Heavy Metals as a Factor Increasing the Functional Genetic Potential of Bacterial Community for Polycyclic Aromatic Hydrocarbon Biodegradation
Abstract
1. Introduction
2. Results
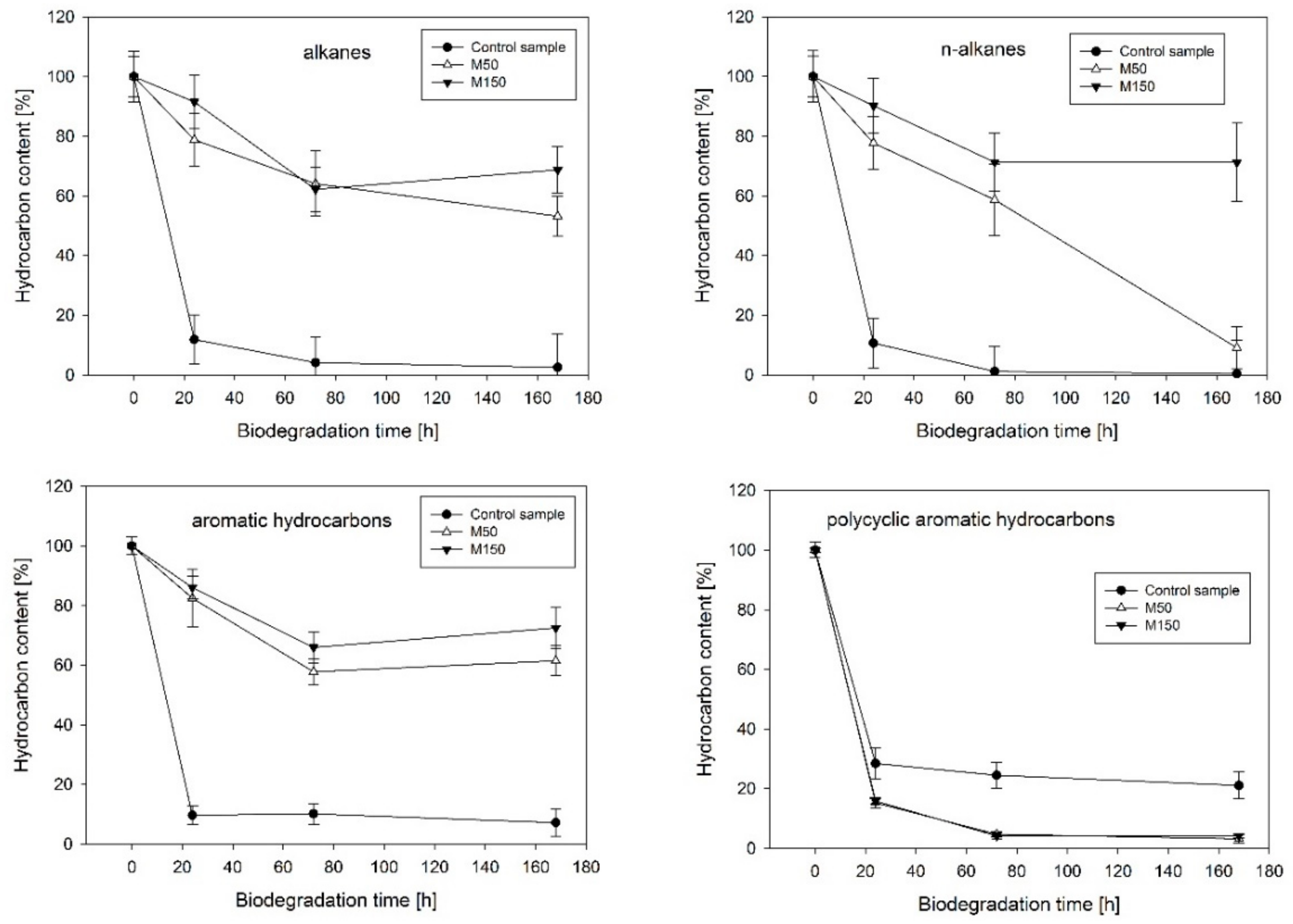
2.1. Hydrocarbon Biodegradation
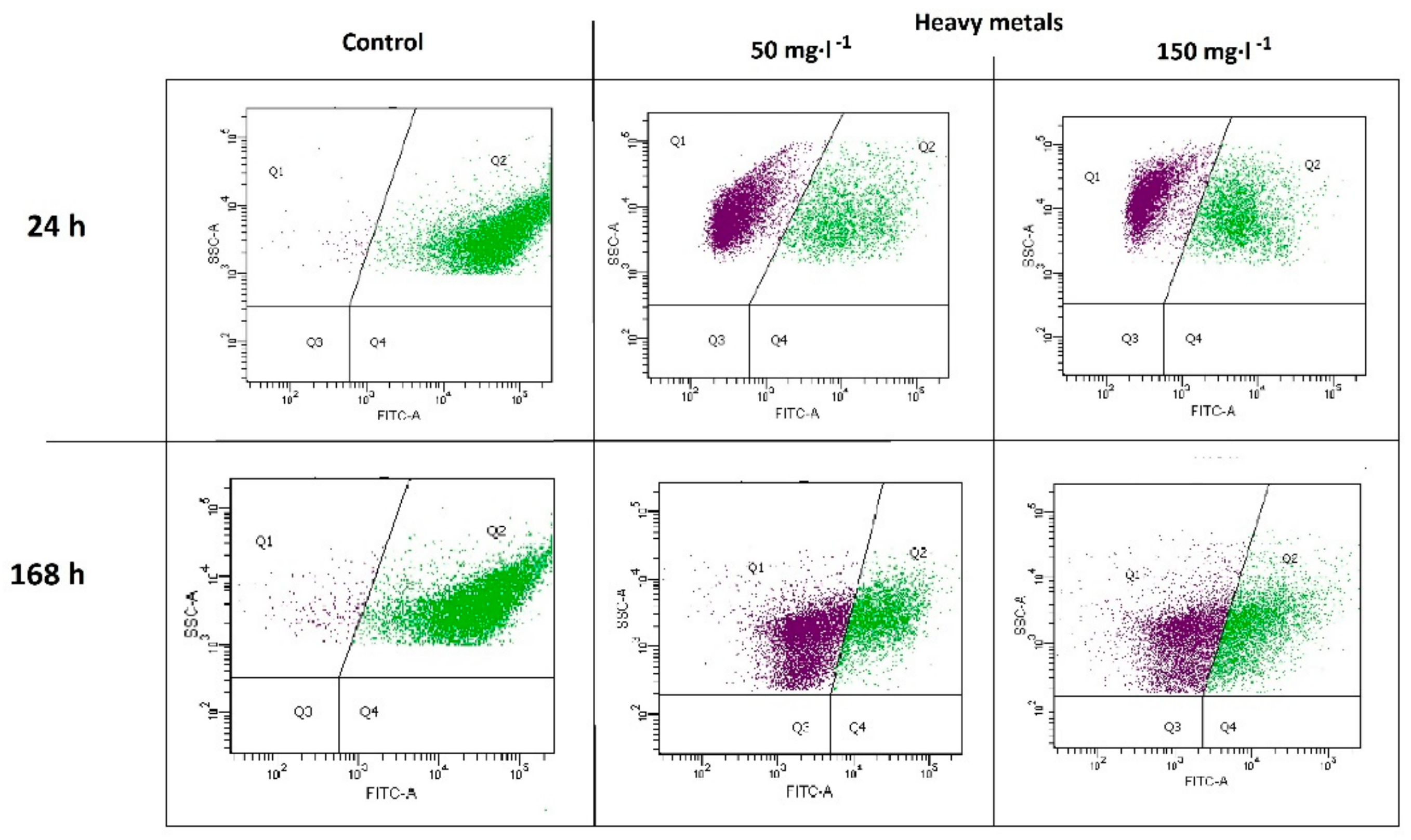
2.2. Metabolic Activity of Microorganisms
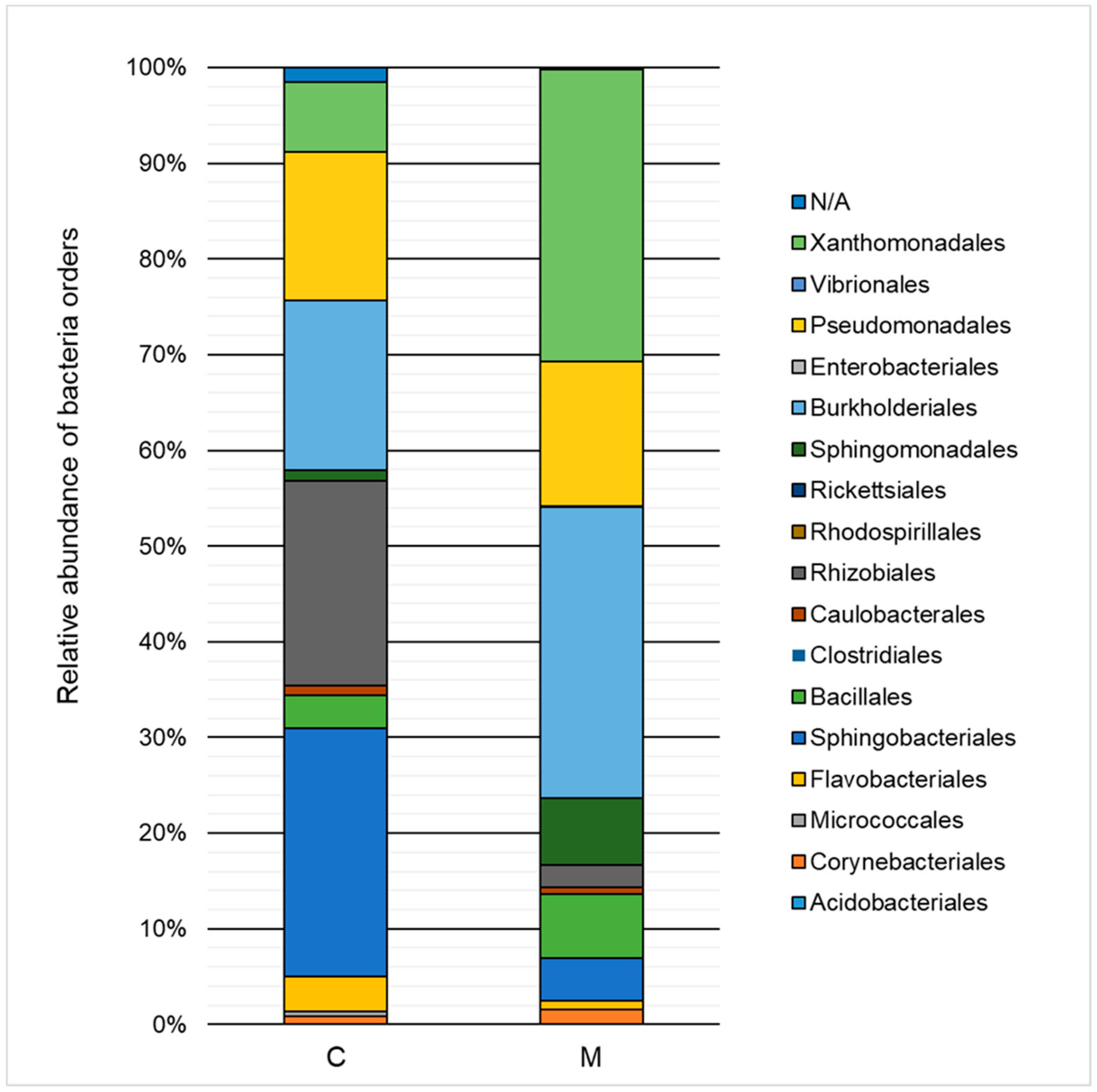
2.3. Taxonomic Structure and Biodiversity
2.4. Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) Analysis
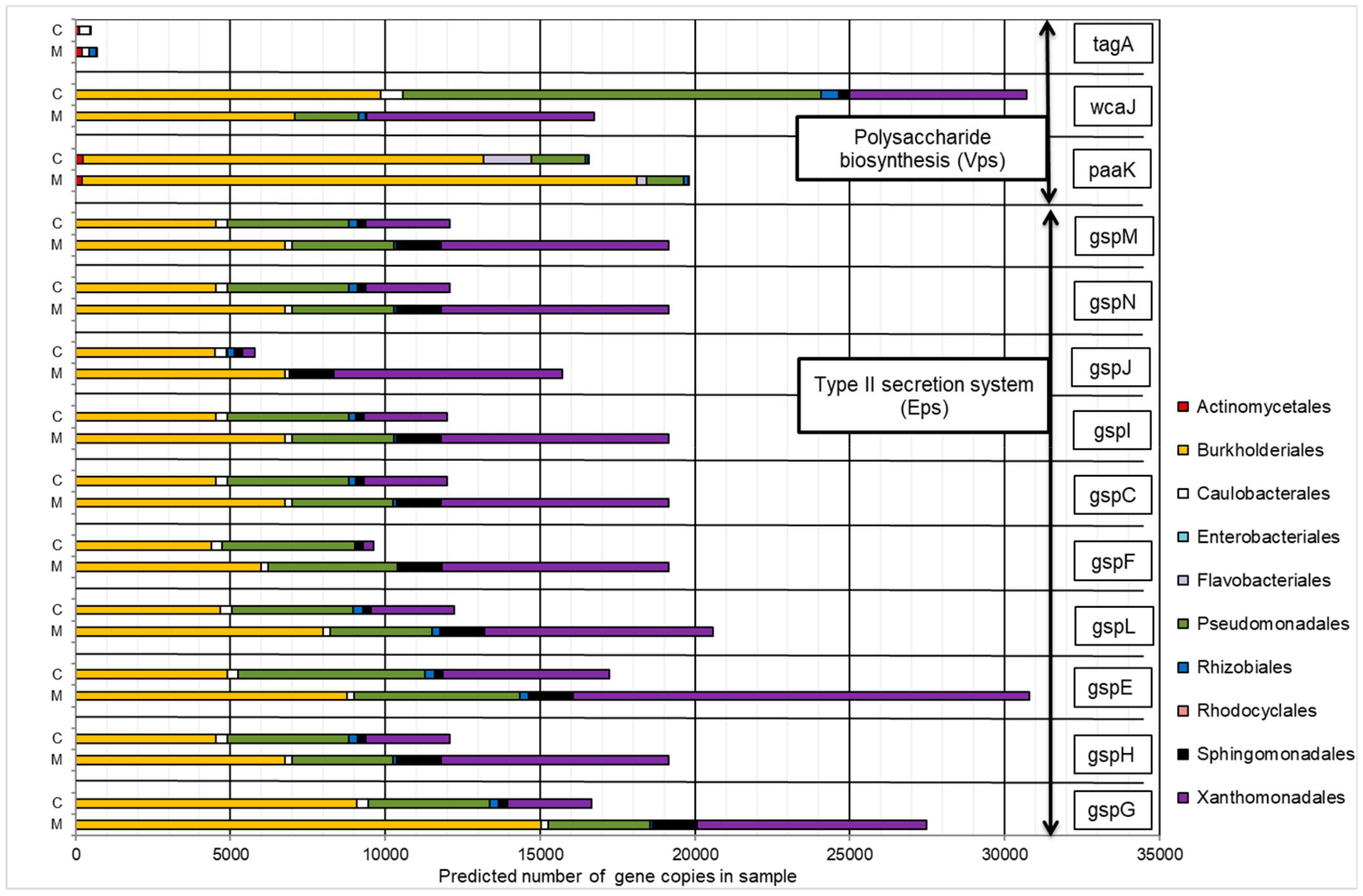
2.4.1. Gene Orthologs Participating in Hydrocarbon Biodegradation
2.4.2. Gene Orthologs Participating in Exopolysaccharides Synthesis
3. Discussion
4. Materials and Methods
4.1. Microbial Community Enrichment
4.2. Biodegradation Experiment
4.3. Analysis of Hydrocarbon Dissipation
4.4. Analysis of Metabolic Activity
4.5. Analysis of Changes in the Taxonomic Structure
4.5.1. Isolation of DNA
4.5.2. Polymerase Chain Reaction (PCR) Amplification and Sequencing
4.5.3. Bioinformatic Analysis
4.6. PICRUSt Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| EPS | exopolysaccharides |
| PICRUSt | Phylogenetic Investigation of Communities by Reconstruction of Unobserved States |
| KO | KEGG Orthologies |
| PAH | polycyclic aromatic hydrocarbons |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| PCR | Polymerase Chain Reaction |
References
- Abioye, O.B. Biological remediation of hydrocarbon and heavy metals contaminated soil. In Soil contamination; Pascucci, S., Ed.; IntechOpen: London, UK, 2011. [Google Scholar] [CrossRef]
- Das, N.; Chandran, P. Microbial degradation of petroleum hydrocarbon contaminants: An overview. Biotechnol. Res. Int. 2011, 2011, 941810. [Google Scholar] [CrossRef]
- Staninska-Pięta, J.; Piotrowska-Cyplik, A.; Juzwa, W.; Zgoła-Grześkowiak, A.; Wolko, Ł.; Sydow, Z.; Kaczorowski, Ł.; Powierska-Czarny, J.; Cyplik, P. The impact of natural and synthetic surfactants on bacterial community during hydrocarbon biodegradation. Int. Biodeterior Biodegrad. 2019, 142, 191–199. [Google Scholar] [CrossRef]
