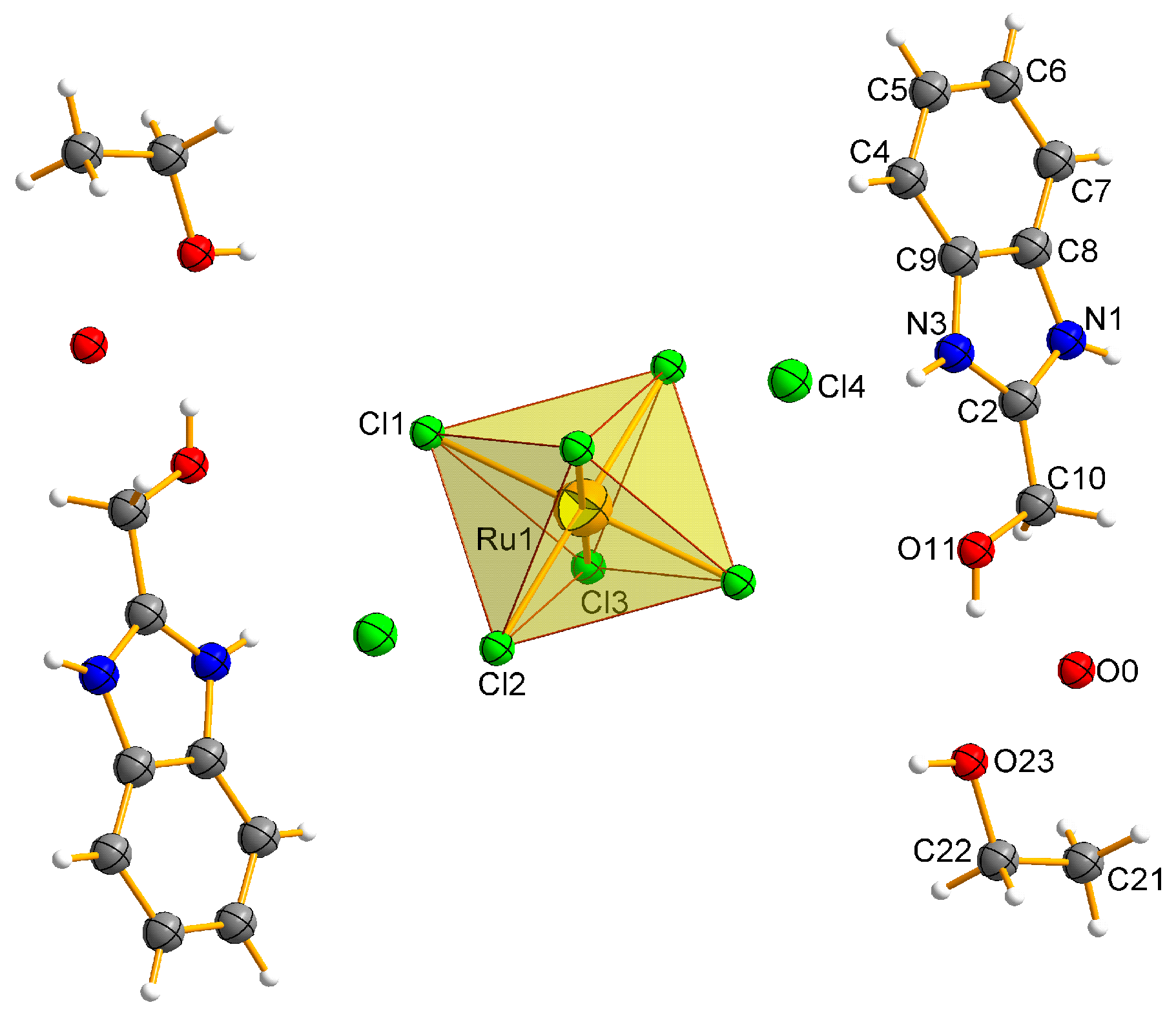
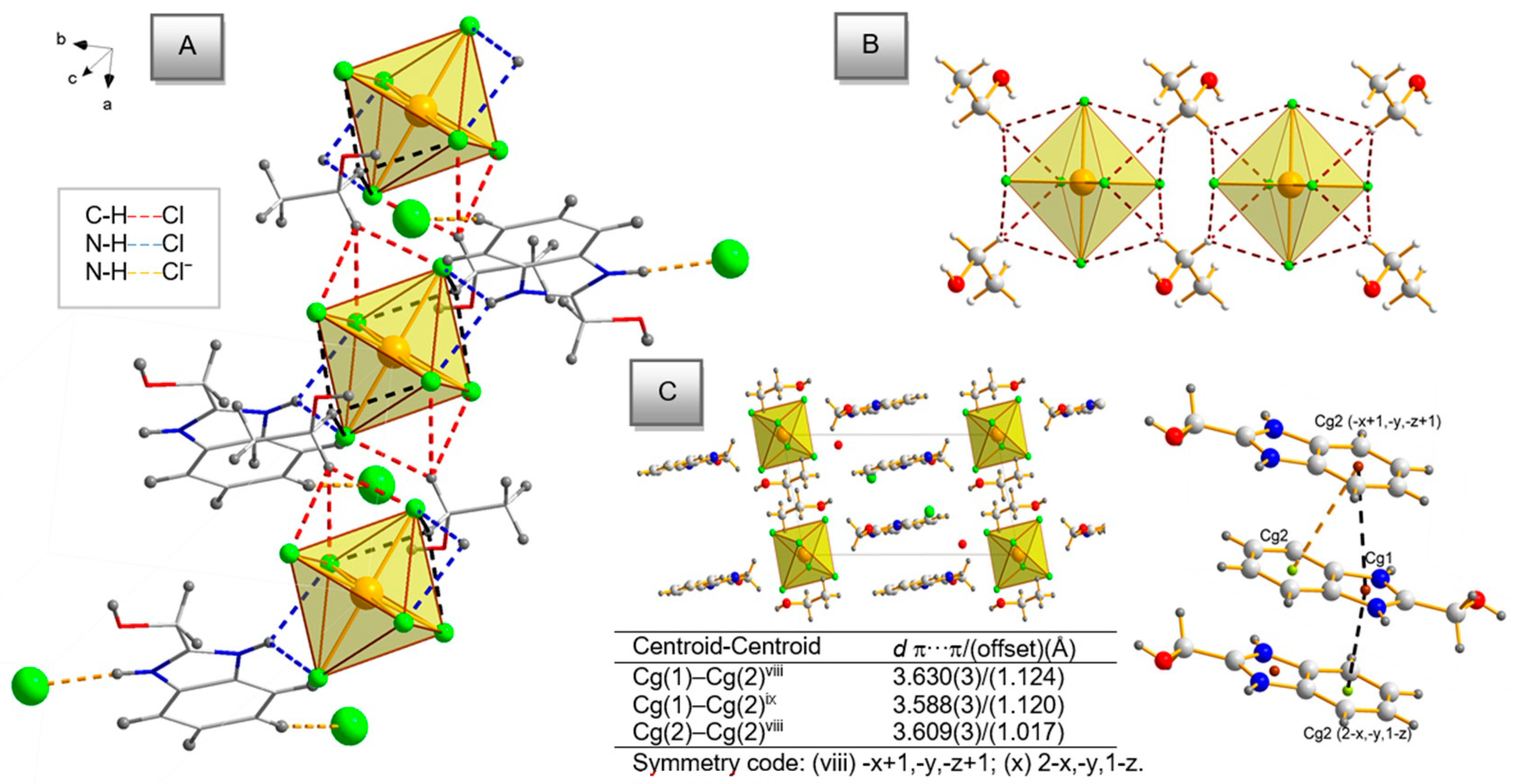
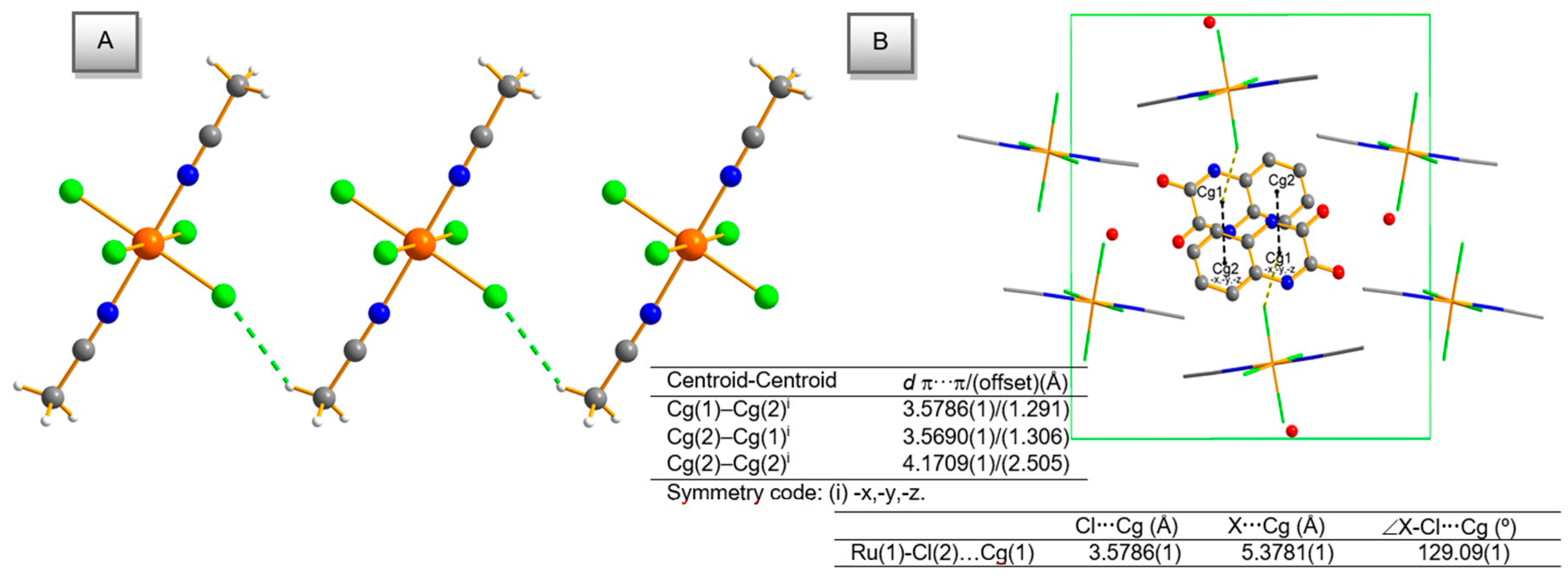
2.2. Description of the Molecular Structures
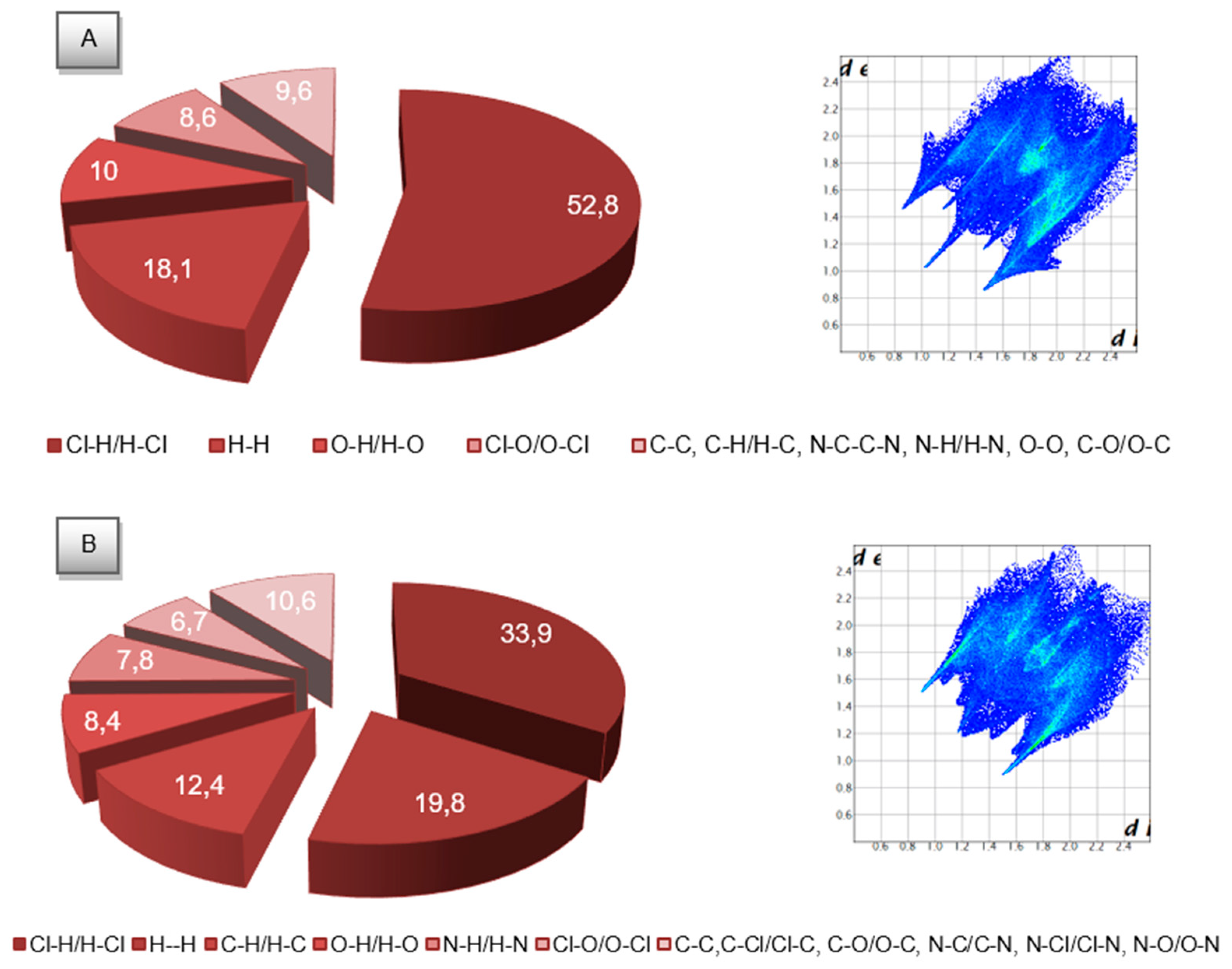
2.3. Molecular Hirshfeld Surfaces
2.4. Spectroscopic, Magnetic and Solution Conductivity Properties
2.4.1. UV-Vis Spectroscopy in Solid-State and Aqueous Solutions
2.4.2. Luminescent Properties
