Ginsenoside Rb3 Inhibits Pro-Inflammatory Cytokines via MAPK/AKT/NF-κB Pathways and Attenuates Rat Alveolar Bone Resorption in Response to Porphyromonas gingivalis LPS
Abstract
1. Introduction
2. Results
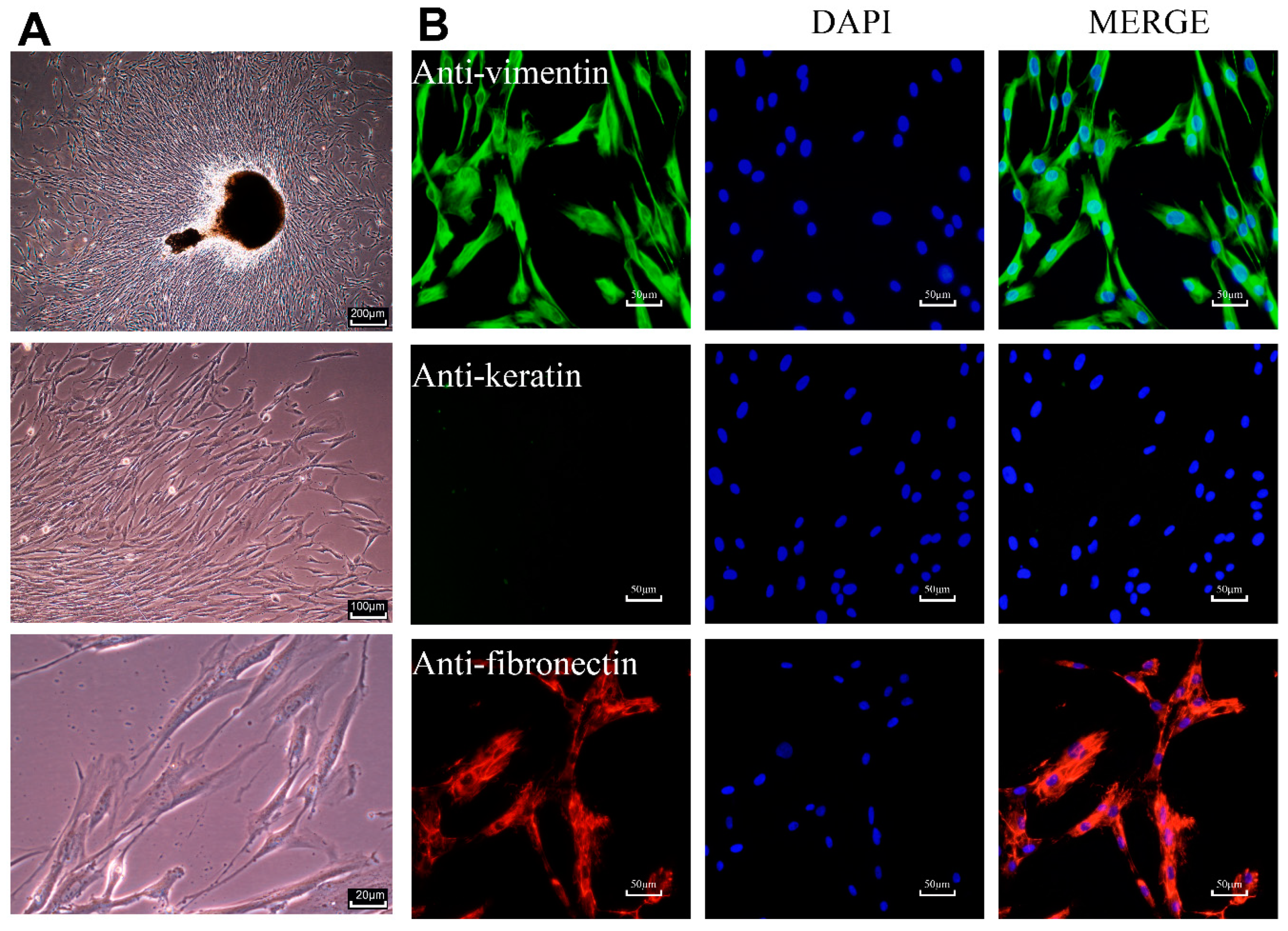
2.1. Characterization of Primary HPLCs
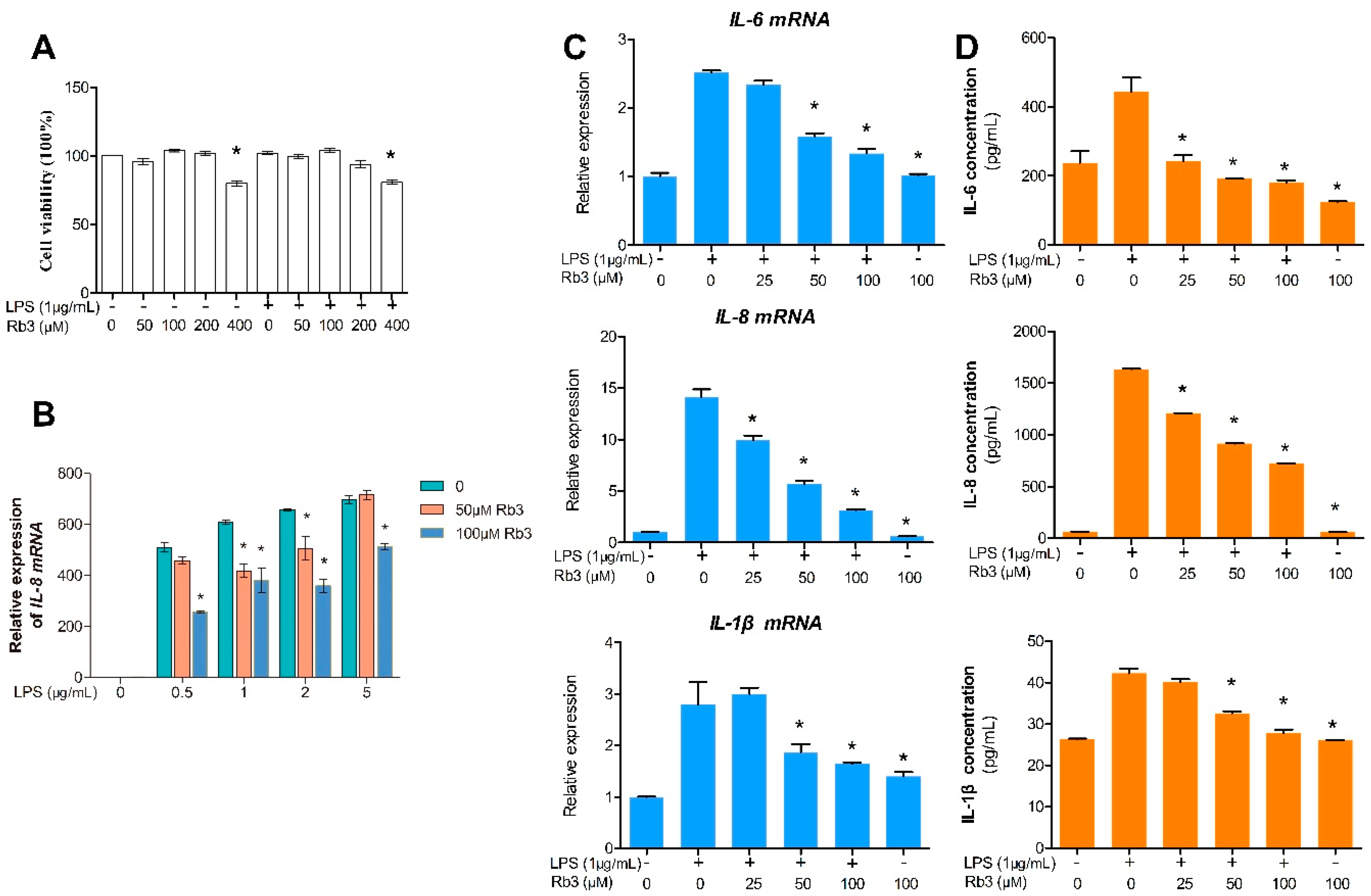
2.2. Rb3 Attenuates the Expression of IL-6, IL-8 and IL-1β in Response to P. gingivalis LPS
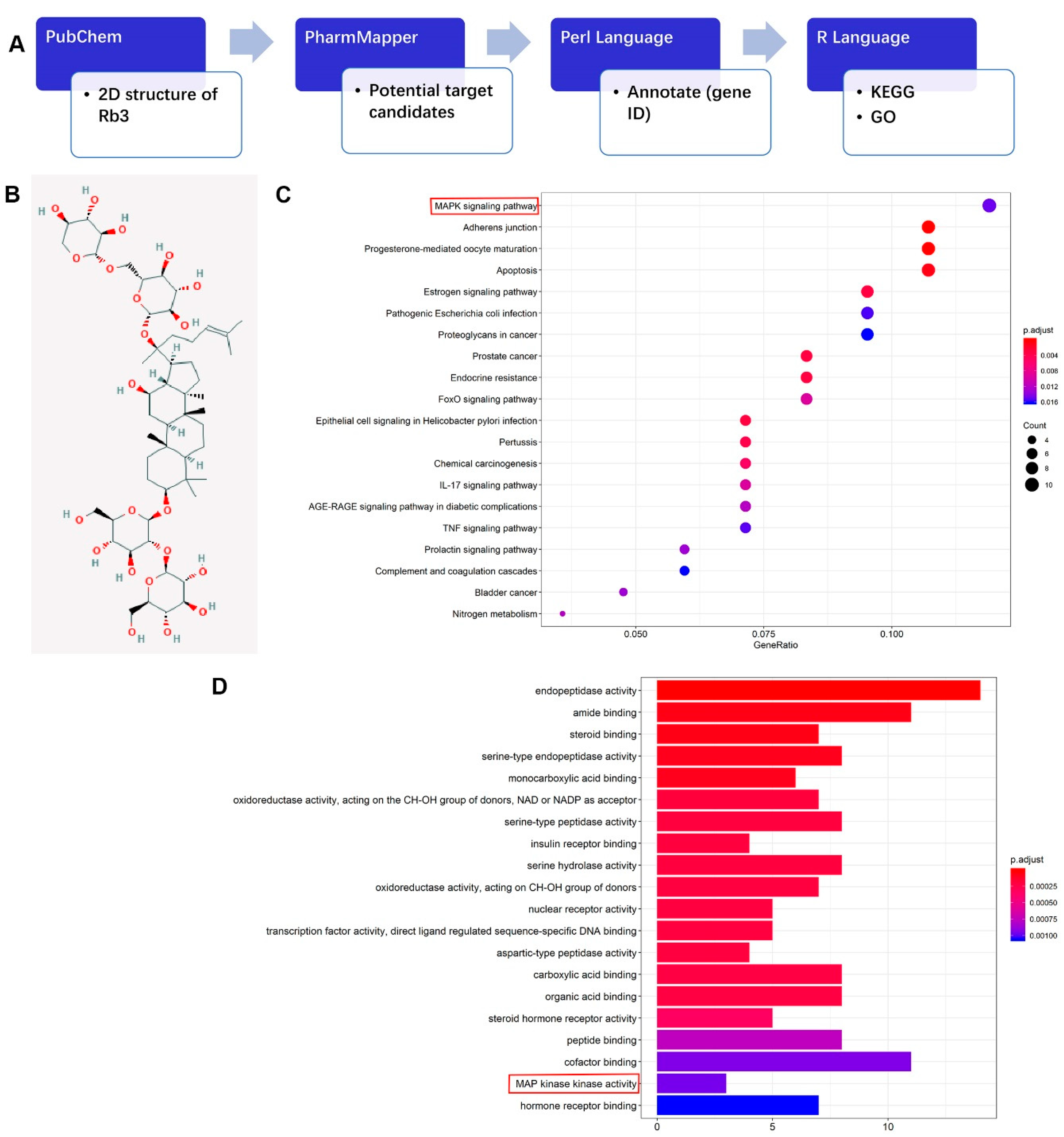
2.3. Potential Target Candidates Analysis of Rb3
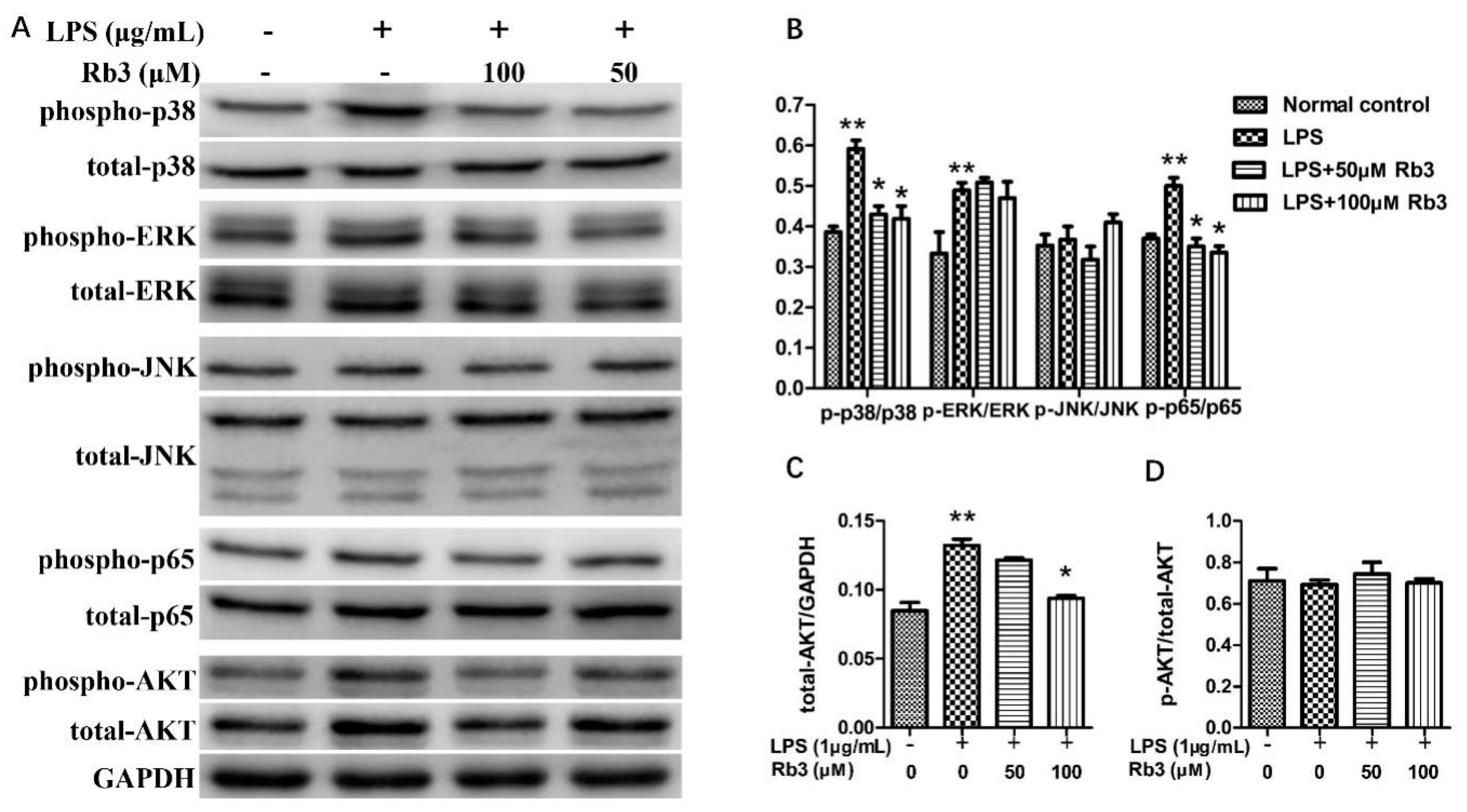
2.4. Rb3 Suppresses Inflammatory Activation of p38 MAPK, AKT and NF-kB
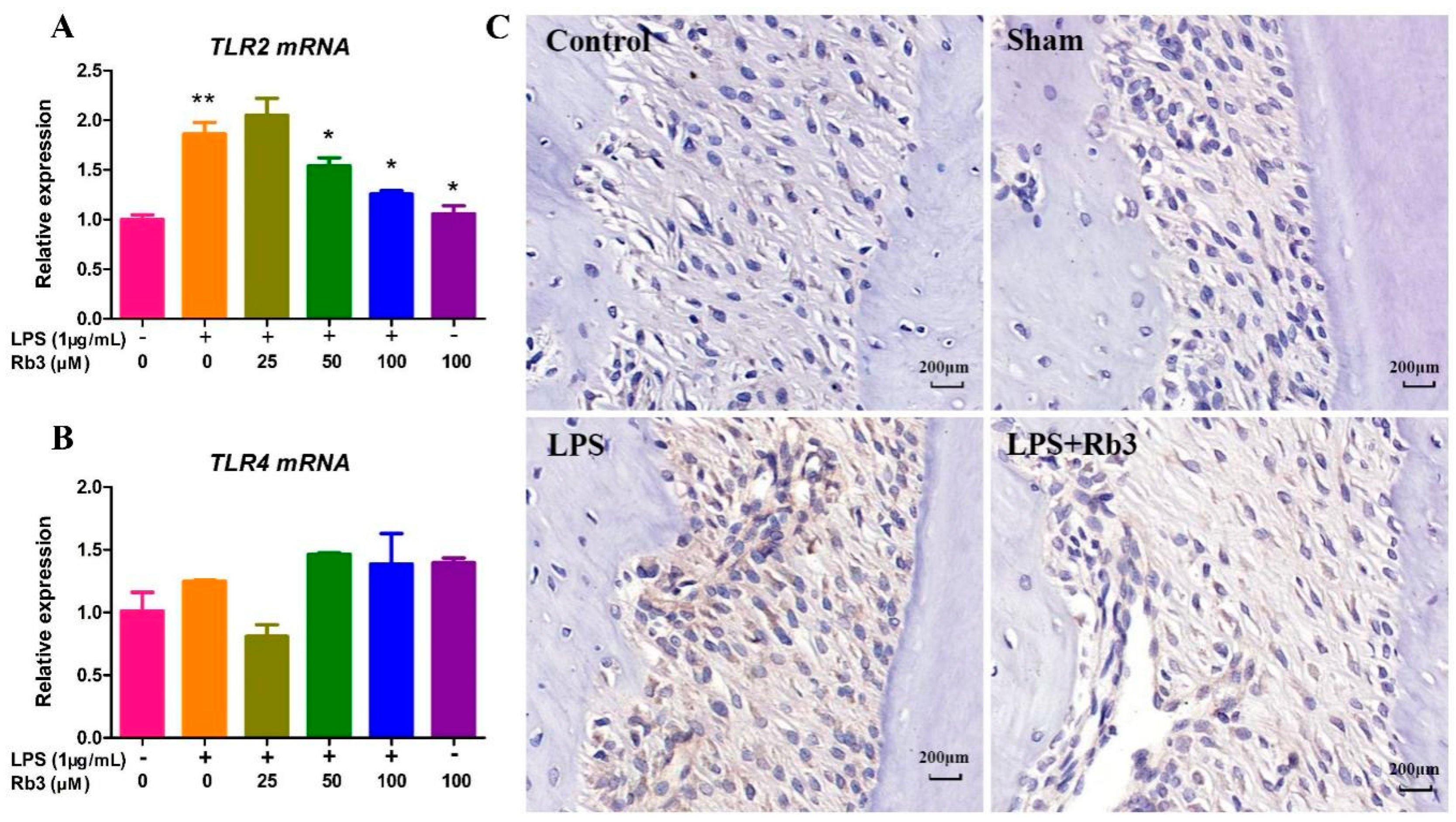
2.5. Rb3 Inhibits Chronic Inflammation by Downregulating the Expression of Toll-like Receptor (TLR)2
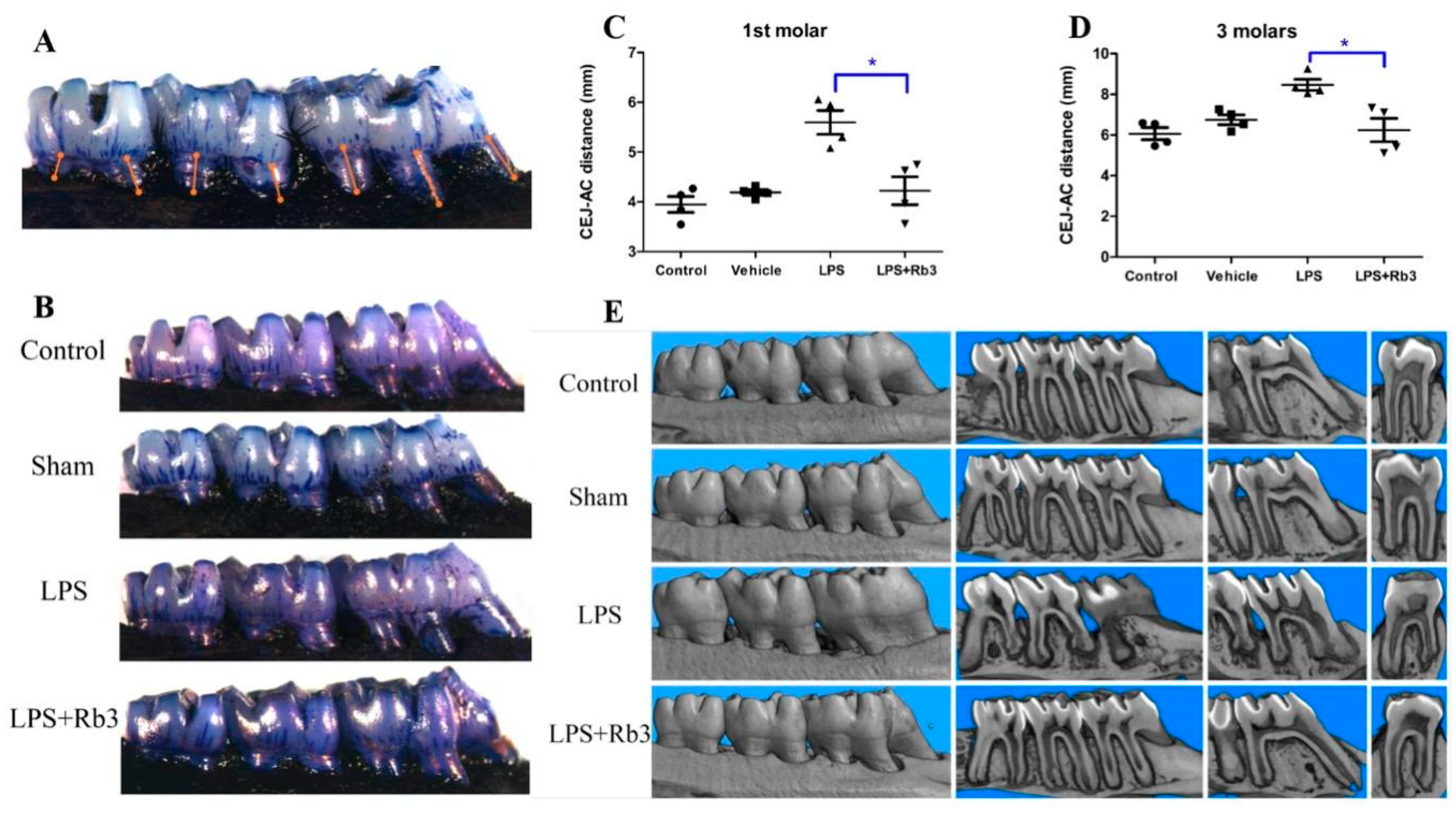
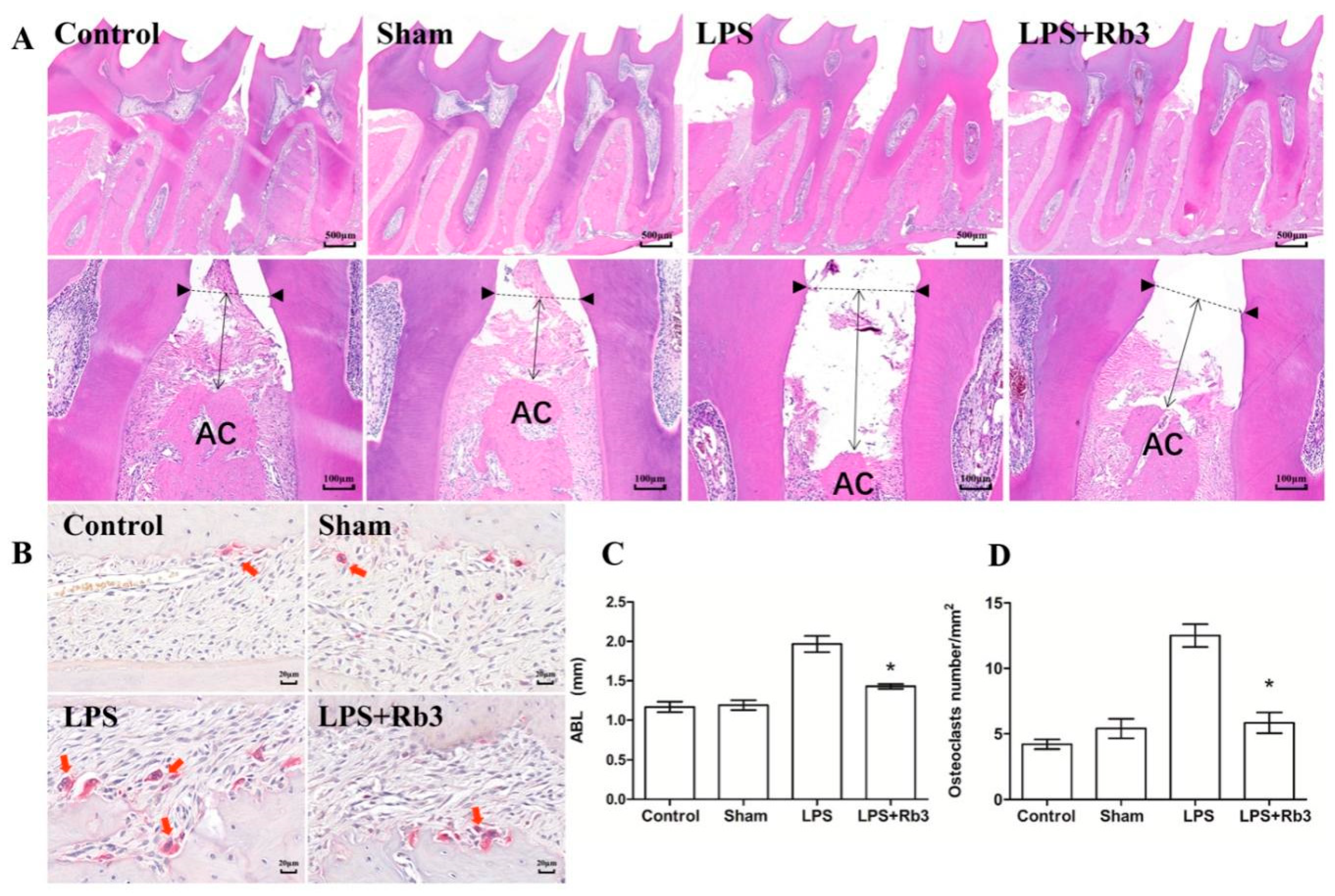
2.6. Rb3 Treatment Ameliorates Alveolar Bone Resorption in Rats
2.7. Rb3 Attenuates P. gingivalis LPS-Induced Osteoclast Differentiation
3. Discussion
4. Materials and Methods
4.1. Primary Culture of HPLCs
4.2. Cell Immunofluorescence Assay
4.3. CCK-8 Assay
4.4. ELISA
4.5. RNA Extraction, Reverse Transcription and qPCR Analysis
4.6. Network Pharmacological Analysis of Rb3
4.7. Western Blot
4.8. Animal and Experimental Design
4.9. Macroscopic Measurement of Alveolar Bone Loss
4.10. Assessment of Bone Resorption Using Micro-CT
4.11. H&E and TRAP Staining
4.12. IHC Assay
4.13. Statistical Analysis for Cell and Animal Studies
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Darveau, R.P. Periodontitis: A polymicrobial disruption of host homeostasis. Nat. Rev. Microbiol. 2010, 8, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Parahitiyawa, N.B.; Jin, L.J.; Leung, W.K.; Yam, W.C.; Samaranayake, L.P. Microbiology of Odontogenic Bacteremia: Beyond Endocarditis. Clin. Microbiol. Rev. 2009, 22, 386. [Google Scholar] [CrossRef]
