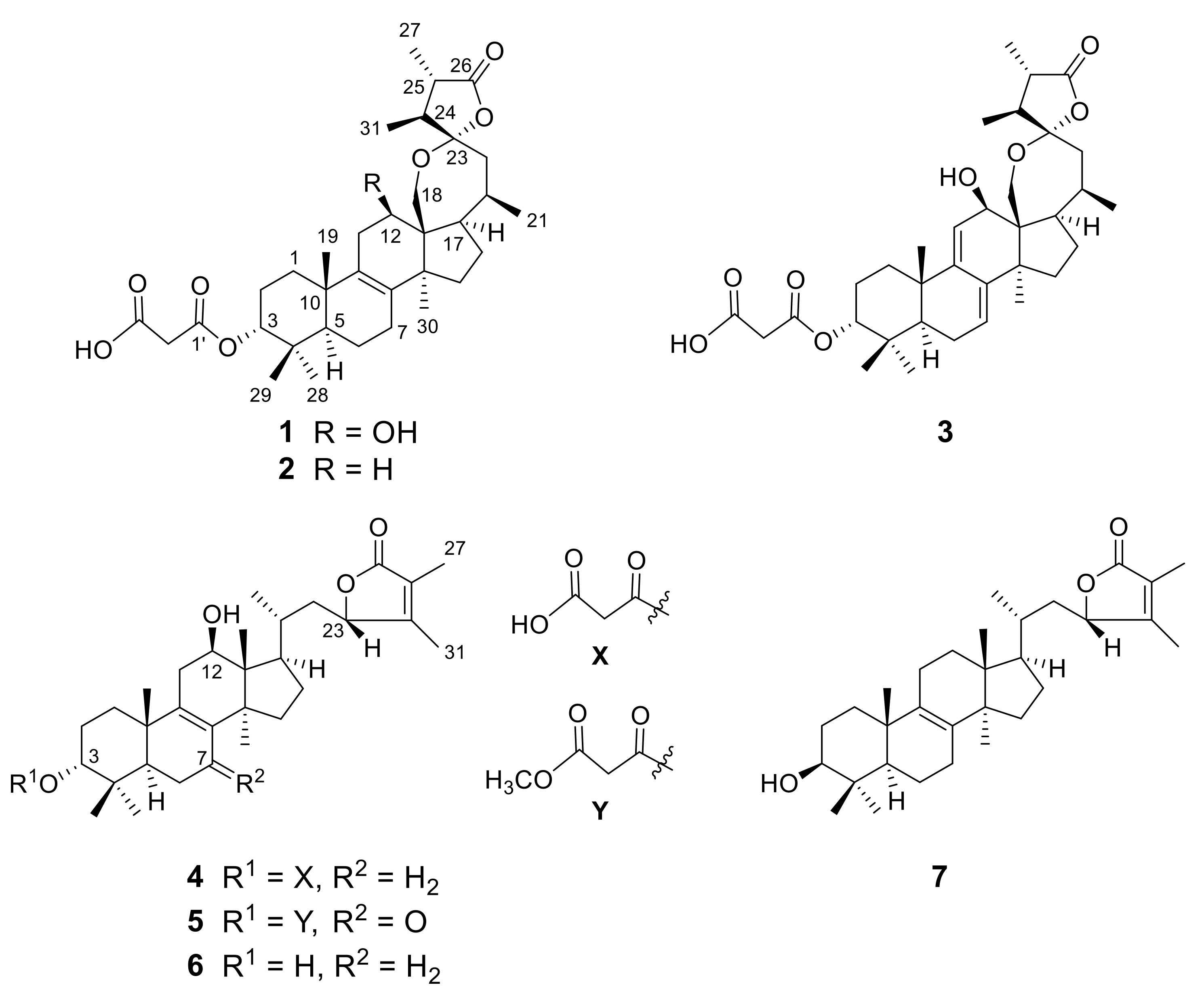
2. Results and Discussion
Compound
1 was isolated as colorless amorphous solid; its molecular formula was identified as C
34H
50O
8 by HR-ESI-MS ion peak at
m/z 609.3382 [M + Na]
+ (calculated for C
34H
50O
8Na, 609.3403) (See
Supplementary Materials). The IR spectrum of 1 showed the absorption bands for hydroxyl (3545 cm
−1) and carboxyl (1779 and 1732 cm
−1), which indicated the presence of a strained lactone unit. The
1H-NMR spectrum (
Table 1) of
1 exhibited four tertiary methyl groups at
δH 1.07, 1.03, 0.96 and 0.90 (each 3H, s), three secondary methyls at
δH 1.18 (3H, d,
J = 6.8 Hz), 1.15 (3H, d,
J = 7.1 Hz) and 0.99 (3H, d,
J = 6.3 Hz), two oxygenated methine protons at
δH 4.68 (brs, H-3) and 4.21 (t,
J = 7.1 Hz, H-12), as well as a oxygenated methylene protons at 3.91 (1H, d,
J = 14.3 Hz) and 3.88 (1H, d,
J = 14.3 Hz). The
13C-NMR (
Table 2) spectra indicated the presence of 33 carbons sorted to seven methyls, nine methylenes, seven methines and 10 quaternary carbons (three of which are carbonyls). It is worth to mention that the NMR data of CH
2-2′ were not observed when they were acquired in CD
3OD, which is consistent with that of officimalonic acid A [
8].