- Sihag, S.; Pathak, H.; Jaroli, D.P. Factors affecting biodegradation of polyaromatic hydrocarbons. Int. J. Pure Appl. Biosci. 2014, 2, 185–202. [Google Scholar] [CrossRef]
- Ghazali, F.M.; Rahman, R.N.Z.A.; Salleh, A.B.; Basri, M. Biodegradation of hydrocarbons in soil by microbial consortium. Int. Biodeterior. Biodegrad. 2004, 54, 61–67. [Google Scholar] [CrossRef]
- Marecik, R.; Wojtera-Kwiczor, J.; Ławniczak, Ł.; Cyplik, P.; Szulc, A.; Piotrowska-Cyplik, A.; Chrzanowski, Ł. Rhamnolipids increase the phytotoxicity of diesel oil towards four common plant species in a terrestial environment. Water Air Soil Pollut. 2012, 223, 4275–4282. [Google Scholar] [CrossRef] [PubMed]
- Piotrowska-Cyplik, A.; Cyplik, P.; Marecik, R.; Czarny, J.; Szymański, A.; Wyrwas, B.; Framski, G.; Chrzanowski, Ł.; Materna, K. Genetic and chemical analyzes of transformations in compost compounds during biodegradation of oiled bleaching earth with waste sludge. Bioresour. Technol. 2012, 114, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Ivshina, I.B.; Kuyukina, M.S.; Krivoruchko, A.V. Hydrocarbon-oxidizing bacteria and their potential. In Microbial Resources From Functional Existence in Nature to Applications, 1st ed.; Kurtböke, I., Ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 121–148. [Google Scholar] [CrossRef]
- Koshlaf, E.; Ball, A.S. Soil bioremediation approaches for petroleum hydrocarbon polluted environments. Aims Microbiol. 2017, 3, 25–49. [Google Scholar] [CrossRef] [PubMed]