- Li, X.; Tse, H.F.; Jin, L. Novel Endothelial Biomarkers: Implications for Periodontal Disease and CVD. J. Dent. Res. 2011, 90, 1062–1069. [Google Scholar] [CrossRef]
- Lalla, E.; Papapanou, P.N. Diabetes mellitus and periodontitis: A tale of two common interrelated diseases. Nat. Rev. Endocrinol. 2011, 7, 738–748. [Google Scholar] [CrossRef]
- Jain, S.; Darveau, R.P. Contribution of Porphyromonas gingivalis lipopolysaccharide to periodontitis. Periodontology 2010, 54, 53–70. [Google Scholar] [CrossRef]
- Yilmaz, Ã.Z. The chronicles of Porphyromonas gingivalis: The microbium, the human oral epithelium and their interplay. Microbiology 2008, 154, 2897–2903. [Google Scholar] [CrossRef]
- Herath, T.D.K.; Wang, Y.; Seneviratne, C.J.; Lu, Q.; Darveau, R.P.; Wang, C.-Y.; Jin, L. Porphyromonas gingivalis lipopolysaccharide lipid A heterogeneity differentially modulates the expression of IL-6 and IL-8 in human gingival fibroblasts. J. Clin. Periodontol. 2011, 38, 694–701. [Google Scholar] [CrossRef]
- Uehara, A.; Takada, H. Functional TLRs and NODs in Human Gingival Fibroblasts. J. Dent. Res. 2007, 86, 249–254. [Google Scholar] [CrossRef]
- Liang, L.; Yu, J.; Zhou, W.; Liu, N.; E, L.; Wang, D.S.; Liu, H. Endothelin-1 Stimulates Proinflammatory Cytokine Expression in Human Periodontal Ligament Cells via Mitogen-Activated Protein Kinase Pathway. J. Periodontol. 2014, 85, 618–626. [Google Scholar] [CrossRef] [PubMed]
- Drabarek, B.; Dymkowska, D.; Szczepanowska, J.; Zabłocki, K. TNFα affects energy metabolism and stimulates biogenesis of mitochondria in EA.hy926 endothelial cells. Int. J. Biochem. Cell Biol. 2012, 44, 1390–1397. [Google Scholar] [CrossRef]
- Schabbauer, G.; Tencati, M.; Pedersen, B.; Pawlinski, R.; Mackman, N. PI3K-Akt Pathway Suppresses Coagulation and Inflammation in Endotoxemic Mice. Arter. Thromb. Vasc. Biol. 2004, 24, 1963–1969. [Google Scholar] [CrossRef] [PubMed]
