Preparation of Chitosan Molecularly Imprinted Polymers and the Recognition Mechanism for Adsorption of Alpha-Lipoic Acid
Abstract
:1. Introduction
2. Results and Discussion
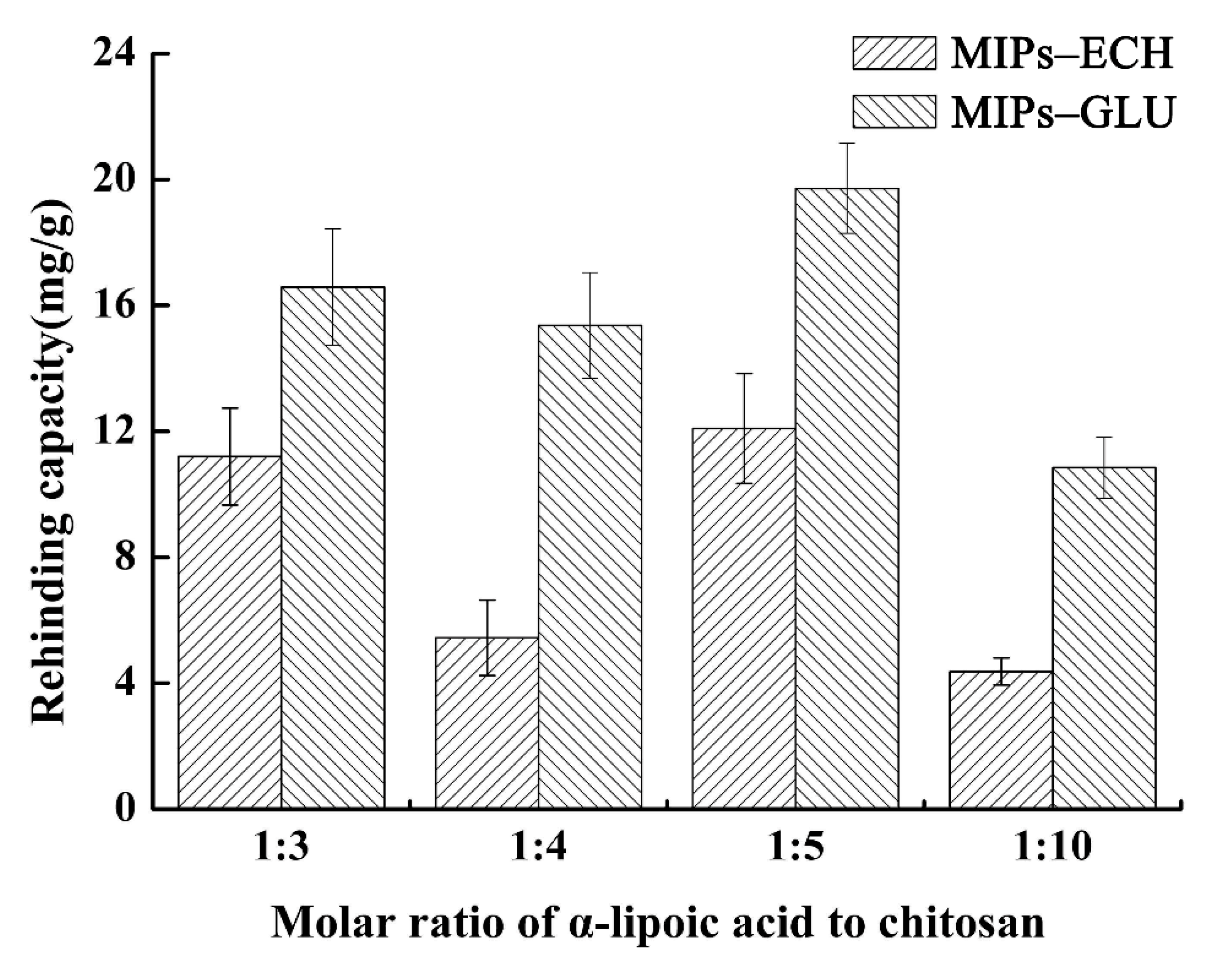
2.1. The Molar Ratio of ALA to Chitosan
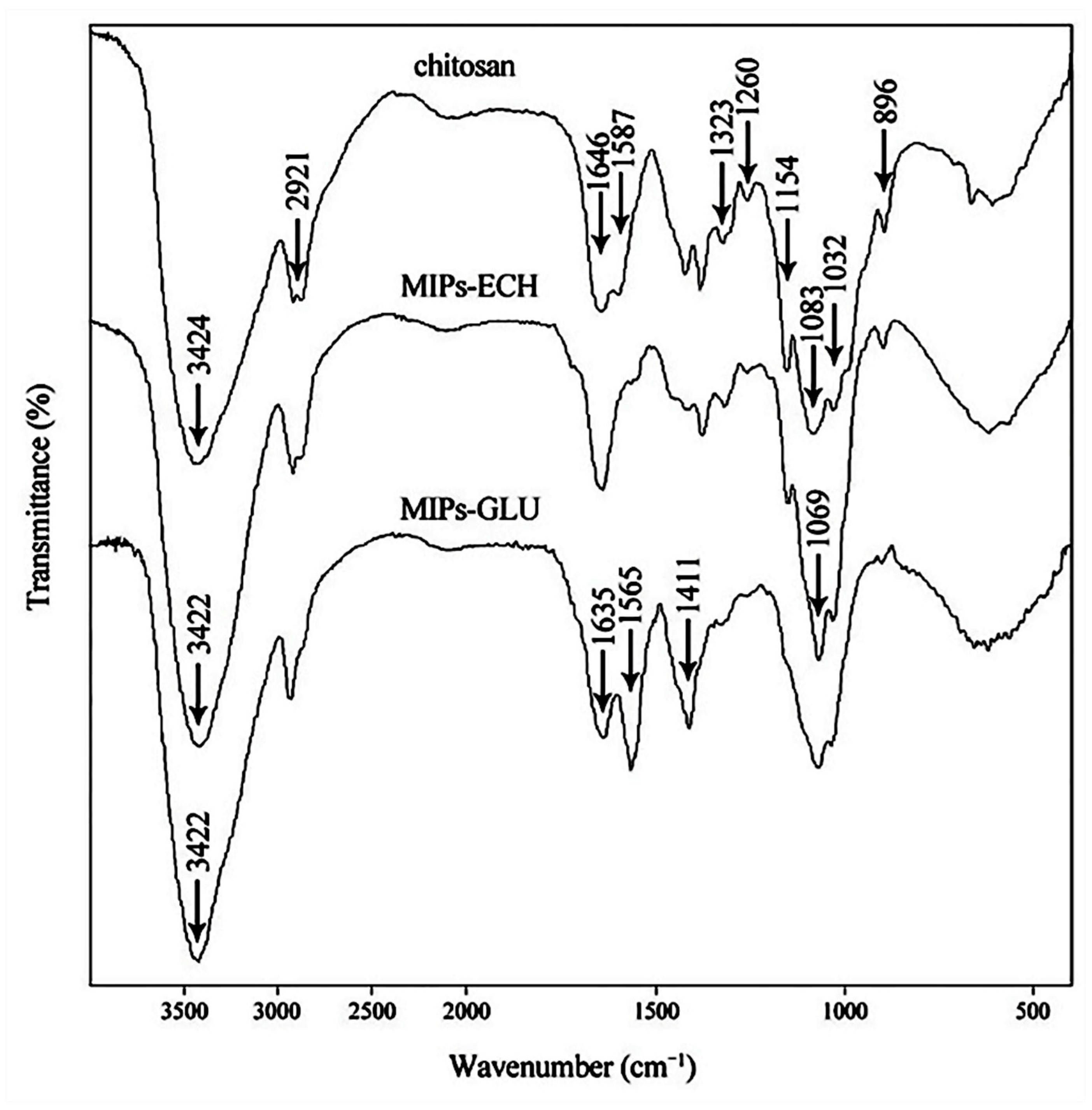
2.2. Characterization of MIPs
2.3. Binding Properties of the MIPs and NIPs
2.3.1. Kinetic Adsorption
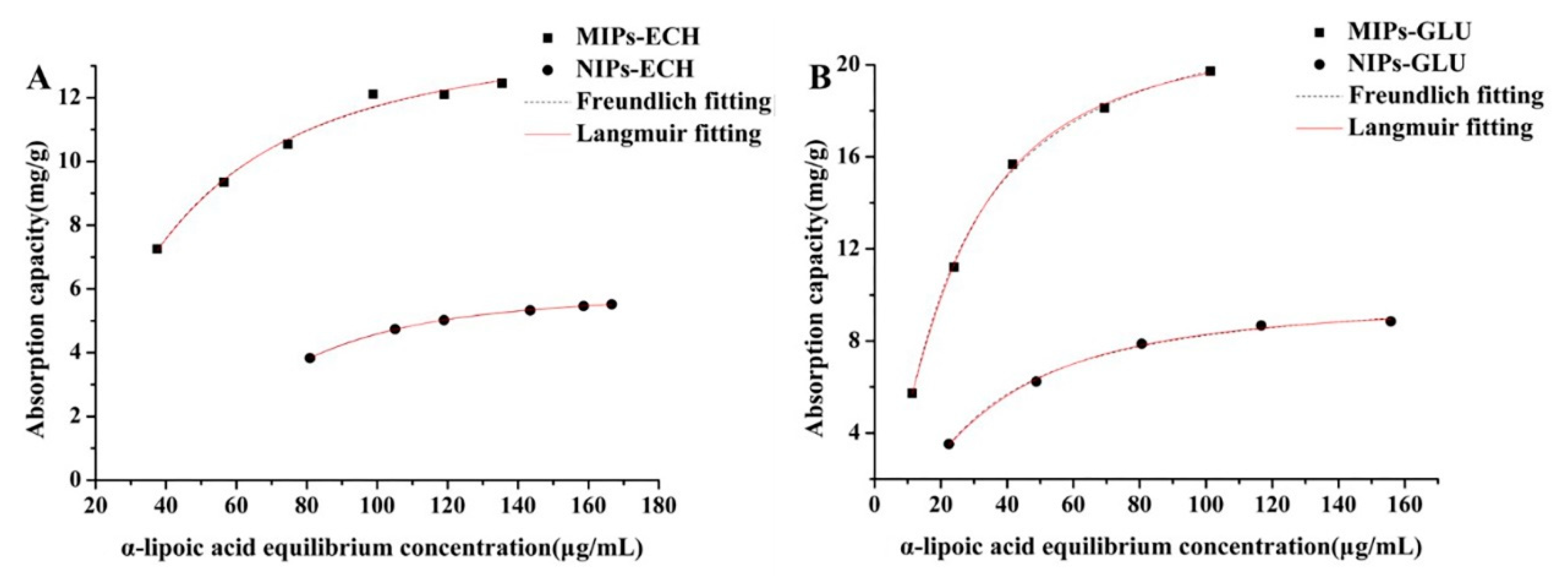
2.3.2. Static Adsorption
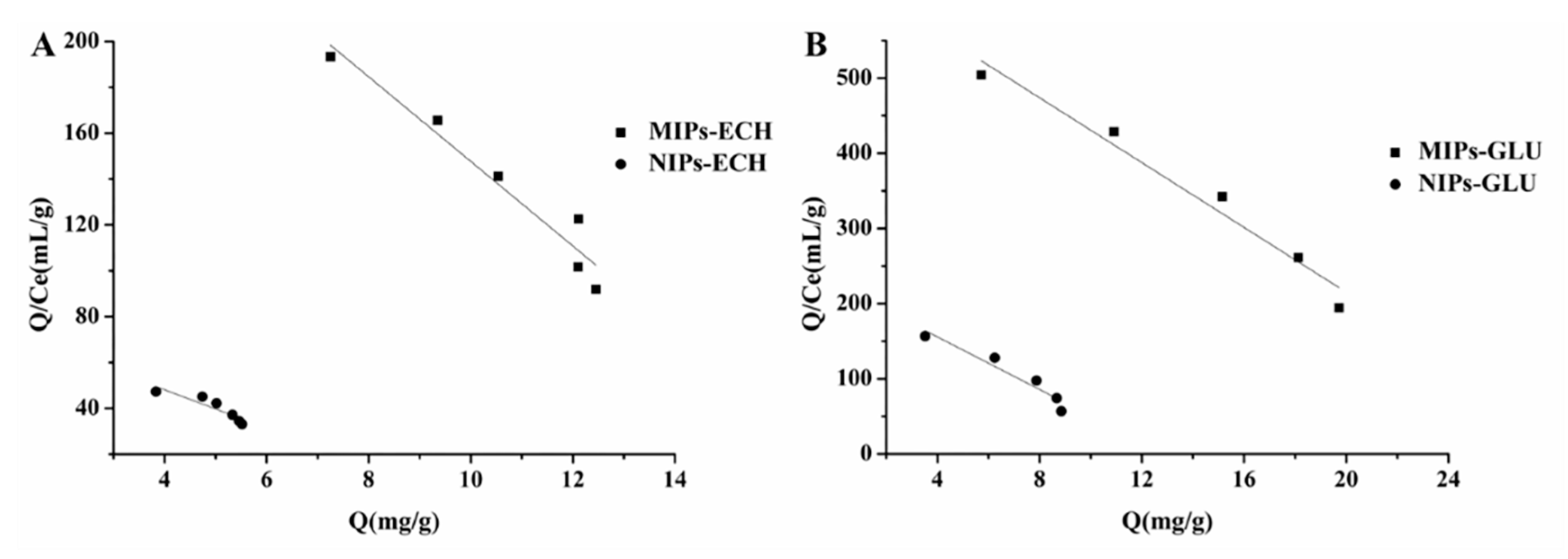
2.4. Adsorption Isotherms
2.5. Scatchard Analysis
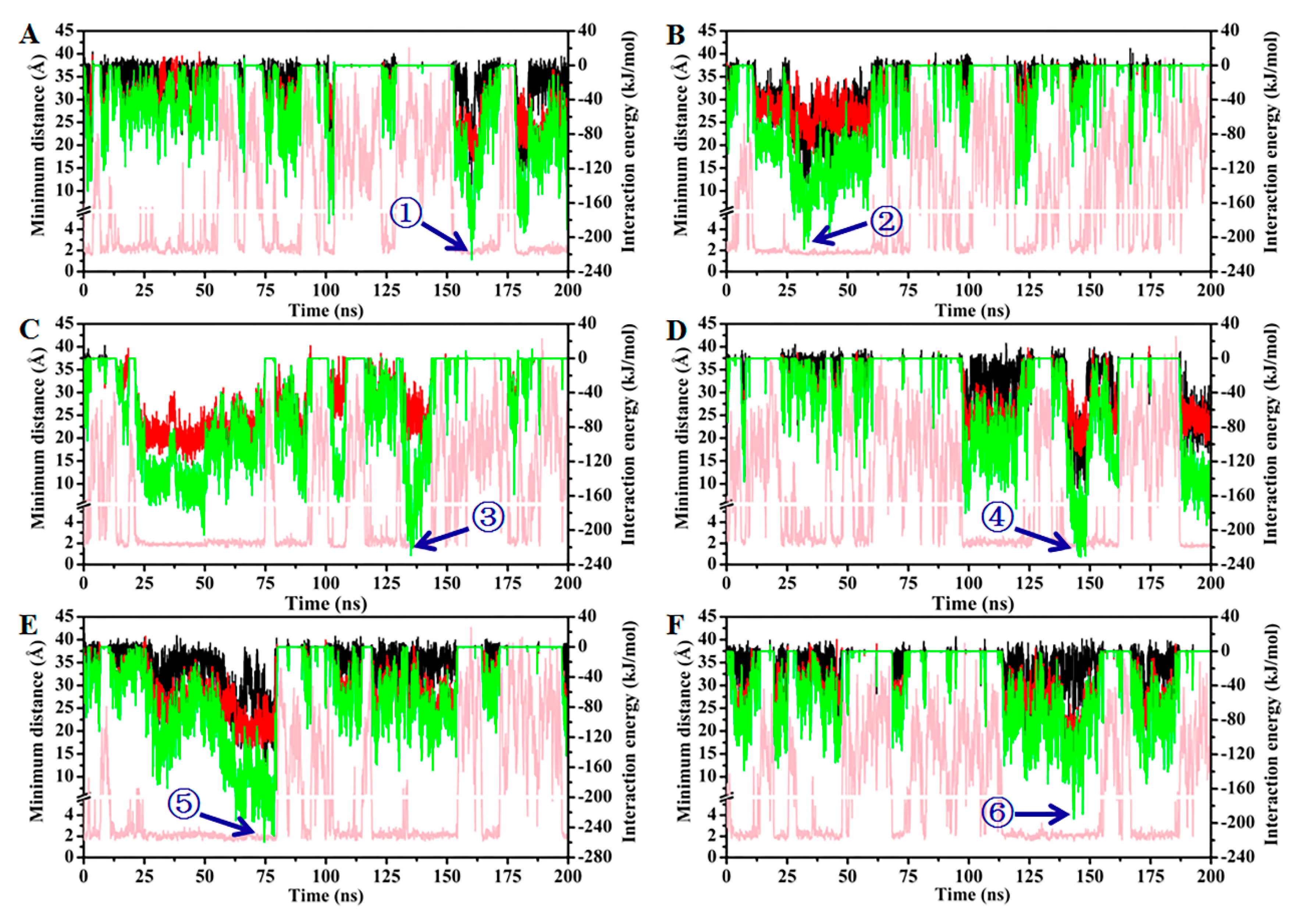
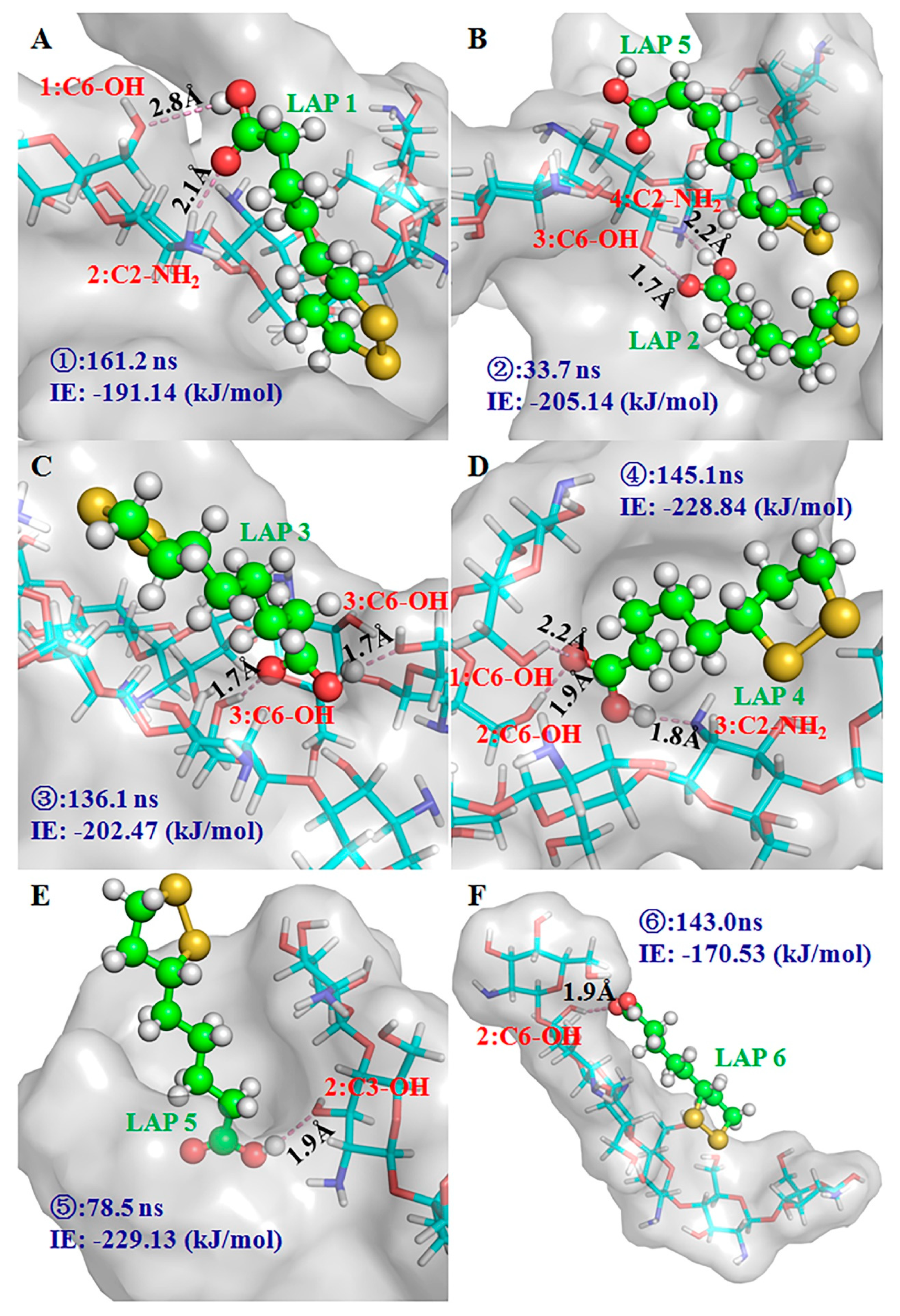
2.6. MD Simulation
3. Materials and Methods
3.1. Chemicals
3.2. Preparation of Chitosan MIPs
3.2.1. ECH as Cross-Linker
3.2.2. GLU as Cross-Linker
3.3. Optimization of Molar Ratio of ALA to Chitosan for MIPs Preparation
3.4. Characterization of MIPs and NIPs
3.5. Kinetic Adsorption
3.6. Computational Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ALA | alpha-lipoic acid |
| ECH | epichlorohydrin |
| FTIR | Fourier transform infrared spectroscopy |
| GLU | glutaraldehyde |
| MD | molecular dynamics |
| MIT | molecular imprinting technology |
| MIPs | molecularly imprinted polymers |
| MIPs–ECH | molecularly imprinted polymers crosslinked by epichlorohydrin |
| MIPs–GLU | molecularly imprinted polymers crosslinked by glutaraldehyde |
| NIPs–ECH | non-imprinted polymers crosslinked by epichlorohydrin |