2.4.3. Magnetic Measurements
2.4.4. Solution Conductivity
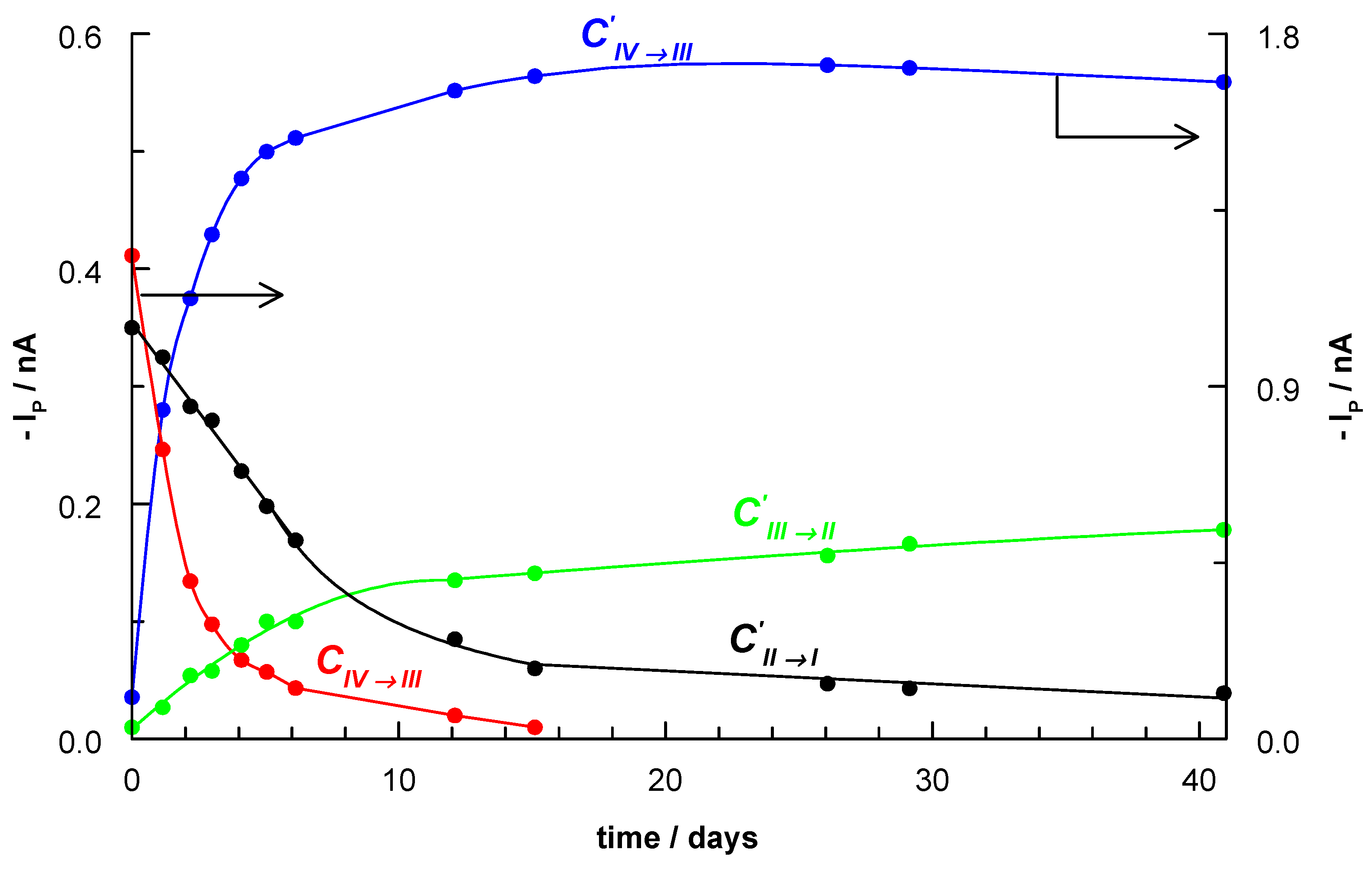
2.5. Electrochemical Studies
- ∙
- The DPV curve recorded on second day shows four signals: CIV→III, C’IV→III, C’III→II and C’II→I. The cathodic C’IV→III peak appeared as a new peak (0.11 V) and this was connected with formation of [RuCl4(CH3CN)2].
- ∙
- The CIV→III peak at −0.01 V corresponding to the existence of the RuCl62- complex gradually disappeared over time. Consequently, this disappearance led to an increase in the intensity of the C’IV→III peak (0.11 V), which in turn is characteristic of the [RuCl4(CH3CN)2] complex. This suggests that +IV oxidation state for ruthenium ions is more stable in the acetonitrile coordination sphere than in the chloride coordination environment.