- Xue, C.C.; Shergis, J.L.; Zhang, A.L.; Worsnop, C.; Fong, H.H.; Story, D.A.; Da Costa, C.; Thien, F.C.K. Panax ginseng C.A Meyer root extract for moderate Chronic Obstructive Pulmonary Disease (COPD): Study protocol for a randomised controlled trial. Trials 2011, 12, 164. [Google Scholar] [CrossRef] [PubMed]
- Shergis, J.L.; Zhang, A.L.; Zhou, W.; Xue, C.C. Panax ginsengin Randomised Controlled Trials: A Systematic Review. Phytother. Res. 2012, 27, 949–965. [Google Scholar] [CrossRef]
- Ma, L.; Liu, H.; Xie, Z.; Yang, S.; Xu, W.; Hou, J.; Yu, B. Ginsenoside Rb3 Protects Cardiomyocytes against Ischemia-Reperfusion Injury via the Inhibition of JNK-Mediated NF-κB Pathway: A Mouse Cardiomyocyte Model. PLoS ONE 2014, 9, e103628. [Google Scholar] [CrossRef]
- Xing, J.; Hou, J.; Ma, Z.; Wang, Z.; Ren, S.; Wang, Y.; Liu, W.; Chen, C.; Li, W. Ginsenoside Rb3 provides protective effects against cisplatin-induced nephrotoxicity via regulation of AMPK-/mTOR-mediated autophagy and inhibition of apoptosis in vitro and in vivo. Cell Prolif. 2019, 52, e12627. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Dong, J.; Liu, P.; Lau, C.W.; Gao, Z.; Zhou, D.; Tang, J.; Ng, C.F.; Huang, Y. Ginsenoside Rb3 attenuates oxidative stress and preserves endothelial function in renal arteries from hypertensive rats. Br. J. Pharmacol. 2014, 171, 3171–3181. [Google Scholar] [CrossRef] [PubMed]
- Aravinthan, A.; Kim, J.H.; Antonisamy, P.; Kang, C.-W.; Choi, J.; Kim, N.S.; Kim, J.-H. Ginseng total saponin attenuates myocardial injury via anti-oxidative and anti-inflammatory properties. J. Ginseng Res. 2014, 39, 206–212. [Google Scholar] [CrossRef]
- Sun, J.; Yu, X.; Huangpu, H.; Yao, F. Ginsenoside Rb3 protects cardiomyocytes against hypoxia/reoxygenation injury via activating the antioxidation signaling pathway of PERK/Nrf2/HMOX1. Biomed. Pharmacother. 2019, 109, 254–261. [Google Scholar] [CrossRef]
- Cui, J.; Jiang, L.; Xiang, H. Ginsenoside Rb3 exerts antidepressant-like effects in several animal models. J. Psychopharmacol. 2011, 26, 697–713. [Google Scholar] [CrossRef]