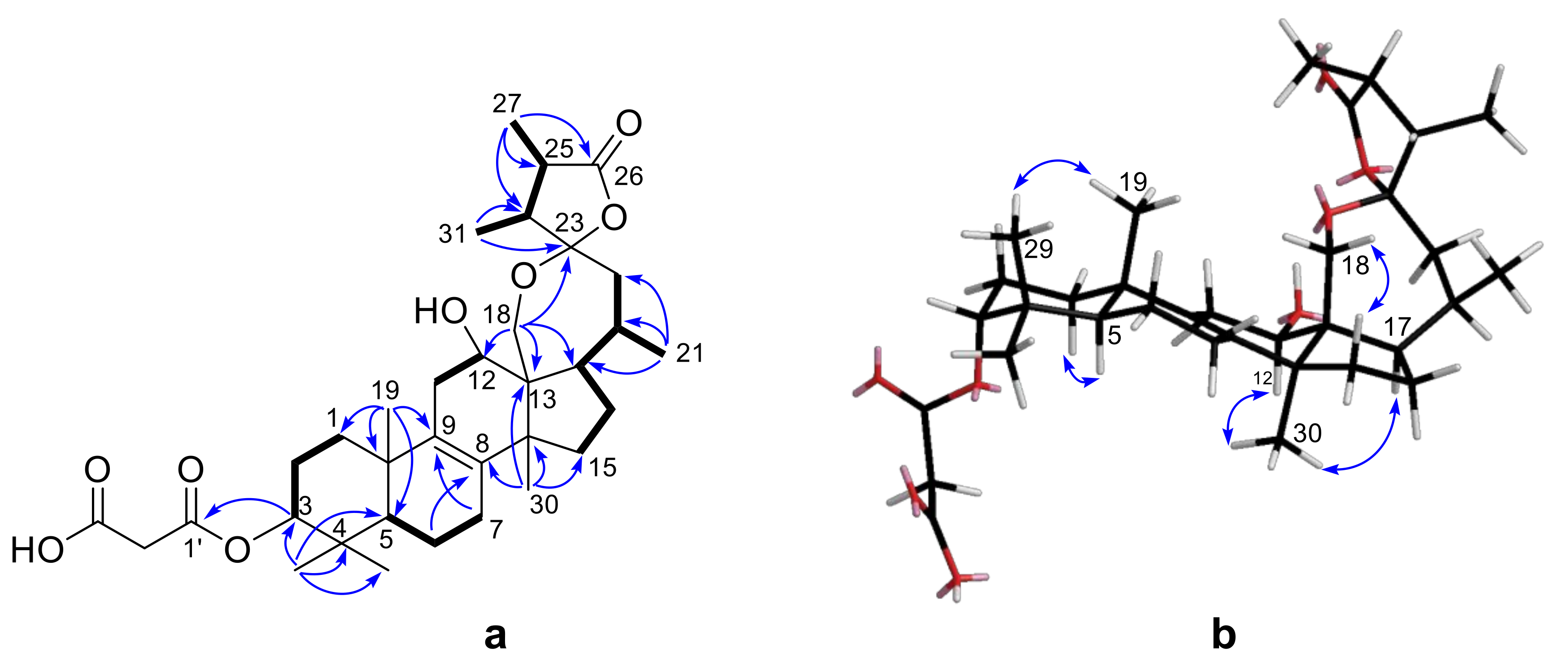
Further analysis of 2D NMR data (
1H-
1H COSY, HSQC, HMBC) revealed that the structure of
1 was similar with that of officimalonic acid C, previously isolated from this species by our group [
8]. The only difference between them is the absence of C-7 carbonyl in
1. The
1H-
1H COSY correlations (
Figure 2a) of H-5/H-6/H-7, and the HMBC correlations (
Figure 2a) from H-6 to C-8 and from H-7 to C-9, confirmed the assignment. The relative configurations of
1 were determined by the NOESY correlations (
Figure 2b) and coupling constant. The H-3 was established as β-configuration by its broad singlet-like signal. The NOESY correlation between H-12 and H
3-30 indicated they were α-configuration. Considering the biogenetic relationship, the absolute configurations of
1 were tentatively assigned as those of officimalonic acid C. Thus, the structure of compound
1 was elucidated as depicted, given the trivial name officimalonic acid I.