| NIPs–GLU | non-imprinted polymers crosslinked by glutaraldehyde |
| RESP | restrained electrostatic potential |
| RMSD | root mean square deviation |
References
- Wollin, S.D.; Jones, P.J.H. α-lipoic acid and cardiovascular disease. J. Nutr. 2003, 133, 3327–3330. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.-J.; Wei, H.; Hagen, T.; Frei, B. α-Lipoic acid attenuates LPS-induced inflammatory responses by activating the phosphoinositide 3-kinase/Akt signaling pathway. Proc. Natl. Acad. Sci. USA 2007, 104, 4077–4082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holmquist, L.; Stuchbury, G.; Berbaum, K.; Muscat, S.; Young, S.; Hager, K.; Engel, J.; Münch, G. Lipoic acid as a novel treatment for Alzheimer’s disease and related dementias. Pharmacol. Ther. 2007, 113, 154–164. [Google Scholar] [CrossRef] [PubMed]
- Farhat, D.; Lincet, H. Lipoic acid a multi-level molecular inhibitor of tumorigenesis. BBA-Rev. Cancer 2020, 1873, 188317. [Google Scholar] [CrossRef] [PubMed]
- TeresaáBes, M.; Peter, W. Application of enzymic Baeyer-Villiger oxidations of 2-substituted cycloalkanones to the total synthesis of (R)-(+)-lipoic acid. J. Chem. Soc. Chem. Commun. 1995, 1563–1564. [Google Scholar]
- Adger, B.; Bes, M.T.; Grogan, G.; McCague, R.; Pedragosa-Moreau, S.; Roberts, S.M.; Villa, R.; Wan, P.W.; Willets, A.J. The synthesis of (R)-(+)-lipoic acid using a monooxygenase-catalysed biotransformation as the key step. Biorg. Med. Chem. 1997, 5, 253–261. [Google Scholar] [CrossRef]