- Wang, M.; Chen, X.; Jin, W.; Xu, X.; Li, X.; Sun, L. Ginsenoside Rb3 exerts protective properties against cigarette smoke extract-induced cell injury by inhibiting the p38 MAPK/NF-κB and TGF-β1/VEGF pathways in fibroblasts and epithelial cells. Biomed. Pharmacother. 2018, 108, 1751–1758. [Google Scholar] [CrossRef]
- Qu, L.; Yu, X.; Xu, L. Effect of ginsenoside Rb3 on cerebral ischemia-reperfusion injury in rats. Chin. J. Gerontol. 2016, 23, 5791–5793. [Google Scholar]
- Garlet, G.P. Destructive and Protective Roles of Cytokines in Periodontitis: A Re-appraisal from Host Defense and Tissue Destruction Viewpoints. J. Dent. Res. 2010, 89, 1349–1363. [Google Scholar] [CrossRef]
- Liu, Y.-C.G.; Lerner, U.H.; Teng, Y.-T.A. Cytokine responses against periodontal infection: Protective and destructive roles. Periodontology 2010, 52, 163–206. [Google Scholar] [CrossRef]
- Seo, T.; Cha, S.; Kim, T.-I.; Lee, J.-S.; Woo, K.M. Porphyromonas gingivalis-derived lipopolysaccharide-mediated activation of MAPK signaling regulates inflammatory response and differentiation in human periodontal ligament fibroblasts. J. Microbiol. 2012, 50, 311–319. [Google Scholar] [CrossRef]
- Pei, Z.; Wang, B.; Zhang, F.; Niu, Z.; Shi, S.; Cannon, R.D.; Mei, L. Response of Human Periodontal Ligament Cells to Baicalin. J. Periodontol. 2014, 85, 1283–1290. [Google Scholar] [CrossRef]
- Yamamoto, T.; Kita, M.; Oseko, F.; Nakamura, T.; Imanishi, J.; Kanamura, N. Cytokine production in human periodontal ligament cells stimulated with Porphyromonas gingivalis. J. Periodontal Res. 2006, 41, 554–559. [Google Scholar] [CrossRef]
- Baggiolini, M.; Walz, A.; Kunkel, S.L. Neutrophil-activating peptide-1/interleukin 8, a novel cytokine that activates neutrophils. J. Clin. Investig. 1989, 84, 1045–1049. [Google Scholar] [CrossRef] [PubMed]