Compound
2 was determined to have the molecular formula C
34H
50O
7 by HR-ESI-MS at
m/z 571.3598 [M + H]
+ (calculated for C
34H
51O
7, 571.3635), 16 mass less than that of
1. The
1H and
13C-NMR (
Table 1 and
Table 2) data of
2 showed very similar results with those of
1. The major difference was that instead of a methylene signal, the signals of oxygenated methine (CH-12) were missed in
2, suggesting that compound
2 is one less hydroxyl than
1. The
1H-
1H COSY correlations of H-11/H-12 and the HMBC correlation from H
2-18 to C-12, supported the assignment. The structure of
2 was thus elucidated as depicted, named officimalonic acid J.
Compound
3 had a molecular formula C
34H
48O
8, which was determined by the HR-ESI-MS at
m/z 607.3218 [M + Na]
+ (calculated for C
34H
48O
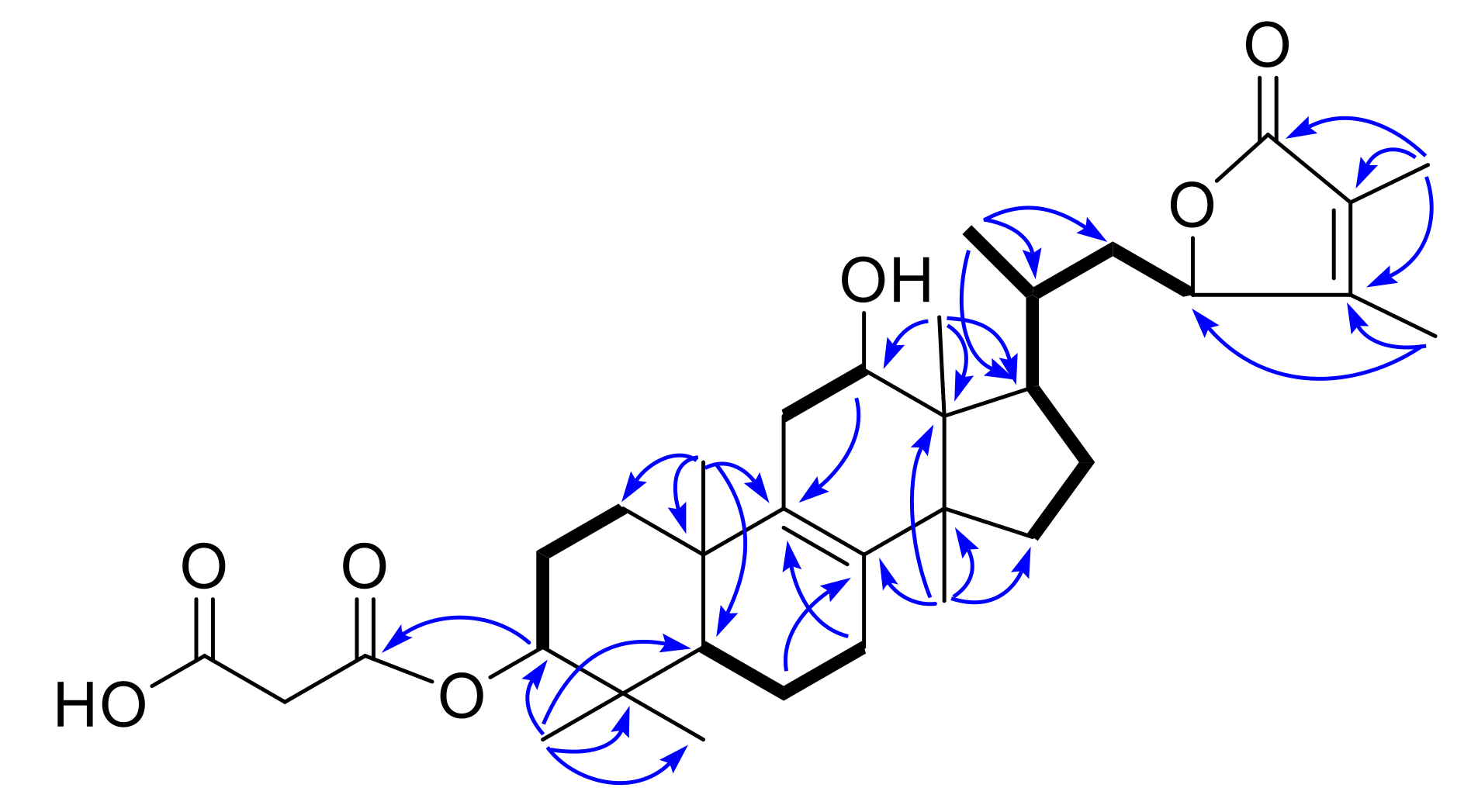
8Na, 607.3247). Analysis of
1H and
13C-NMR (
Table 1 and
Table 2) data of
3 indicated that it had a similar structure with
1. The obvious differences between them were the presence of two double bonds (Δ
7 and Δ
9(11)) in
3 instead of one double bond (Δ
8) present in
1. The two olefinic protons signals at
δH 5.57 (brs, H-7) and 5.40 (brs, H-11) supported the assignment. The
1H-
1H COSY correlations of H-5/H-6/H-7 and H-11/H-12, together with the HMBC correlations from H-7 to C-9, and from H-11 to C-8, confirmed the above assignment. Therefore, the structure of
3 was elucidated as depicted, named officimalonic acid K.
Compound
4 was obtained as a colorless amorphous solid; its HR-ESI-MS displayed a quasi-molecular ion peak at
m/z 615.3512 [M + HCOO]
−, corresponding to the molecular formula C
34H
50O
7. Analysis of
1H and
13C-NMR (
Table 1 and
Table 2) data of
4 revealed it shared the close structure similarity with that of officimalonic acid B, previously isolated from the same species [