- Sydow, M.; Owsianiak, M.; Framki, G.; Woźniak-Karczewska, M.; Piotrowska-Cyplik, A.; Ławniczak, Ł.; Szulc, A.; Zgoła-Grześkowiak, A.; Heipieper, H.J.; Chrzanowski, Ł. Biodiversity of soil bacteria exposed to sub-lethal concentrations of phosphonium-based ionic liquids: Effects of toxicity and biodegradation. Ecotoxicol. Env. Saf. 2018, 149, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Woźniak-Karczewska, M.; Lisiecki, P.; Białas, W.; Owsianiak, W.; Piotrowska-Cyplik, A.; Wolko, Ł.; Ławniczak, Ł.; Heipieper, H.J.; Gutierrez, T.; Chrzanowski, Ł. Effect of bioaugmentation on long-term biodegradation of diesel/biodiesel blends in soil microcosms. Sci. Total Env. 2019, 671, 948–958. [Google Scholar] [CrossRef]
- Marecik, R.; Chrzanowski, Ł.; Piotrowska-Cyplik, A.; Juzwa, W.; Biegańska-Marecik, R. Rhizosphere as a tool to introduce a soil-isolated hydrocarbon-degrading bacterial consortium into a wetland environment. Int. Biodeterior. Biodegrad. 2015, 97, 135–142. [Google Scholar] [CrossRef]
- Sydow, M.; Owsianiak, M.; Szczepaniak, Z.; Framski, G.; Smets, B.F.; Ławniczak, ł.; Lisiecki, P.; Cyplik, P.; Chrzanowski, Ł. Evaluating robustness of a diesel-degrading bacterial consortium isolated from contaminated soil. N. Biotechnol. 2016, 33, 852–859. [Google Scholar] [CrossRef] [PubMed]
- Abioye, O.P.; Aina, P.F.; Ijah, J.U.J.; Aransiola, A.S. Effects of cadmium and lead on the biodegradation of diesel-conaminated soil. JTUSCI 2019, 13, 628–638. [Google Scholar] [CrossRef]
- Kuperman, R.G.; Carreiro, M.M. Soil heavy metal concentrations, microbial biomass and enzyme activities in a contaminated grassland ecosystem. Soil Biol. Biochem. 1997, 29, 179–190. [Google Scholar] [CrossRef]
- Piotrowska-Cyplik, A.; Czarnecki, Z. Phytoextraction of heavy metals by hemp during anaerobic sewage sludge management in the non-industrial sites. Pol. J. Env. Stud. 2003, 12, 779–784. [Google Scholar]
- Ma, X.K.; Li, T.T.; Peterson, C.E.; Zhao, W.W.; Guo, W.; Zhou, B. The influence of heavy metals on the bioremediation of polycyclic aromatic hydrocarbons in aquatic system by a bacterial-fungal consortium. Environ. Technol. 2018, 39, 2128–2137. [Google Scholar] [CrossRef] [PubMed]