- Graves, D.T.; Cochran, D. The Contribution of Interleukin-1 and Tumor Necrosis Factor to Periodontal Tissue Destruction. J. Periodontol. 2003, 74, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, Y.; Hosokawa, I.; Ozaki, K.; Nakanishi, T.; Nakae, H.; Matsuo, T. Tea polyphenols inhibit IL-6 production in tumor necrosis factor superfamily 14-stimulated human gingival fibroblasts. Mol. Nutr. Food Res. 2010, 54, S151–S158. [Google Scholar] [CrossRef]
- Kim, W.-H.; An, H.-J.; Kim, J.-Y.; Gwon, M.-G.; Gu, H.; Park, J.-B.; Sung, W.J.; Kwon, Y.-C.; Park, K.-D.; Han, S.M.; et al. Bee Venom Inhibits Porphyromonas gingivalis Lipopolysaccharides-Induced Pro-Inflammatory Cytokines through Suppression of NF-κB and AP-1 Signaling Pathways. Molecules 2016, 21, 1508. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, S.; Xiao, Y.; Zhang, W.; Wu, S.; Qin, T.; Yue, Y.; Qian, W.; Li, L. NLRP3 Inflammasome and Inflammatory Diseases. Oxidative Med. Cell. Longev. 2020. [Google Scholar] [CrossRef] [PubMed]
- Burns, E.; Bachrach, G.; Shapira, L.; Nussbaum, G. Cutting Edge: TLR2 Is Required for the Innate Response toPorphyromonas gingivalis: Activation Leads to Bacterial Persistence and TLR2 Deficiency Attenuates Induced Alveolar Bone Resorption. J. Immunol. 2006, 177, 8296–8300. [Google Scholar] [CrossRef] [PubMed]
- Eskan, M.A.; Hajishengallis, G.; Kinane, D.F. Differential Activation of Human Gingival Epithelial Cells and Monocytes by Porphyromonas gingivalis Fimbriae. Infect. Immun. 2006, 75, 892–898. [Google Scholar] [CrossRef] [PubMed]
- Hirschfeld, M.; Weis, J.J.; Toshchakov, V.; Salkowski, C.A.; Cody, M.J.; Ward, D.C.; Qureshi, N.; Michalek, S.M.; Vogel, S.N. Signaling by Toll-Like Receptor 2 and 4 Agonists Results in Differential Gene Expression in Murine Macrophages. Infect. Immun. 2001, 69, 1477–1482. [Google Scholar] [CrossRef]
- Martin, M.; Katz, J.; Vogel, S.N.; Michalek, S.M. Differential Induction of Endotoxin Tolerance by Lipopolysaccharides Derived fromPorphyromonas gingivalisandEscherichia coli. J. Immunol. 2001, 167, 5278–5285. [Google Scholar] [CrossRef] [PubMed]
- Teng, Y.-T. Protective and Destructive Immunity in the Periodontium: Part 1—Innate and Humoral Immunity and the Periodontium. J. Dent. Res. 2006, 85, 198–208. [Google Scholar] [CrossRef]
- Wara-Aswapati, N.; Chayasadom, A.; Surarit, R.; Pitiphat, W.; Boch, J.A.; Nagasawa, T.; Ishikawa, I.; Izumi, Y. Induction of Toll-Like Receptor Expression by Porphyromonas gingivalis. J. Periodontol. 2013, 84, 1010–1018. [Google Scholar] [CrossRef]
- Brightbill, H.D. Host Defense Mechanisms Triggered by Microbial Lipoproteins through Toll-Like Receptors. Science 1999, 285, 732–736. [Google Scholar] [CrossRef]
- Darveau, R.P.; Arbabi, S.; Garcia, I.; Bainbridge, B.; Maier, R.V. Porphyromonas gingivalis Lipopolysaccharide Is Both Agonist and Antagonist for p38 Mitogen-Activated Protein Kinase Activation. Infect. Immun. 2002, 70, 1867–1873. [Google Scholar] [CrossRef]
- Coats, S.R.; Reife, R.A.; Bainbridge, B.W.; Pham, T.T.-T.; Darveau, R.P. Porphyromonas gingivalis lipopolysaccharide antagonizes Escherichia coli lipopolysaccharide at toll-like receptor 4 in human endothelial cells. Infect. Immun. 2003, 71, 6799–6807. [Google Scholar] [CrossRef]
- Zhang, P.; Liu, J.; Xu, Q.; Harber, G.; Feng, X.; Michalek, S.M.; Katz, J. TLR2-dependent Modulation of Osteoclastogenesis by Porphyromonas gingivalis through Differential Induction of NFATc1 and NF-κB. J. Biol. Chem. 2011, 286, 24159–24169. [Google Scholar] [CrossRef] [PubMed]
- Ossola, C.A.; Surkin, P.N.; Mohn, C.E.; Elverdín, J.C.; Solari, J.F. Anti-Inflammatory and Osteoprotective Effects of Cannabinoid-2 Receptor Agonist HU-308 in a Rat Model of Lipopolysaccharide-Induced Periodontitis. J. Periodontol. 2016, 87, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Ossola, C.A.; Surkin, P.N.; Pugnaloni, A.; Mohn, C.E.; Elverdín, J.C.; Solari, J.F. Long-term treatment with methanandamide attenuates LPS-induced periodontitis in rats. Inflamm. Res. 2012, 61, 941–948. [Google Scholar] [CrossRef] [PubMed]
- Llavaneras, A.; Ramamurthy, N.S.; Heikkilä, P.; Teronen, O.; Salo, T.; Rifkin, B.R.; Ryan, M.E.; Golub, L.M.; Sorsa, T. A Combination of a Chemically Modified Doxycycline and a Bisphosphonate Synergistically Inhibits Endotoxin-Induced Periodontal Breakdown in Rats. J. Periodontol. 2001, 72, 1069–1077. [Google Scholar] [CrossRef] [PubMed]
- Gürkan, A.; Emingil, G.; Nizam, N.; Doganavsargil, B.; Sezak, M.; Kutukculer, N.; Atilla, G. Therapeutic Efficacy of Vasoactive Intestinal Peptide inEscherichia coliLipopolysaccharide-Induced Experimental Periodontitis in Rats. J. Periodontol. 2009, 80, 1655–1664. [Google Scholar] [CrossRef]
- Crawford, J.M.; Taubman, M.A.; Smith, D.J. The natural history of periodontal bone loss in germfree and gnotobiotic rats infected with Periodontopathic microorganisms. J. Periodontal Res. 1978, 13, 316–325. [Google Scholar] [CrossRef]
- Terrizzi, A.R.; Solari, J.F.; Lee, C.M.; Bozzini, C.; Mandalunis, P.M.; Elverdín, J.C.; Conti, M.I.; Martínez, M.P. Alveolar bone loss associated to periodontal disease in lead intoxicated rats under environmental hypoxia. Arch. Oral Biol. 2013, 58, 1407–1414. [Google Scholar] [CrossRef]
- Mester, A.; Ciobanu, L.; Taulescu, M.; Apostu, D.; Lucaciu, O.; Filip, G.A.; Feldrihan, V.; Licarete, E.; Ilea, A.; Piciu, A.; et al. Periodontal disease may induce liver fibrosis in an experimental study on Wistar rats. J. Periodontol. 2019, 90, 911–919. [Google Scholar] [CrossRef]
- Bi, J.; Koivisto, L.; Pang, A.; Li, M.; Jiang, G.; Aurora, S.; Wang, Z.; Owen, G.R.; Dai, J.; Shen, Y.; et al. Suppression of αvβ6 Integrin Expression by Polymicrobial Oral Biofilms in Gingival Epithelial Cells. Sci. Rep. 2017, 7, 4411. [Google Scholar] [CrossRef]
- Li, C.H.; Amar, S. Morphometric, Histomorphometric, and Microcomputed Tomographic Analysis of Periodontal Inflammatory Lesions in a Murine Model. J. Periodontol. 2007, 78, 1120–1128. [Google Scholar] [CrossRef]
| Primer | Sequence | Amplicon Size (bp) | NCBI Gene ID |