8]. The difference between these two compounds is the absence of a carbonyl group signal and the presence of a methylene in
4. The
1H-
1H COSY correlations (
Figure 3) of H-5/H-6/H-7, and the HMBC correlations (
Figure 3) from H-6 to C-8 and from H-7 to C-9, indicated that compound
4 possess methylene at C-7 in
4 instead of a carbonyl group in officimalonic acid B. The structure of
3 was thus elucidated as depicted, gave a trivial name officimalonic acid L.
Compound
5 had a molecular formula C
35H
50O
8, which was established by its HR-ESI-MS at
m/z 621.3384 [M + Na]
+ (calculated for C
35H
50O
8Na, 621.3403). The
1H and
13C-NMR (
Table 1 and
Table 2) data of
5 showed almost the same as those of officimalonic acid B, the only difference being the presence of one more methoxyl signal in
5. The methoxy group was attached to C-3′ of malonate half-ester by the HMBC correlation from
δH 3.68 (3H, s) to C-3′ (
δC 167.4). Therefore, the structure of
5 was determined as depicted, named officimalonic acid M.
Compound
6 had a molecular formula of C
31H
48O
4 as determined on the basis of HR-ESI-MS at
m/z 507.3427 [M + Na]
+ (calculated for C
35H
50O
8Na, 507.3450). Analysis of
1H and
13C-NMR (
Table 1 and
Table 2) data of
6 revealed it had a similar structure with that of
4. The difference is that the signals for malonate half-ester at C-3 are not observed in
6, correspondingly, its proton signal of H-3 is up-field shifted to
δH 3.36 (brs), indicated that compound
6 had hydroxy group at C-3, rather than carboxyacetyloxy group at C-3. The HMBC correlations from H
3-28 (or H
3-29) to C-3, C-4, and C-5, supported the assignment. The structure of
6 was thus elucidated as depicted, given the trivial name officimalonic acid N.