- Olaniran, A.O.; Balgobind, A.; Pillay, B. Bioavailability of heavy metals in soil: Impact on microbial biodegradation of organic compounds and possible improvement stratefies. Int. J. Mol. Sci. 2013, 14, 10197–10228. [Google Scholar] [CrossRef] [PubMed]
- Giwa, O.E.; Ibitoye, F.O. Bioremediation of heavy metal in crude oil contaminated soil using isolated Indigenous microorganism cultured with E coli DE3 BL21. IJEAS 2017, 4, 2394–3661. [Google Scholar]
- Kazy, S.K.; Sar, P.; Singh, S.P.; Sen, A.K.; D’Souza, S.F. Extracellular polysaccharides of a copper-sensitive and a copper-resistant Pseudomonas aeruginosa strain: Synthesis, chemical nature and copper binding. World J. Microbiol. Biotechnol. 2002, 18, 583–588. [Google Scholar] [CrossRef]
- González, A.G.; Shirokova, L.S.; Pokrovsky, O.S.; Emnova, E.E.; Martínez, R.E.; Santana-Casiano, J.M.; González-Dávila, M.; Pokrovski, G.S. Adsorption of copper on Pseudomonas aureofaciens: Protective role of surface exopolysaccharides. J. Colloid Interface Sci. 2010, 350, 305–314. [Google Scholar] [CrossRef]
- Mohite, B.V.; Koli, S.; Patil, S.V. Heavy metal stress and its consequences on exopolysaccharide (EPS)-producing Pantoea agglomerans. Appl. Biochem. Biotechnol. 2018, 186, 199–216. [Google Scholar] [CrossRef]
- Martins, P.S.O.; Almeida, N.F.; Leite, S.G.F. Application of a bacterial extracellular polymeric substance in heavy metal adsorption in a co-contaminated aqueous system. Braz. J. Microbiol. 2008, 39, 780–786. [Google Scholar] [CrossRef]
- Mangwani, N.; Kumari, S.; Das, S. Marine bacterial biofilms in bioremediation of polycyclic aromatic hydrocarbons (PAHs) under terrestrial condition in a soil microcosm. Pedosphere 2017, 27, 548–558. [Google Scholar] [CrossRef]