- Fadnavis, N.; Babu, R.L.; Vadivel, S.K.; Deshpande, A.A.; Bhalerao, U. Lipase catalyzed regio- and stereospecific hydrolysis: Chemoenzymatic synthesis of both (R)-and (S)-enantiomers of α-lipoic acid. Tetrahedron Asymmetry 1998, 9, 4109–4112. [Google Scholar] [CrossRef]
- Zhang, S.; Chen, X.; Zhang, J.; Wang, W.; Duan, W. An enantioselective formal synthesis of (+)-(R)-α-lipoic acid by an L-proline-catalyzed aldol reaction. Synthesis 2008, 3, 383–386. [Google Scholar]
- Panchgalle, S.P.; Jogdand, G.F.; Chavan, S.P.; Kalkote, U.R. Enantioselective synthesis of (R)-(+)-α-lipoic acid via proline-catalyzed sequential alpha-aminoxylation and HWE olefination of aldehyde. Tetrahedron Lett. 2010, 51, 3587–3589. [Google Scholar] [CrossRef]
- Chavan, S.P.; Pawar, K.P.; Praveen, C.; Patil, N.B. Chirality induction and chiron approaches to enantioselective total synthesis of alpha-lipoic acid. Tetrahedron 2015, 71, 4213–4218. [Google Scholar] [CrossRef]
- Olbrich, M.; Gewald, R. Process for the Enantioselective Reduction of 8-Chloro-6-Oxo-Octanoic Acid Alkyl Esters. U.S. Patent 7,135,328 B2, 14 November 2006. [Google Scholar]
- Müller, M.; Sauer, W.; Laban, G. Method for the Production of (R)-and (S)-8-Chloro-6-Hydroxyoctanic Acid Alkyl Esters by Enzymatic Reduction. U.S. Patent 7,157,253 B2, 2 January 2007. [Google Scholar]