Compound
7 had a molecular formula of C
31H
48O
3 by HR-ESI-MS at
m/z 469.3651 [M + H]
+ (calculated for C
31H
49O
3, 469.3682), 16 mass less than that of
6. The
1H and
13C-NMR (
Table 1 and
Table 2) data of
7 showed very similar signals with those of
6. The only two oxygenated methines signals [
δH 3.22 (dd,
J = 11.6, 4.4 Hz),
δC 79.0;
δH 4.71 (brd,
J = 8.0 Hz),
δC 83.2] were observed in
7, suggesting that it had one less hydroxy group than that of
6. Further analysis 2D NMR spectra permitted to determine the structure. The HMBC correlations from H
3-28 (or H
3-29) to C-3 (
δC 79.0), C-4 and C-5, indicated there was a hydroxy group at C-3, while the HMBC correlations from H
3-18 to C-12 (
δC 30.9), C-13, C-14 and C-17, suggested hydroxyl at C-12 in
6 disappeared in
7. Notably, coupling constant of H-3 [
9] of compound
7 is different with that of compounds
1–
6, implying its stereochemistry is not β-orientation, which is confirmed by the NOESY correlation between H-3 and H-5. Therefore, the structure of
7 was determined as depicted, named officimalonic acid O.
All of the new compounds had their inhibitory activities tested against NO production in lipopolysaccharide (LPS)-induced RAW264.7 cells. Their cell viabilities were firstly examined by the CCK-8 method. As compounds 2 and 4 showed strong cytotoxic effects against RAW 264.7 cells at the concentration of 60 μM, they would not be conducted, following anti-inflammation evaluation. Except compounds 2 and 4, other tested compounds showed NO inhibition lower than 60 μM. The results indicated that compounds 3 and 5 exhibited significant inhibitory activities against NO production in LPS-induced RAW 264.7 cells with IC50 values at 33.0 and 25.4 μM, compared to that of positive control dexamethasone (IC50 = 20.35 μM).
In addition, the cyclooxygenase (COX-2) inhibitory activities of these new compounds were evaluated using the in vitro assay kit [
10]. Interestingly, the similar results were observed as those of NO production inhibition assay, only compounds
3 and
5 showed COX-2 inhibitory activities with IC
50 values at 30.1 and 42.3 μM, compared to that of positive control naproxen (IC
50 = 8.2 μM).
3. Materials and Methods
3.1. General Experimental Procedures
IR spectra were recorded on a Nicolet 380 FT-IR spectrometer. The optical rotations were measured on an Auto Pol IV automatic polarimeter (Rudolph Research, Flanders, NJ, USA) at room temperature. 1D and 2D NMR data were recorded on a Varian 400 MHz instrument with TMS as internal standard. HR-ESI-MS data were acquired using a Triple TOF 6600 mass spectrometer (AB Sciex, Framingham, MA, USA). Semi-preparative HPLC separations were performed on a Hitachi Chromaster system consisting of a 5110 pump, 5210 autosampler, 5310 column oven, 5430 diode array detector, and a Phenomenex Luna C18 column (250 × 10 mm, S-5 μm), all operated using EZChrom Elite software. All solvents were of ACS or HPLC grade, and were obtained from Tansoole (Shanghai, China), Sigma-Aldrich (St. Louis, MO, USA), respectively. Silica gel (300−400 mesh), C18 reverse-phased silica gel (150–200 mesh, Merck, city, country), and MCI gel (CHP20P, 75−150 μM, Mitsubishi Chemical Industries Ltd., Tokyo, Japan) were used for column chromatography (CC), and pre-coated silica gel GF254 plates (Qingdao Marine Chemical Plant, Qingdao, China) were used for TLC.