- Xie, Y.; Fan, J.; Zhu, W.; Amombo, E.; Lou, Y.; Chen, L.; Fu, J. Effect of heavy metals pollution on soil microbial diversity and bermudgrass genetic variation. Front. Plant Sci. 2016, 7, 755. [Google Scholar] [CrossRef] [PubMed]
- Šmejkalová, M.; Mikanová, O.; Borůvka, L. Effect of heavy metal concentrations on biological activity of soil micro-organisms. Plant Soil Env. 2003, 49, 321–326. [Google Scholar] [CrossRef]
- Chodak, M.; Gołębiewski, M.; Norawska-Płoskonka, J.; Kurdyk, K.; Niklińska, M. Diversity of microorganisms from forest soils differently polluted with heavy metals. Appl. Soil Ecol. 2013, 64, 7–14. [Google Scholar] [CrossRef]
- Ivanieva, O.D. Heavy metal resistance mechanisms in bacteria. Mikrobiol. Z. 2009, 71, 54–65. [Google Scholar]
- Lenart-Boroń, A.; Boroń, P. The effect of industrial heavy metal pollution on microbial abundance and diversity in soils-a review. In Environmental Risk Assesment of Soil Contamination; Hernandez-Soriano, M.C., Ed.; InTech Open: London, UK, 2014. [Google Scholar] [CrossRef]
- Yao, H.; Xu, J.; Huang, C. Substrate utilization pattern, biomass and activity of microbial communities in a sequence of heavy metal-polluted paddy soils. Goderma 2003, 115, 139–148. [Google Scholar] [CrossRef]
- Kou, W.; Zhang, J.; Lu, X.; Ma, Y.; Mou, X.; Wu, L. Identification of bacterial communities in sediments of Poyang Lake, the largest freshwater lake in China. Singerplus 2016, 5, 401. [Google Scholar] [CrossRef]
- Bruins, M.R.; Kapil, S.; Oehme, F.O. Microbial resistance to metals in the environment. Ecotoxicol. Environ. Saf. 2000, 45, 198–207. [Google Scholar] [CrossRef]
- Tewari, S.; Ramteke, P.W.; Tripathi, M.; Kumar, S.; Kumar-Garg, S. Plasmid mediated transfer of antibiotic resistance and heavy metal tolerance in thermotolerant water borne coliforms. Afr. J. Microbiol. Res. 2013, 7, 130–136. [Google Scholar] [CrossRef]
- Abbasian, F.; Lockington, R.; Mallavarpu, M.; Naidu, R. A comprehensive review of aliphatic hydrocarbon biodegradation by bacteria. Appl. Biochem. Biotechnol. 2015, 176, 670–699. [Google Scholar] [CrossRef] [PubMed]
- Czarny, J.; Staninska-Pięta, J.; Piotrowska-Cyplik, A.; Juzwa, W.; Wolniewicz, A.; Marecik, R.; Ławniczak, Ł.; Chrzanowski, Ł. Acinetobacter sp. as the key player in diesel oil degrading community exposed to PAHs and heavy metals. J. Hazar. Mater. 2020, 383, 121168. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; Diwan, B. Bacterial exopolysaccharide mediated heavy metal removal: A review on biosynthesis, mechanism and remediation strategies. Biotechnol. Rep. 2017, 13, 58–71. [Google Scholar] [CrossRef] [PubMed]
- Marchal, M.; Briandet, R.; Halter, S.D.; Koechler, S.; DuBow, M.S.; Lett, M.-C.; Bertin, P.N. Subinhibitory arsenite concentrations lead to population dispersal in Thiomonas sp. PLoS ONE 2011, 6, e23181. [Google Scholar] [CrossRef]
- Prabhakaran, P.; Ashraf, M.A.; Aqma, W.S. Microbial stress response to heavy metals in the environment. RSC Adv. 2016, 6, 109862–109877. [Google Scholar] [CrossRef]
- Wang, S.; Teng, S.; Fan, M. Interaction between heavy metals and aerobic granular sludge. In Environmental Management; Kumar, S.S., Ed.; Sciyo: Rijeka, Croatia, 2010; pp. 173–188. [Google Scholar] [CrossRef]
- Mangwani, N.; Shukla, S.K.; Kumari, S.; Das, S.; Rao, T.S. Effect of biofilm parameters and extracellular polymeric substance composition on polycyclic aromatic hydrocarbon degradation. RSC Adv. 2016, 6, 57540–57551. [Google Scholar] [CrossRef]
- Szczepaniak, Z.; Cyplik, P.; Juzwa, W.; Czarny, J.; Staninska, J.; Piotrowska-Cyplik, A. Antibacterial effect of the Trichoderma viride fungi on soil microbiome during PAH’s biodegradation. Int. Biodeterior Biodegrad. 2015, 104, 170–177. [Google Scholar] [CrossRef]
- Cyplik, P.; Juzwa, W.; Marecik, R.; Powierska-Czarny, J.; Piotrowska-Cyplik, A.; Czarny, J.; Drożdżyńska, A.; Chrzanowski, Ł. Denitrification of industrial wastewater: Influence of glycerol addition on metabolic activity and community shifts in a microbial consortium. Chemosphere 2013, 93, 2823–2831. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Brg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betly, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. Isme J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef]
- DeSantis, Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [PubMed]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |


| Number of OTU | Chao1 | Phylogenetic Diversity | |
|---|---|---|---|
| Control sample | 95 | 107 | 4.41 |
| M150 | 85 | 96 | 4.16 |
| KO Number | Gene Name | Definition | EC Number |
|---|---|---|---|
| PAH biodegradation | |||
| K00449 | pcaH | protocatechuate 3,4-dioxygenase, beta subunit | EC: 1.13.11.3 |
| K00448 | pcaG | protocatechuate 3,4-dioxygenase, alpha subunit | EC: 1.13.11.3 |
| K00480 | E1.14.13.1 | salicylate hydroxylase | EC: 1.14.13.1 |
| K14578 | nahAb | naphthalene 1,2-dioxygenase ferredoxin component | EC: 1.14.12.12 |
| K14579 | nahAc | naphthalene 1,2-dioxygenase subunit alpha | EC: 1.14.12.12 |
| K04102 | pht5 | 4,5-dihydroxyphthalate decarboxylase | EC: 4.1.1.55 |
| K04100 | ligA | protocatechuate 4,5-dioxygenase, alpha chain | EC: 1.13.11.8 |
| EPS synthesis | |||
| K02456 | gspG | general secretion pathway protein G | EC: 7.4.2.8 |
| K02457 | gspH | general secretion pathway protein H | EC: 7.4.2.8 |
| K02454 | gspE | general secretion pathway protein E | EC: 7.4.2.8 |
| K02455 | gspF | general secretion pathway protein F | EC: 7.4.2.8 |
| K02452 | gspC | general secretion pathway protein C | EC: 7.4.2.8 |
| K02453 | gspD | general secretion pathway protein D | EC: 7.4.2.8 |
| K02458 | gspI | general secretion pathway protein I | EC: 7.4.2.8 |
| K02459 | gspJ | general secretion pathway protein J | EC: 7.4.2.8 |
| K02463 | gspN | general secretion pathway protein N | EC: 7.4.2.8 |
| K02462 | gspM | general secretion pathway protein M | EC: 7.4.2.8 |
| K02461 | gspL | general secretion pathway protein L | EC: 7.4.2.8 |
| K02460 | gspK | general secretion pathway protein K | EC: 7.4.2.8 |
| K01912 | paaK | phenylacetate-CoA ligase | EC: 6.2.1.30 |
| K03646 | wcaJ | putative colanic acid biosynthesis UDP-glucose lipid carrier transferase | EC: 2.7.8.31 |
| K05946 | tagA | N-acetylglucosaminyldiphosphoundecaprenol N-acetyl-beta-D-mannosaminyltransferase | EC: 2.4.1.187 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Staninska-Pięta, J.; Czarny, J.; Piotrowska-Cyplik, A.; Juzwa, W.; Wolko, Ł.; Nowak, J.; Cyplik, P. Heavy Metals as a Factor Increasing the Functional Genetic Potential of Bacterial Community for Polycyclic Aromatic Hydrocarbon Biodegradation. Molecules 2020, 25, 319. https://doi.org/10.3390/molecules25020319
Staninska-Pięta J, Czarny J, Piotrowska-Cyplik A, Juzwa W, Wolko Ł, Nowak J, Cyplik P. Heavy Metals as a Factor Increasing the Functional Genetic Potential of Bacterial Community for Polycyclic Aromatic Hydrocarbon Biodegradation. Molecules. 2020; 25(2):319. https://doi.org/10.3390/molecules25020319
Chicago/Turabian StyleStaninska-Pięta, Justyna, Jakub Czarny, Agnieszka Piotrowska-Cyplik, Wojciech Juzwa, Łukasz Wolko, Jacek Nowak, and Paweł Cyplik. 2020. "Heavy Metals as a Factor Increasing the Functional Genetic Potential of Bacterial Community for Polycyclic Aromatic Hydrocarbon Biodegradation" Molecules 25, no. 2: 319. https://doi.org/10.3390/molecules25020319
APA StyleStaninska-Pięta, J., Czarny, J., Piotrowska-Cyplik, A., Juzwa, W., Wolko, Ł., Nowak, J., & Cyplik, P. (2020). Heavy Metals as a Factor Increasing the Functional Genetic Potential of Bacterial Community for Polycyclic Aromatic Hydrocarbon Biodegradation. Molecules, 25(2), 319. https://doi.org/10.3390/molecules25020319