- Gopalan, A.S.; Jacobs, H.K. Bakers’ yeast reduction of alkyl 6-chloro-3-oxohexanoates: Synthesis of (R)-(+)-α-lipoic acid. J. Chem. Soc. Perkin Trans. 1 1990, 7, 1897–1900. [Google Scholar] [CrossRef]
- Zhou, W.-J.; Ni, Y.; Zheng, G.-W.; Chen, H.-H.; Zhu, Z.-R.; Xu, J.-H. Enzymatic resolution of a chiral chlorohydrin precursor for (R)-α-lipoic acid synthesis via lipase catalyzed enantioselective transacylation with vinyl acetate. J. Mol. Catal. B Enzym. 2014, 99, 102–107. [Google Scholar] [CrossRef]
- Zhang, Y.-J.; Zhang, W.-X.; Zheng, G.-W.; Xu, J.-H. Identification of an ε-keto ester reductase for the efficient synthesis of an (R)-α-lipoic acid precursor. Adv. Synth. Catal. 2015, 357, 1697–1702. [Google Scholar] [CrossRef]
- Chavan, S.P.; Praveen, C.; Ramakrishna, G.; Kalkote, U. Enantioselective synthesis of R-(+)-α and S-(−)-α-lipoic acid. Tetrahedron Lett. 2004, 45, 6027–6028. [Google Scholar] [CrossRef]
- Chen, L.X.; Wang, X.Y.; Lu, W.H.; Wu, X.Q.; Li, J.H. Molecular imprinting: Perspectives and applications. Chem. Soc. Rev. 2016, 45, 2137–2211. [Google Scholar] [CrossRef]
- Ji, W.; Ma, X.; Zhang, J.; Xie, H.; Liu, F.; Wang, X. Preparation of the high purity gingerols from ginger by dummy molecularly imprinted polymers. J. Chromatogr. A 2015, 1387, 24–31. [Google Scholar] [CrossRef]
- Ji, W.; Chen, L.; Ma, X.; Wang, X.; Gao, Q.; Geng, Y.; Huang, L. Molecularly imprinted polymers with novel functional monomer for selective solid-phase extraction of gastrodin from the aqueous extract of Gastrodia elata. J. Chromatogr. A 2014, 1342, 1–7. [Google Scholar] [CrossRef]
- Theodoridis, G.; Lasakova, M.; Skerikova, V.; Tegou, A.; Giantsiou, N.; Jandera, P. Molecular imprinting of natural flavonoid antioxidants: Application in solid-phase extraction for the sample pretreatment of natural products prior to HPLC analysis. J. Sep. Sci. 2006, 29, 2310–2321. [Google Scholar] [CrossRef]
- Zeng, H.; Wang, Y.Z.; Liu, X.J.; Kong, J.H.; Nie, C. Preparation of molecular imprinted polymers using bi-functional monomer and bi-crosslinker for solid-phase extraction of rutin. Talanta 2012, 93, 172–181. [Google Scholar] [CrossRef]
- Chen, L.X.; Xu, S.F.; Li, J.H. Recent advances in molecular imprinting technology: Current status, challenges and highlighted applications. Chem. Soc. Rev. 2011, 40, 2922–2942. [Google Scholar] [CrossRef]
- Xu, L.; Huang, Y.-A.; Zhu, Q.-J.; Ye, C. Chitosan in molecularly-imprinted polymers: Current and future prospects. Int. J. Mol. Sci. 2015, 16, 18328–18347. [Google Scholar] [CrossRef]
- Xu, L.; Huang, Y.-A.; Zhu, Q.-J.; Ye, C. Preparation and application of molecularly imprinted polymers based on chitosan. Chem. Ind. Eng. Prog. 2016, 35, 847–855. [Google Scholar]
- Yu, Q.; Deng, S.; Yu, G. Selective removal of perfluorooctane sulfonate from aqueous solution using chitosan-based molecularly imprinted polymer adsorbents. Water Res. 2008, 42, 3089–3097. [Google Scholar] [CrossRef]
- Liu, B.; Wang, D.; Li, H.; Xu, Y.; Zhang, L. As(III) removal from aqueous solution using α-Fe2O3 impregnated chitosan beads with As(III) as imprinted ions. Desalination 2011, 272, 286–292. [Google Scholar] [CrossRef]
- Gupta, K.C.; Jabrail, F.H. Glutaraldehyde and glyoxal cross-linked chitosan microspheres for controlled delivery of centchroman. Carbohydr. Res. 2006, 341, 744–756. [Google Scholar] [CrossRef]
- Monier, M.; Ayad, D.; Wei, Y.; Sarhan, A. Preparation of cross-linked chitosan/glyoxal molecularly imprinted resin for efficient chiral resolution of aspartic acid isomers. Biochem. Eng. J. 2010, 51, 140–146. [Google Scholar] [CrossRef]
- Wu, H.; Zhao, Y.-Y.; Yu, Y.-X.; Jiang, Z.-Y. Molecularly imprinted chitosan membrane for chiral resolution of phenylalanine isomers. J. Funct. Polym. 2007, 19, 262–266. [Google Scholar]
- Ogunlaja, A.S.; Coombes, M.J.; Torto, N.; Tshentu, Z.R. The adsorptive extraction of oxidized sulfur-containing compounds from fuels by using molecularly imprinted chitosan materials. React. Funct. Polym. 2014, 81, 61–76. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, E.; Wu, Z.; Li, H.; Zhu, Z.; Zhu, X.; Dong, Y. Synthesis of chitosan molecularly imprinted polymers for solid-phase extraction of methandrostenolone. Carbohydr. Polym. 2014, 101, 517–523. [Google Scholar] [CrossRef]
- Ma, X.; Chen, R.; Zheng, X.; Youn, H.; Chen, Z. Preparation of molecularly imprinted CS membrane for recognizing naringin in aqueous media. Polym. Bull. 2011, 66, 853–863. [Google Scholar] [CrossRef]
- Huang, Y.A.; Xu, L.; Yang, B.; Zhu, Q. Self-assembly system of α-lipoic acid as an antioxidant and preparation of molecularly imprinted polymers for its selective adsorption. Food Science 2018, 39, 8–14. [Google Scholar]
- Ho, Y.-S.; McKay, G. Pseudo-second order model for sorption processes. Process Biochem. 1999, 34, 451–465. [Google Scholar] [CrossRef]
- Chiou, M.; Li, H. Adsorption behavior of reactive dye in aqueous solution on chemical cross-linked chitosan beads. Chemosphere 2003, 50, 1095–1105. [Google Scholar] [CrossRef]
- Wong, Y.; Szeto, Y.; Cheung, W.; McKay, G. Equilibrium studies for acid dye adsorption onto chitosan. Langmuir 2003, 19, 7888–7894. [Google Scholar] [CrossRef]
- Shaikh, H.; Memon, N.; Khan, H.; Bhanger, M.; Nizamani, S. Preparation and characterization of molecularly imprinted polymer for di (2-ethylhexyl) phthalate: Application to sample clean-up prior to gas chromatographic determination. J. Chromatogr. A 2012, 1247, 125–133. [Google Scholar] [CrossRef]
- Kirschner, K.N.; Yongye, A.B.; Tschampel, S.M.; González-Outeiriño, J.; Daniels, C.R.; Foley, B.L.; Woods, R.J. GLYCAM06: A generalizable biomolecular force field. Carbohydrates. J. Comput. Chem. 2008, 29, 622–655. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Revision D. 01; Gaussian. Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef] [Green Version]
Sample Availability: Not Available. |

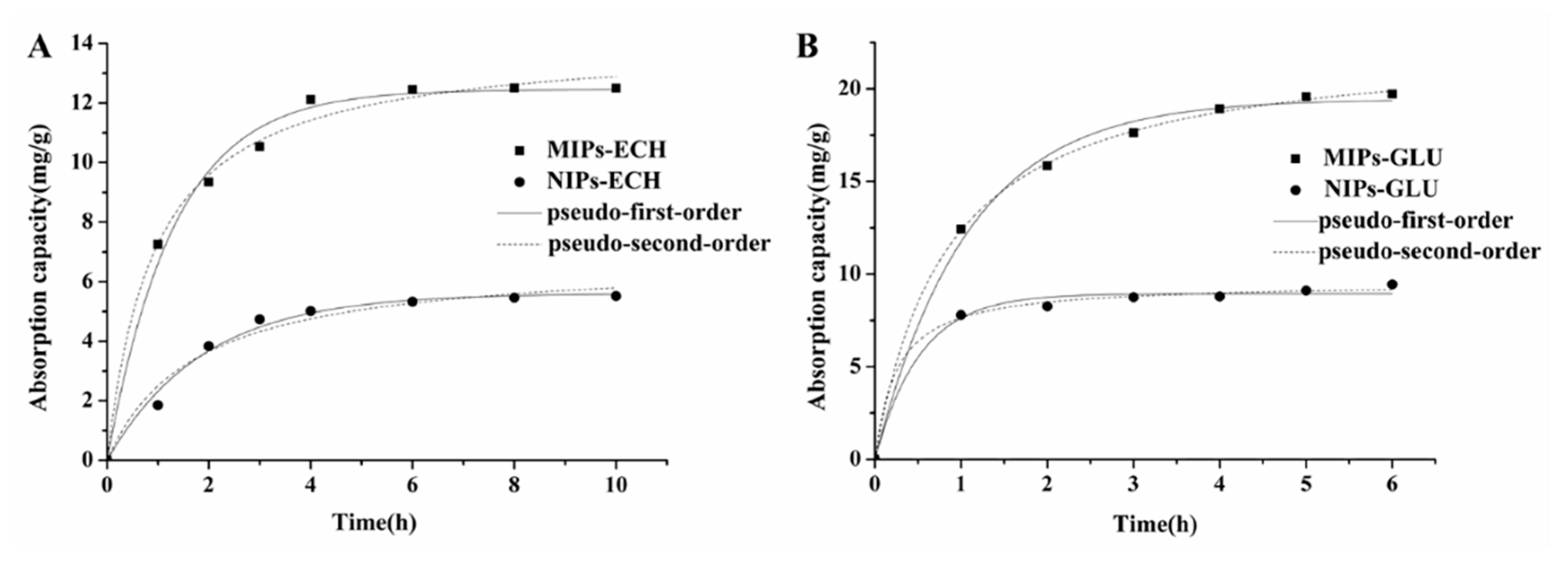
| Polymers | Qe(mg/g) | Pseudo-First-Order | Pseudo-Second-Order | ||
|---|---|---|---|---|---|
| k1 | R2 | k2 | R2 | ||
| MIPs–ECH | 12.50 | 0.9200 | 0.9670 | 0.1079 | 0.9973 |
| NIPs–ECH | 5.50 | 0.5698 | 0.9933 | 0.1297 | 0.9786 |
| MIPs–GLU | 19.72 | 0.9141 | 0.9631 | 0.0703 | 0.9995 |
| NIPs–GLU | 9.45 | 0.5134 | 0.8617 | 0.3145 | 0.9979 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, L.; Zhao, Z.-X.; Huang, Y.-A.; Zhu, Q.-J. Preparation of Chitosan Molecularly Imprinted Polymers and the Recognition Mechanism for Adsorption of Alpha-Lipoic Acid. Molecules 2020, 25, 312. https://doi.org/10.3390/molecules25020312
Xu L, Zhao Z-X, Huang Y-A, Zhu Q-J. Preparation of Chitosan Molecularly Imprinted Polymers and the Recognition Mechanism for Adsorption of Alpha-Lipoic Acid. Molecules. 2020; 25(2):312. https://doi.org/10.3390/molecules25020312
Chicago/Turabian StyleXu, Long, Ze-Xin Zhao, Yun-An Huang, and Qiu-Jin Zhu. 2020. "Preparation of Chitosan Molecularly Imprinted Polymers and the Recognition Mechanism for Adsorption of Alpha-Lipoic Acid" Molecules 25, no. 2: 312. https://doi.org/10.3390/molecules25020312
APA StyleXu, L., Zhao, Z.-X., Huang, Y.-A., & Zhu, Q.-J. (2020). Preparation of Chitosan Molecularly Imprinted Polymers and the Recognition Mechanism for Adsorption of Alpha-Lipoic Acid. Molecules, 25(2), 312. https://doi.org/10.3390/molecules25020312