3.2. Fugnal Material
Fruiting bodies of F. officinalis were bought from Xinjiang Uyghur Medicine Hospital (Xinjiang, China), and identified by Prof. Xinping Yang (Institute of Applied Microbiology, Xinjiang Academy of Agricultural Sciences). A voucher specimen (FO-201411) is deposited in the Key Laboratory of Plant Resources and Chemistry of Arid Zone, Xinjiang Technical Institute of Physics and Chemistry, Chinese Academy of Sciences (Xinjiang, China).
3.3. Extraction and Isolation
Air-dried ground powder of F. officinalis fruiting bodies (1.0 kg) was extracted with MeOH (4.5 L × 3) by maceration at room temperature (seven days each time) to afford a crude methanol extract (575.9 g). The crude extract was suspended in distilled H2O and then extracted successively with EtOAc and n-BuOH. The EtOAc fraction (135.8 g) was then applied to a column of MCI gel (MeOH-H2O, 50:50 to 100:0, v/v) to yield five fractions (A−E). Fraction B (8.4 g) was then subjected to silica gel CC eluted with a CHCl3: MeOH (100:1 to 1:1 v/v) gradient to obtain seven fractions (B1−B7). Fraction B1 (1.3 g) was subjected to RP-18 silica gel CC (MeOH-H2O, 60/40 to 100:0, v/v) to give four sub-fractions (B1a−B1d). Purification of B1b (87.0 mg) by semi-preparative HPLC, eluting with MeOH-H2O (0–25 min: 70:30 to 78:22; 25–26 min: 78:22 to 100:0; 26–27 min: 100:0; 27–28 min: 100:0 to 70:30; 28–35 min: 70:30; v/v, 3 mL/min), yielded compounds 2 (6.4 mg) and 3 (7.5 mg). Fraction B3 (2.5 g) was separated on RP-18 silica gel CC (MeOH-H2O, 60/40 to 100:0, v/v) to give four sub-fractions (B3a−B3d). Sub-fraction B3a (562.4 mg) was subjected to silica gel CC eluted with a CHCl3: MeOH (100:1 to 1:1 v/v) gradient to obtain two fractions (B3a1−B3a2). Fraction B3a1 (172.7 mg) was purified by semi-preparative HPLC, eluting with isocratic MeOH-H2O (65:35, v/v, 3 mL/min) to yield the compound 1 (10.6 mg). Fraction D (16.8 g) was subjected to silica gel CC eluted with a CHCl3: MeOH (100:1 to 1:1 v/v) gradient to obtain three fractions (D1−D3). Fraction D1 (3.6 g) was separated on RP-18 silica gel CC (MeOH-H2O, 80/20 to 100:0, v/v) to give four sub-fractions (D1a−D1d). Sub-fraction D1b (2.31 g) was subjected to silica gel CC eluted with a CHCl3: MeOH (100:1 to 1:1 v/v) gradient to obtain seven fractions (D1b2−D1b7). Purification of D1b5 (149.2 mg) by silica gel CC eluted with a CHCl3: MeOH (60:1 to 50:1 v/v) gradient to yield compound 4 (12.3 mg). Fraction D3 (2.4 g) was subjected to a Sephadex LH-20 column eluted with MeOH to obtain three sub-fractions (D3a−D3c). Purification of D3a (1.8 g) by semi-preparative HPLC, eluting with MeOH-H2O (0–25 min: 83:17 to 85:15; 25–26 min: 85:15 to 100:0; 26–27 min: 100:0; 27–28 min: 100:0 to 83:17; 28–35 min: 83:17; v/v, 3 mL/min), yielded compounds 5 (6.3 mg) and 6 (34.2 mg). Fraction E (42.5 g) was subjected to silica gel CC eluted with a petroleum ether: EtOAc (50:1 to 1:1 v/v) gradient to obtain four fractions (E1−E4). Fraction E4 (3.4 g) was separated by silica gel CC eluted with a petroleum ether: EtOAc (20:1 to 0:1 v/v) gradient to obtain five fractions (E4a−E4e). Fraction E4c (1.4 g) was subjected to RP-18 silica gel CC (MeOH-H2O, 70:30 to 100:0, v/v) to give three sub-fractions (E4c1−E4c3). Purification of E4c3 (654.1 mg) by semi-preparative HPLC, eluting with MeOH-H2O (0-20 min: 90:10; 21–40 min: 90:10 to 94:6; 40–41 min: 94:6 to 100:0; 41–42 min: 100:0; 42–43 min: 100:0 to 90:10; 43–50 min: 90:10; v/v, 3 mL/min), yielded compounds 7 (6.4 mg).