|---|---|---|---|
| GAPDH | Forward 5′-CAGGAGGCATTGCTGATGAT-3′ | 138 | 2597 |
| Reverse 5′-GAAGGCTGGGGCTCATTT-3′ | |||
| IL-6 | Forward 5′-CACTGGTCTTTTGGAGTTTGAG-3′ | 101 | 3569 |
| Reverse 5′-GGACTTTTGTACTCATCTGCAC-3′ | |||
| IL-1β | Forward 5′-GCCAGTGAAATGATGGCTTATT-3′ | 85 | 3553 |
| Reverse 5′-AGGAGCACTTCATCTGTTTAGG-3′ | |||
| IL-8 | Forward 5′-AACTGAGAGTGATTGAGAGTGG-3′ | 147 | 3576 |
| Reverse 5′-ATGAATTCTCAGCCCTCTTCAA-3′ | |||
| TLR2 | Forward 5′-AAGCAGCATATTTTACTGCTGG-3′ | 184 | 7097 |
| Reverse 5′-CCTGAAACAAACTTTCATCGGT-3′ | |||
| TLR4 | Forward 5′-GACTGGGTAAGGAATGAGCTAG-3′ | 143 | 7099 |
| Reverse 5′-ACCTTTCGGCTTTTATGGAAAC-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, M.; Ji, Y.; Li, Z.; Chen, R.; Zhou, S.; Liu, C.; Du, M. Ginsenoside Rb3 Inhibits Pro-Inflammatory Cytokines via MAPK/AKT/NF-κB Pathways and Attenuates Rat Alveolar Bone Resorption in Response to Porphyromonas gingivalis LPS. Molecules 2020, 25, 4815. https://doi.org/10.3390/molecules25204815
Sun M, Ji Y, Li Z, Chen R, Zhou S, Liu C, Du M. Ginsenoside Rb3 Inhibits Pro-Inflammatory Cytokines via MAPK/AKT/NF-κB Pathways and Attenuates Rat Alveolar Bone Resorption in Response to Porphyromonas gingivalis LPS. Molecules. 2020; 25(20):4815. https://doi.org/10.3390/molecules25204815
Chicago/Turabian StyleSun, Minmin, Yaoting Ji, Zhen Li, Rourong Chen, Shuhui Zhou, Chang Liu, and Minquan Du. 2020. "Ginsenoside Rb3 Inhibits Pro-Inflammatory Cytokines via MAPK/AKT/NF-κB Pathways and Attenuates Rat Alveolar Bone Resorption in Response to Porphyromonas gingivalis LPS" Molecules 25, no. 20: 4815. https://doi.org/10.3390/molecules25204815
APA StyleSun, M., Ji, Y., Li, Z., Chen, R., Zhou, S., Liu, C., & Du, M. (2020). Ginsenoside Rb3 Inhibits Pro-Inflammatory Cytokines via MAPK/AKT/NF-κB Pathways and Attenuates Rat Alveolar Bone Resorption in Response to Porphyromonas gingivalis LPS. Molecules, 25(20), 4815. https://doi.org/10.3390/molecules25204815