- ∙
- It is worth noting that the CIV→III signal (0.15 V) in complex 2 is observed at a very similar potential value.
- ∙
- The C’III→II and C’II→I peaks in comparison to the CIII→II and CII→I peaks are shifted to more negative potentials (Table S7). Additionally, the C’II→I peak current was decreased in favor of an increase in the C’III→II peak.
- ∙
- The exchange of chloride ions for acetonitrile molecules resulted in a decrease in the electron density at the metal centre and made it easier reduce Ru(IV) ion.
2.6. Biological Studies of Ruthenium(IV) Complexes
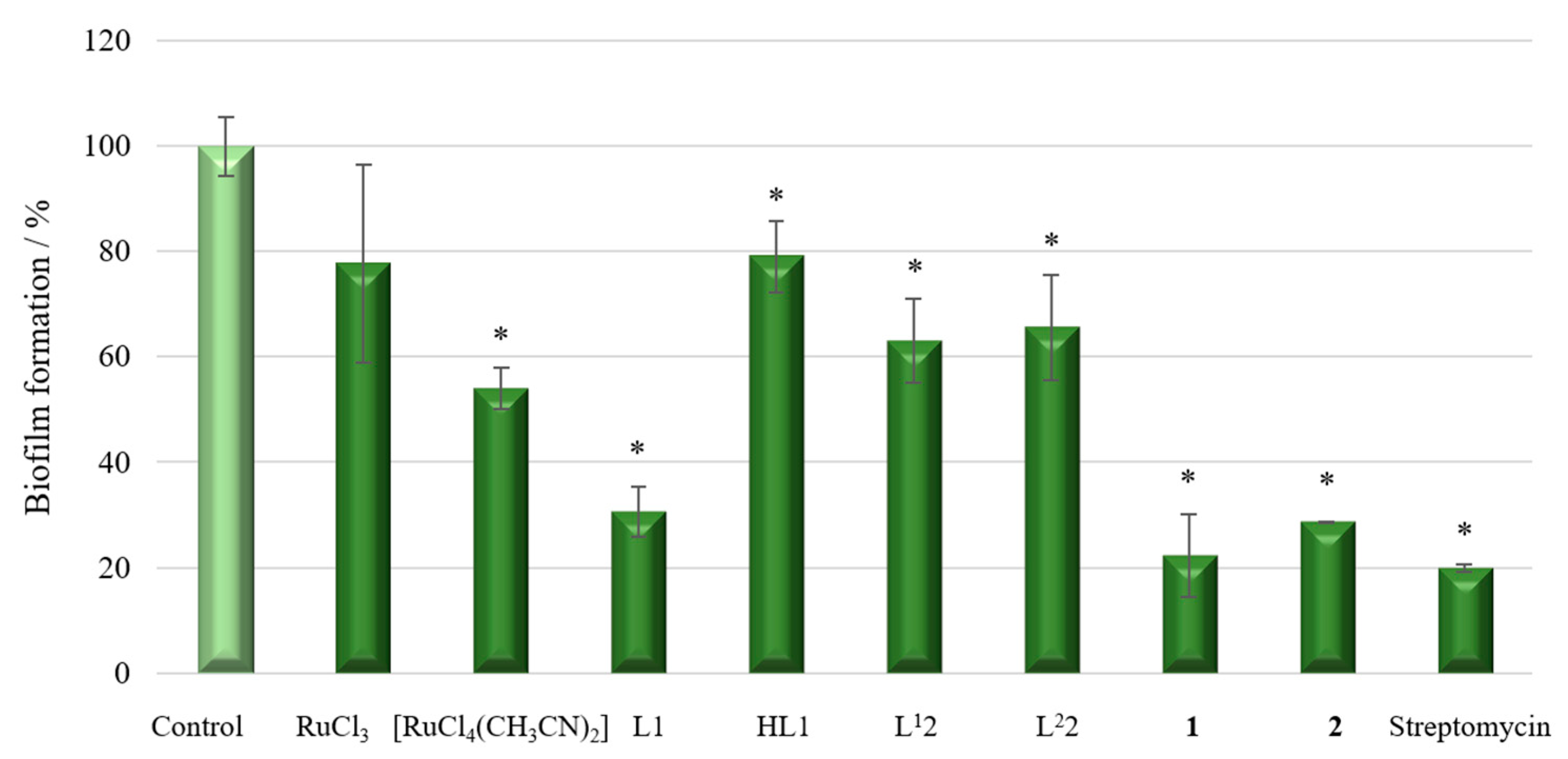
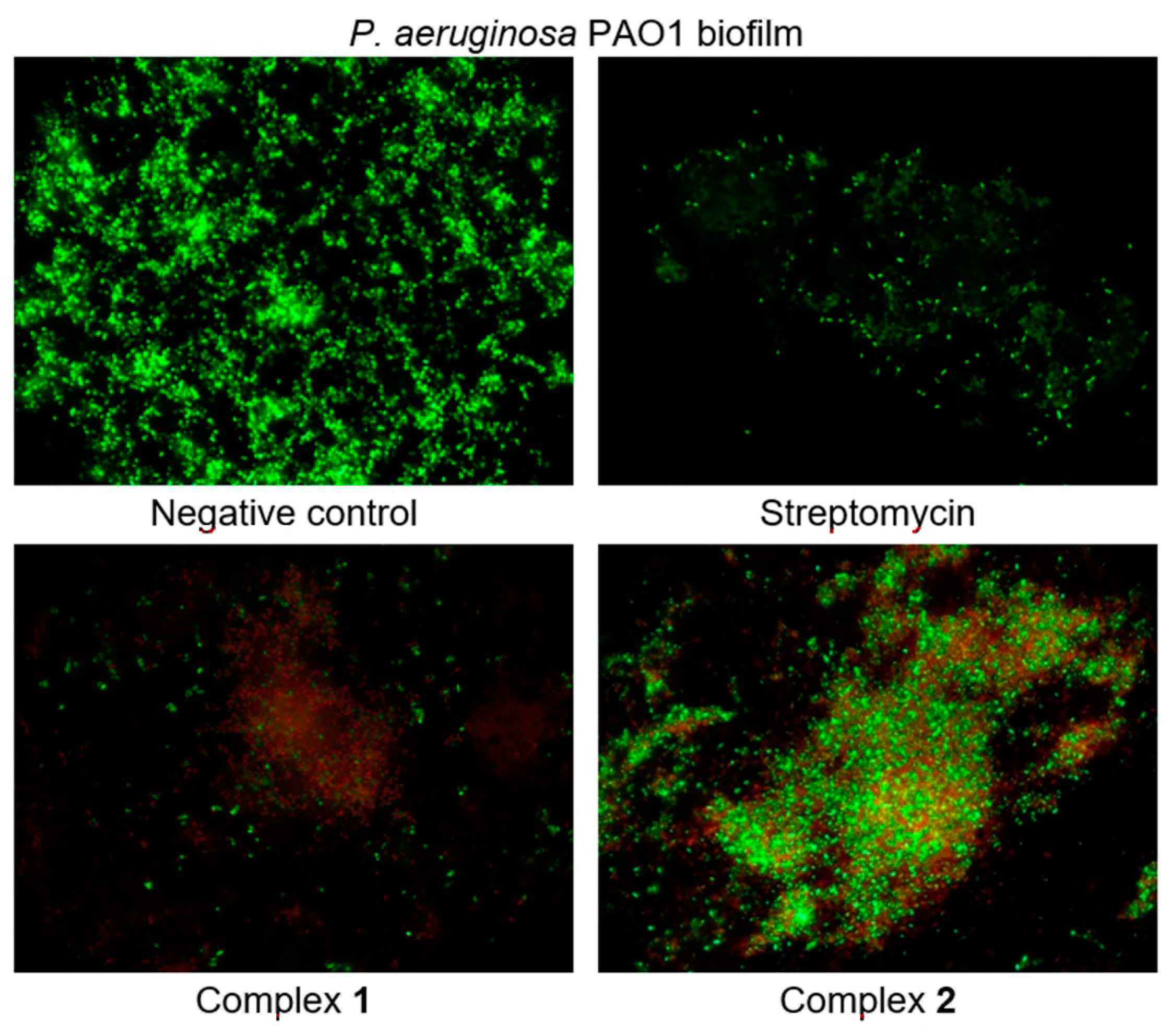
2.6.1. Anti-Biofilm and Antibacterial Activity
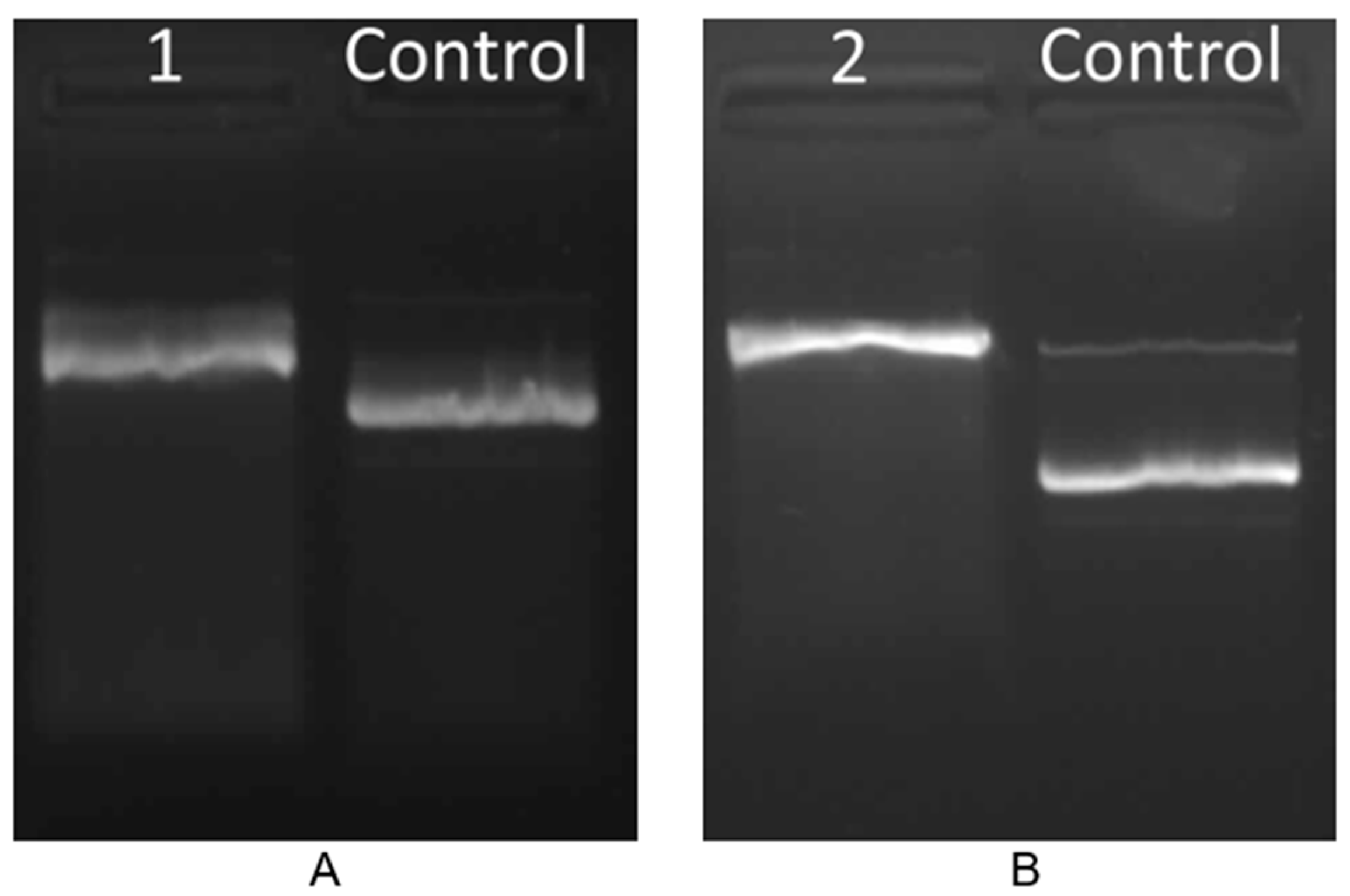
2.6.2. Estimation of Oxidative Damage Based on Digestion of Plasmid DNA with Fpg Protein
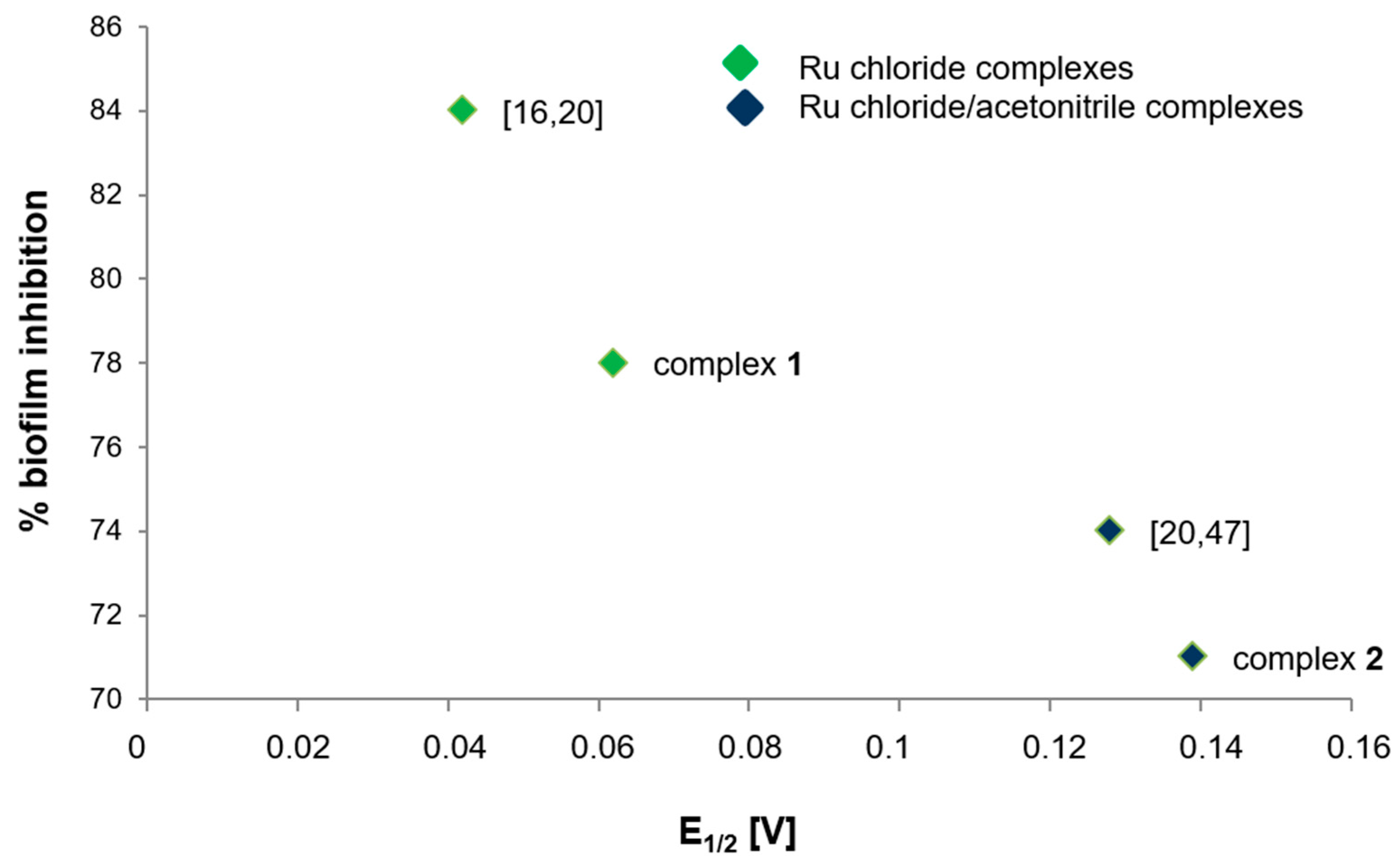
2.7. Regularity between Electrochemical Properties and Biological Activity
3. Materials and Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- The World Health Organization. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics; The World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Aloush, V.; Navon-Venezia, S.; Seigman-Igra, Y.; Cabili, S.; Carmeli, Y. Multidrug-resistant Pseudomonas aeruginosa: Risk factors and clinical impact. Antimicrob. Agents Chemother. 2006, 50, 43–48. [Google Scholar] [CrossRef]
- Hirsch, E.B.; Tam, V.H. Impact of multidrug-resistant Pseudomonas aeruginosa infection on patient outcomes. Expert Rev. Pharmacoecon. Outcomes Res. 2010, 10, 441–451. [Google Scholar] [CrossRef]
- Strateva, T.; Yordanov, D. Pseudomonas aeruginosa—A phenomenon of bacterial resistance. J. Med. Microbiol. 2009, 58, 1133–1148. [Google Scholar] [CrossRef]
- Sadikot, R.T.; Blackwell, T.S.; Christman, J.W.; Prince, A.S. Pathogen–host interactions in Pseudomonas aeruginosa pneumonia. Am. J. Respir. Crit. Care Med. 2005, 171, 1209–1223. [Google Scholar] [CrossRef]
- Davies, D. Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discov. 2003, 2, 114–122. [Google Scholar] [CrossRef]
- Høiby, N.; Bjarnsholt, T.; Givskov, M.; Molin, S.; Ciofu, O. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 2010, 35, 322–332. [Google Scholar] [CrossRef]
- Paraje, M.G. Antimicrobial resistance in biofilms. In Science Against Microbial Pathogens: Communicating Current Research and Technological Advances; Méndez-Vilas, A., Ed.; Formatex Research Center: Badajoz, Spain, 2011; Volume 2, pp. 736–744. ISBN 978-84-939843-2-8. [Google Scholar]
- Donlan, R.M.; Costerton, J.W. Biofilms: Survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. 2002, 15, 167–193. [Google Scholar] [CrossRef]
- Đurić, S.; Vojnovic, S.; Pavic, A.; Mojicevic, M.; Wadepohl, H.; Savić, N.D.; Popsavin, M.; Nikodinovic-Runic, J.; Djuran, M.I.; Glišić, B.Đ. New polynuclear 1,5-naphthyridine-silver(I) complexes as potential antimicrobial agents: The key role of the nature of donor coordinated to the metal center. J. Inorg. Biochem. 2020, 203, 110872. [Google Scholar] [CrossRef]
- Glišić, B.Đ.; Senerovic, L.; Comba, P.; Wadepohl, H.; Veselinovic, A.; Milivojevic, D.R.; Djuran, M.I.; Nikodinovic-Runic, J. Silver(I) complexes with phthalazine and quinazoline as effective agents against pathogenic Pseudomonas aeruginosa strains. J. Inorg. Biochem. 2016, 155, 115–128. [Google Scholar] [CrossRef]
- Glišić, B.Đ.; Aleksic, I.; Comba, P.; Wadepohl, H.; Ilic-Tomic, T.; Nikodinovic-Runic, J.; Djuran, M.I. Copper(II) complexes with aromatic nitrogen-containing heterocycles as effective inhibitors of quorum sensing activity in Pseudomonas aeruginosa. RSC Adv. 2016, 6, 86695–86709. [Google Scholar] [CrossRef]
- Beeton, M.L.; Aldrich-Wright, J.R.; Bolhuis, A. The antimicrobial and antibiofilm activities of copper(II) complexes. J. Inorg. Biochem. 2014, 140, 167–172. [Google Scholar] [CrossRef]
- Rzhepishevska, O.; Hakobyan, S.; Ekstrand-Hammarström, B.; Nygren, Y.; Karlsson, T.; Bucht, A.; Elofsson, M.; Boily, J.-F.; Ramstedt, M. The gallium(III)–salicylidene acylhydrazide complex shows synergistic anti-biofilm effect and inhibits toxin production by Pseudomonas aeruginosa. J. Inorg. Biochem. 2014, 138, 1–8. [Google Scholar] [CrossRef]
- Reiss, A.; Chifiriuc, M.C.; Amzoiu, E.; Spînu, C.I. Transition metal(II) complexes with cefotaxime-derived Schiff base: Synthesis, characterization, and antimicrobial studies. Bioinorg. Chem. Appl. 2014, 2014, 1–17. [Google Scholar] [CrossRef]
- Czerwonka, G.; Gmiter, D.; Guzy, A.; Rogala, P.; Jabłońska-Wawrzycka, A.; Borkowski, A.; Cłapa, T.; Narożna, D.; Kowalczyk, P.; Syczewski, M.; et al. A benzimidazole-based ruthenium(IV) complex inhibits Pseudomonas aeruginosa biofilm formation by interacting with siderophores and the cell envelope, and inducing oxidative stress. Biofouling 2019, 35, 59–74. [Google Scholar] [CrossRef]
- Rogala, P.; Czerwonka, G.; Michałkiewicz, S.; Hodorowicz, M.; Barszcz, B.; Jabłońska-Wawrzycka, A. Synthesis, structural characterization and antimicrobial evaluation of ruthenium complexes with heteroaromatic carboxylic acids. Chem. Biodivers. 2019, 16, e1900403. [Google Scholar] [CrossRef]
- Keppler, B.K.; Henn, M.; Juhl, U.M.; Berger, M.R.; Niebl, R.; Wagner, F.E. New ruthenium complexes for the treatment of cancer. In Ruthenium and Other Non-Platinum Metal Complexes in Cancer Chemotherapy. Progress in Clinical Biochemistry and Medicine; Baulieu, E., Forman, D.T., Ingelman-Sundberg, M., Jaenicke, L., Kellen, J.A., Nagai, Y., Springer, G.F., Träger, L., Will-Shahab, L., Wittliff, J.L., Eds.; Progress in Clinical Biochemistry and Medicine; Springer: Berlin/Heidelberg, Germany, 1989; Volume 10, pp. 41–69. ISBN 978-3-642-74762-5. [Google Scholar]
- Groessl, M.; Tsybin, Y.O.; Hartinger, C.G.; Keppler, B.K.; Dyson, P.J. Ruthenium versus platinum: Interactions of anticancer metallodrugs with duplex oligonucleotides characterised by electrospray ionisation-mass spectrometry. J. Biol. Inorg. Chem. 2010, 15, 677–688. [Google Scholar] [CrossRef]
- Jabłońska-Wawrzycka, A.; Rogala, P.; Michałkiewicz, S.; Hodorowicz, M.; Barszcz, B. Ruthenium complexes in different oxidation states: Synthesis, crystal structure, spectra and redox properties. Dalton Trans. 2013, 42, 6092–6101. [Google Scholar] [CrossRef]
- Morgan, K.J. The infrared spectra of some simple benzimidazoles. J. Chem. Soc. 1961, 455, 2343–2347. [Google Scholar] [CrossRef]
- Halpern, J.; Kemp, A.L.W. The decarbonylation of formic acid by ruthenium(II) chloride. J. Am. Chem. Soc. 1966, 88, 5147–5150. [Google Scholar] [CrossRef]
- Katryniok, B.; Paul, S.; Dumeignil, F. Highly efficient catalyst for the decarbonylation of lactic acid to acetaldehyde. Green Chem. 2010, 12, 1910–1913. [Google Scholar] [CrossRef]
- Priimagi, A.; Cavallo, G.; Metrangolo, P.; Resnati, G. The halogen bond in the design of functional supramolecular materials: Recent advances. Acc. Chem. Res. 2013, 46, 2686–2695. [Google Scholar] [CrossRef] [PubMed]
- Aullón, G.; Bellamy, D.; Brammer, L.; Bruton, E.A.; Guy Orpen, A. Metal-bound chlorine often accepts hydrogen bonds. Chem. Commun. 1998, 653–654. [Google Scholar] [CrossRef]
- Oxtoby, N.S.; Blake, A.J.; Champness, N.R.; Wilson, C. Water superstructures within organic arrays; hydrogen-bonded water sheets, chains and clusters. Chem. Eur. J. 2005, 11, 4643–4654. [Google Scholar] [CrossRef] [PubMed]
- Abu-El-Halawah, R.; Ali, B.F.; Ibrahim, M.M.; Zahra, J.A.; Frey, W. 1,4-Dihydroxyquinoxaline-2,3(1H,4H)-dione. Acta Crystallogr. Sect. E Struct. Rep. Online 2008, 64, o571–o572. [Google Scholar] [CrossRef] [PubMed]
- Akkurt, M.; Öztürk, S.; Küçükbay, H.; Orhan, E.; Büyükgüngör, O. 1-Ethyl-4-phenylethyl-1,4-dihydroquinoxaline-2,3-dione. Acta Crystallogr. Sect. E Struct. Rep. Online 2004, 60, o1266–o1268. [Google Scholar] [CrossRef]
- Mustaphi, N.E.; Ferfra, S.; Essassi, E.M.; Pierrot, M. 1,4-Diallylquinoxaline-2,3(1H,4H)-dione. Acta Crystallogr. Sect. E Struct. Rep. Online 2001, 57, o176–o177. [Google Scholar] [CrossRef]
- Allen, F.H.; Kennard, O.; Watson, D.G.; Brammer, L.; Orpen, A.G.; Taylor, R. Tables of bond lengths determined by X-ray and neutron diffraction. Part 1. Bond lengths in organic compounds. J. Chem. Soc. Perkin Trans. 1987, 2, S1–S19. [Google Scholar] [CrossRef]
- Giese, M.; Albrecht, M.; Rissanen, K. Experimental investigation of anion-π interactions—Applications and biochemical relevance. Chem. Commun. 2016, 52, 1778–1795. [Google Scholar] [CrossRef]
- Estarellas, C.; Frontera, A.; Quiñonero, D.; Deyã, P.M. Relevant anion-π interactions in biological systems: The case of urate oxidase. Angew. Chemie Int. Ed. 2011, 50, 415–418. [Google Scholar] [CrossRef]
- Bauzá, A.; Quiñonero, D.; Deyà, P.M.; Frontera, A. Long-Range effects in anion-π interactions: Their crucial role in the inhibition mechanism of Mycobacterium tuberculosis malate synthase. Chem. Eur. J. 2014, 20, 6985–6990. [Google Scholar] [CrossRef]
- Geary, W.J. The use of conductivity measurements in organic solvents for the characterisation of coordination compounds. Coord. Chem. Rev. 1971, 7, 81–122. [Google Scholar] [CrossRef]
- Marken, F.; Neudeck, A.; Bond, A.M.; Stojek, Z. Cyclic voltammetry; Pulse voltammetry. In Electroanalytical Methods. Guide to Experiments and Applications; Scholz, F., Ed.; Springer: Berlin, Germany, 2010; pp. 57–119. ISBN 978-3-642-02914-1. [Google Scholar]
- Kukavica-Ibrulj, I.; Bragonzi, A.; Paroni, M.; Winstanley, C.; Sanschagrin, F.; O’Toole, G.A.; Levesque, R.C. In vivo growth of Pseudomonas aeruginosa strains PAO1 and PA14 and the hypervirulent strain LESB58 in a rat model of chronic lung infection. J. Bacteriol. 2008, 190, 2804–2813. [Google Scholar] [CrossRef] [PubMed]
- Wagner, V.E.; Iglewski, B.H. P. aeruginosa biofilms in CF infection. Clin. Rev. Allergy Immunol. 2008, 35, 124–134. [Google Scholar] [CrossRef]
- Cussac, C.; Laval, F. Reduction of the toxicity and mutagenicity of aziridine in mammalian cells harboring the Escherichia coli fpg gene. Nucleic Acids Res. 1996, 24, 1742–1746. [Google Scholar] [CrossRef] [PubMed]
- Boiteux, S.; O’Connor, T.R.; Laval, J. Formamidopyrimidine-DNA glycosylase of Escherichia coli: Cloning and sequencing of the fpg structural gene and overproduction of the protein. EMBO J. 1987, 6, 3177–3183. [Google Scholar] [CrossRef] [PubMed]
- Tudek, B.; VanZeeland, A.A.; Kusmierek, J.T.; Laval, J. Activity of Escherichia coli DNA-glycosylases on DNA damaged by methylating and ethylating agents and influence of 3-substituted adenine derivatives. Mutat. Res. 1998, 407, 169–176. [Google Scholar] [CrossRef]
- Halliwell, B. Effect of diet on cancer development: Is oxidative DNA damage a biomarker? Free Radic. Biol. Med. 2002, 32, 968–974. [Google Scholar] [CrossRef]
- Bailly, V.; Derydt, M.; Verly, W.G. δ-Elimination in the repair of AP (apurinic/apyrimidinic) sites in DNA. Biochem. J. 1989, 261, 707–713. [Google Scholar] [CrossRef]
- Graves, R.J.; Felzenszwalb, I.; Laval, J.; O’Connor, T.R. Excision of 5′-terminal deoxyribose phosphate from damaged DNA is catalyzed by the Fpg protein of Escherichia coli. J. Biol. Chem. 1992, 267, 14429–14435. [Google Scholar]
- De Abreu, F.C.; de, L. Ferraz, P.A.; Goulart, M.O.F. Some applications of electrochemistry in biomedical chemistry. Emphasis on the correlation of electrochemical and bioactive properties. J. Braz. Chem. Soc. 2002, 13, 19–35. [Google Scholar] [CrossRef]
- Dryhurst, G. Electrochemistry of Biological Molecules, 1st ed.; Academic Press: New York, NY, USA, 1977; ISBN 9780323144520. [Google Scholar]
- Verduzco-Ramírez, A.; Manzanilla-Dávila, S.G.; Morales-Guillén, M.E.; García-Ramos, J.C.; Toledano-Magaña, Y.; Marin-Becerra, A.; Flores-Álamo, M.; Ortiz-Frade, L.A.; Olguín-Contreras, L.F.; Ruiz-Azuara, L. Essential metal-based drugs: Correlation between redox potential and biological activity of M2+ with a N2O2 ligand. J. Mex. Chem. Soc. 2017, 61, 109–119. [Google Scholar] [CrossRef]
- Jabłońska-Wawrzycka, A.; Rogala, P.; Czerwonka, G. Unpublished data.




Sample Availability: Samples of the compounds 1, 2 and L22 are available from the authors. | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jabłońska-Wawrzycka, A.; Rogala, P.; Czerwonka, G.; Michałkiewicz, S.; Hodorowicz, M.; Kowalczyk, P. Ruthenium(IV) Complexes as Potential Inhibitors of Bacterial Biofilm Formation. Molecules 2020, 25, 4938. https://doi.org/10.3390/molecules25214938
Jabłońska-Wawrzycka A, Rogala P, Czerwonka G, Michałkiewicz S, Hodorowicz M, Kowalczyk P. Ruthenium(IV) Complexes as Potential Inhibitors of Bacterial Biofilm Formation. Molecules. 2020; 25(21):4938. https://doi.org/10.3390/molecules25214938
Chicago/Turabian StyleJabłońska-Wawrzycka, Agnieszka, Patrycja Rogala, Grzegorz Czerwonka, Sławomir Michałkiewicz, Maciej Hodorowicz, and Paweł Kowalczyk. 2020. "Ruthenium(IV) Complexes as Potential Inhibitors of Bacterial Biofilm Formation" Molecules 25, no. 21: 4938. https://doi.org/10.3390/molecules25214938
APA StyleJabłońska-Wawrzycka, A., Rogala, P., Czerwonka, G., Michałkiewicz, S., Hodorowicz, M., & Kowalczyk, P. (2020). Ruthenium(IV) Complexes as Potential Inhibitors of Bacterial Biofilm Formation. Molecules, 25(21), 4938. https://doi.org/10.3390/molecules25214938