Officimalonic acid I(1): white amorphous powder; [α]
20D = −45 (
c 0.100, MeOH); IR ν
max 3544, 2956, 1779, 1732, 1456, 1376, 1142, 1062 cm
−1;
1H-NMR and
13C-NMR data, see
Table 1 and
Table 2; HR-ESI-MS:
m/z 609.3382 [M + Na]
+ (calculated for C
34H
50O
8Na, 609.3403).
Officimalonic acid J(2): white amorphous powder; [α]
20D = −32 (
c 0.05, MeOH); IR ν
max 3523, 2953, 1775, 1723, 1456, 1375, 1269, 1155, 918 cm
−1;
1H-NMR and
13C-NMR data, see
Table 1 and
Table 2; HR-ESI-MS:
m/z 571.3598 [M + H]
+ (calculated for C
34H
51O
7, 571.3635).
Officimalonic acid K(3): white amorphous powder; [α]
20D = −20 (
c 0.05, MeOH); IR ν
max 3400, 2933, 1780, 1683, 1455, 1376, 1207, 1140, 926 cm
−1;
1H-NMR and
13C-NMR data, see
Table 1 and
Table 2; HR-ESI-MS:
m/z 607.3218 [M + Na]
+ (calculated for C
34H
48O
8Na, 607.3247).
Officimalonic acid L(4): white amorphous powder; [α]
20D = −6 (
c 0.100, MeOH); IR ν
max 3392, 2948, 1733, 1594, 1456, 1374, 1207, 1033 cm
−1;
1H-NMR and
13C-NMR data, see
Table 1 and
Table 2; HR-ESI-MS:
m/z 615.3512 [M + HCOO]
- (calculated for C
35H
51O
9, 615.3533).
Officimalonic acid M(5): white amorphous powder; [α]
20D = −36 (
c 0.05, MeOH); IR ν
max 3420, 2953, 1733, 1683, 1457, 1376, 1204, 1140, 1031 cm
−1;
1H-NMR and
13C-NMR data, see
Table 1 and
Table 2; HR-ESI-MS:
m/z 621.3384 [M + Na]
+ (calculated for C
35H
50O
8Na, 621.3403).
Officimalonic acid N(6): white amorphous powder; [α]
20D = −17 (
c 0.05, MeOH); IR ν
max 3446, 2923, 1734, 1683, 1457, 1375, 1042 cm
−1;
1H-NMR and
13C-NMR data, see
Table 1 and
Table 2; HR-ESI-MS:
m/z 507.3427 [M + Na]
+ (calculated for C
35H
50O
8Na, 507.3450).
Officimalonic acid O(7): white amorphous powder; [α]
20D = +6 (
c 0.05, MeOH); IR ν
max 3260, 2930, 1745, 1683, 1447, 1370, 1087, 1030 cm
−1;
1H-NMR and
13C-NMR data, see
Table 1 and
Table 2; HR-ESI-MS:
m/z 469.3651 [M + H]
+ (calculated for C
31H
49O
3, 469.3682).
3.4. NO Production Inhibition Assay
Detection of accumulated nitrites was performed using Griess reagent (Beyotime, Shanghai, China) as described previously [
8]. Briefly, the RAW264.7 cells at approximately 1.5 × 10
4 cells/well were incubated for another 24 h, then cultured with or without LPS (1.0 μg/mL) in the presence or absence of the test compounds or positive control (10 μL) for another 24 h, the culture supernatant (50 μL) and Griess reagent (100 μL) were mixed to measure the optical density at a wavelength of 540 nm. Dexamethasone was used as positive control.
3.5. Cyclooxygenase (COX-2) Inhibition Assay
The COX-2 inhibitory activity study was carried out using ovine recombinant COX-2 enzyme by a COX colorimetric inhibitor screening assay kit from Cayman Chemical Co. (Ann Arbor, MI, USA). The assay was performed following the protocol provided by the manufacturer of assay kit. Non-selective inhibitor, naproxen, was used as a positive control. The COX inhibitory activity was expressed as % inhibition and was calculated as following Equation: