The Effect of GD1a Ganglioside-Expressing Bacterial Strains on Murine Norovirus Infectivity
Abstract
1. Introduction
2. Results
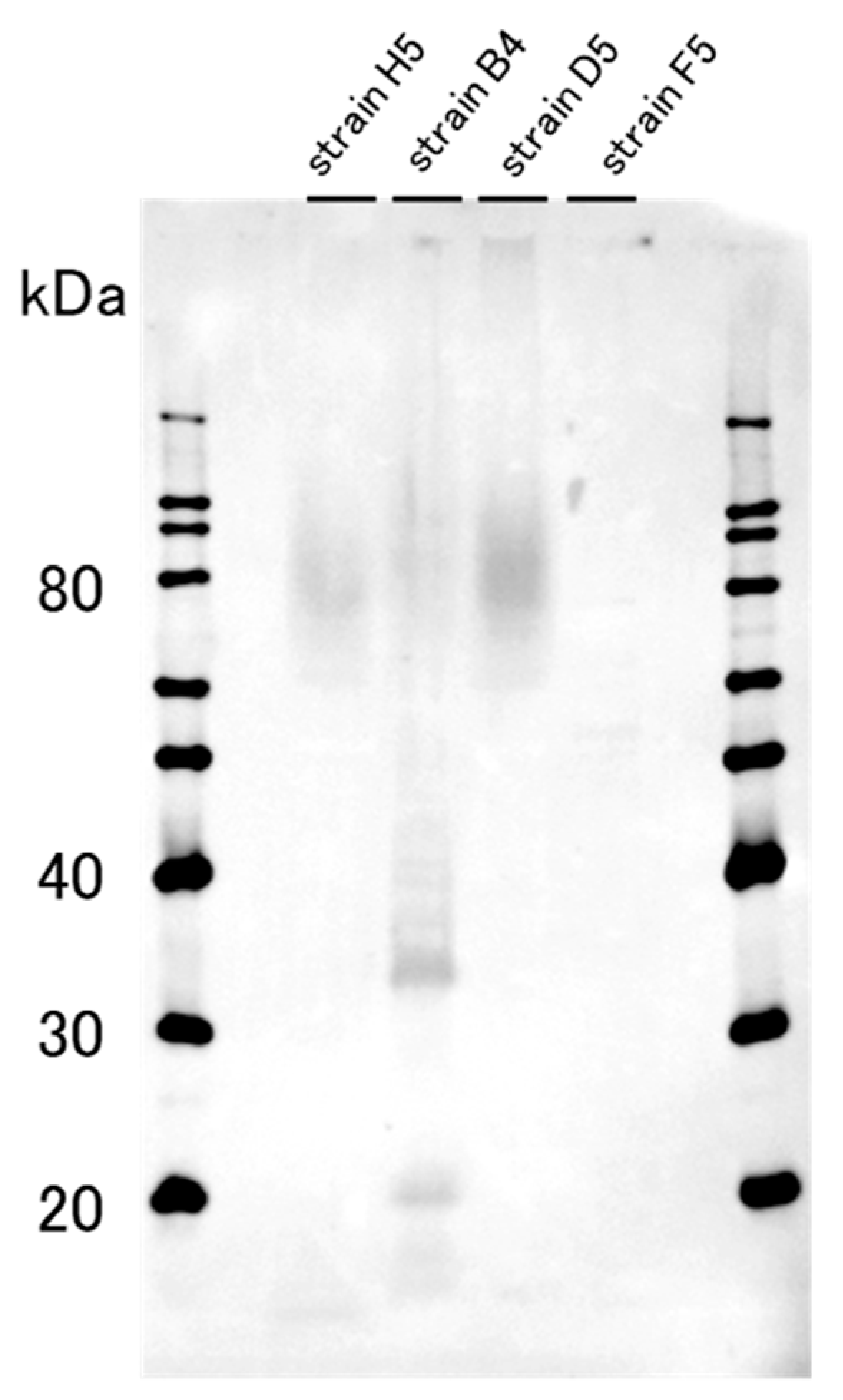
2.1. The Western Blotting Result for GD1a Expression Confirmation
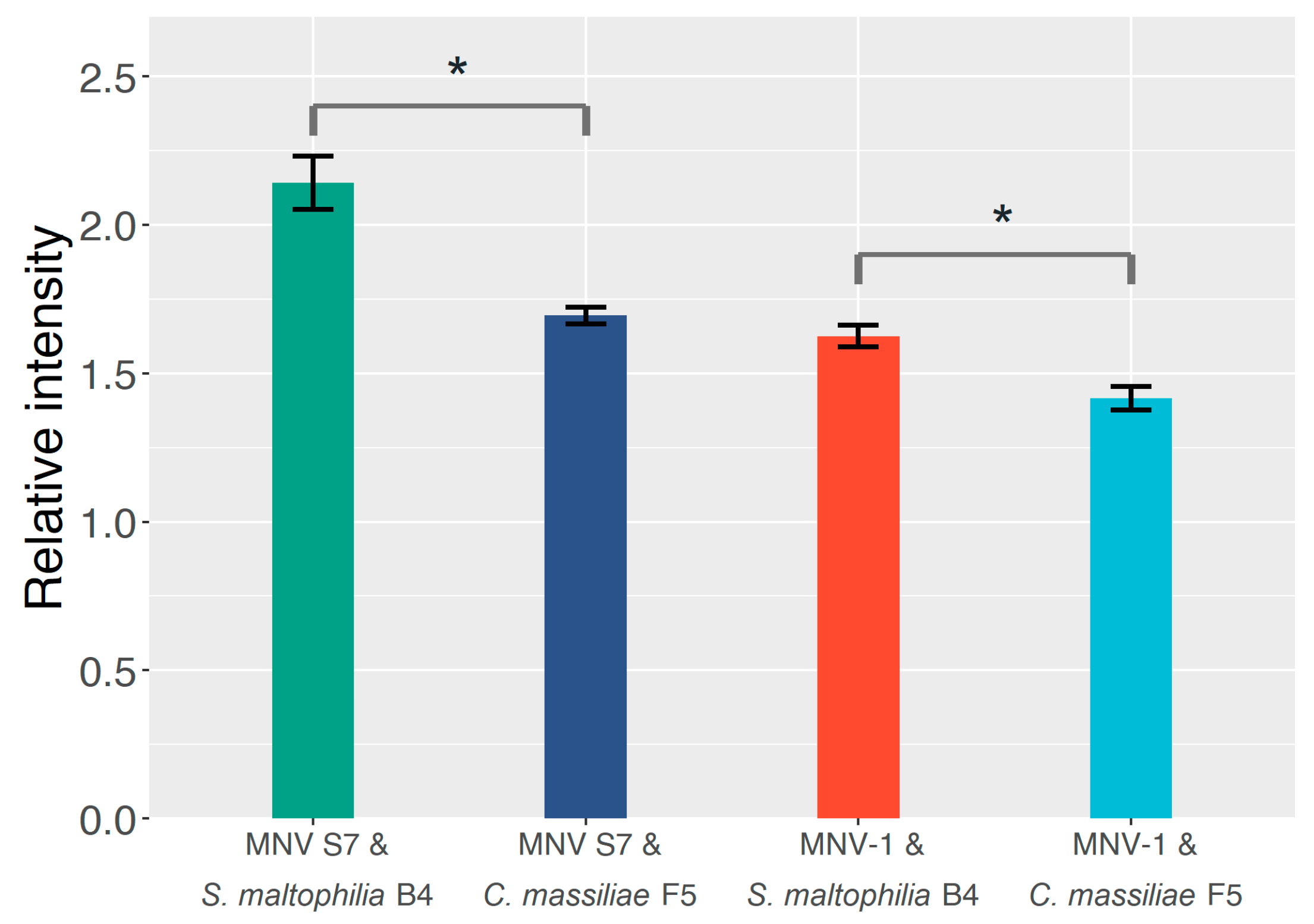
2.2. The Effect of GD1a-Expressing Bacterial Strains on the MNV Binding Profile
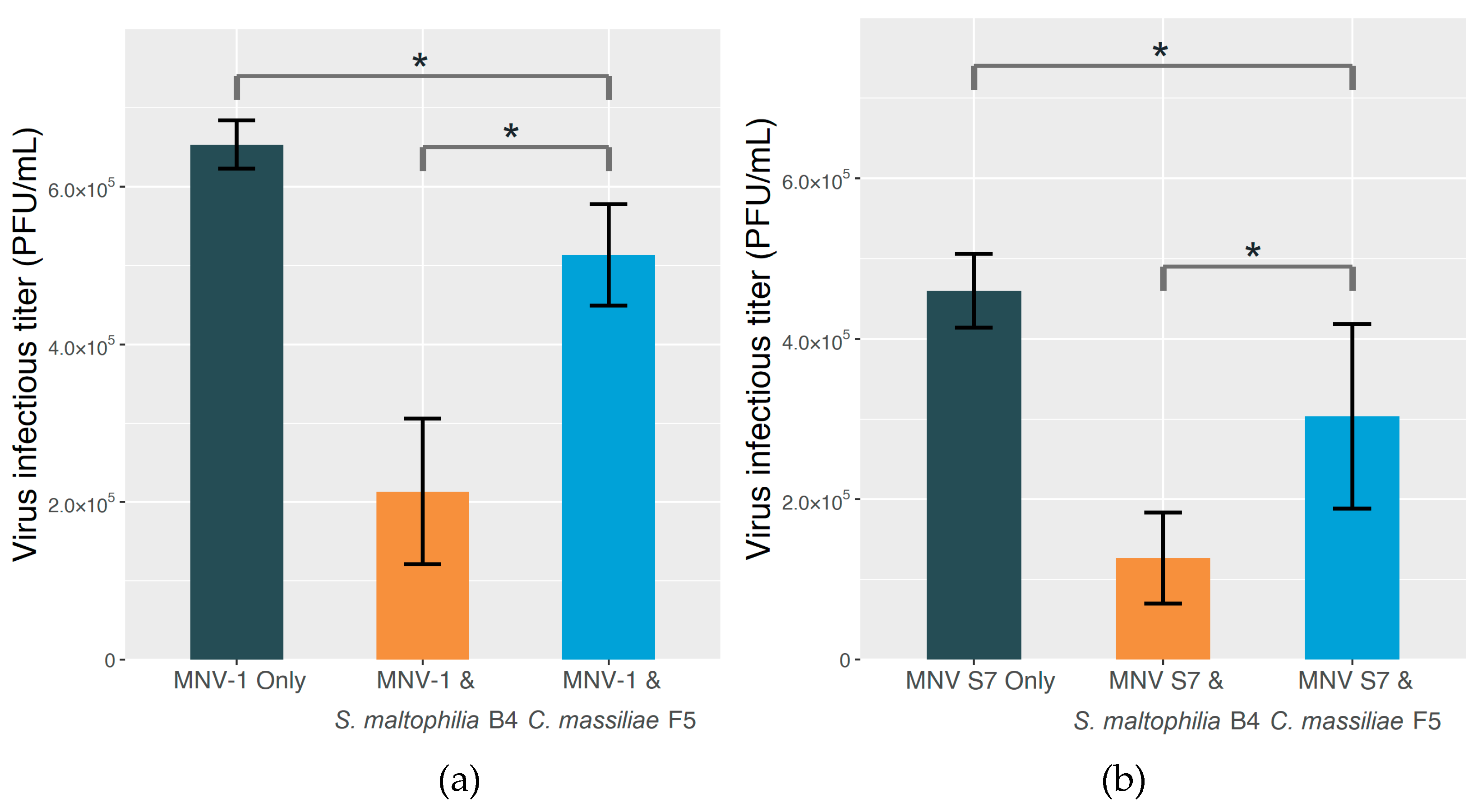
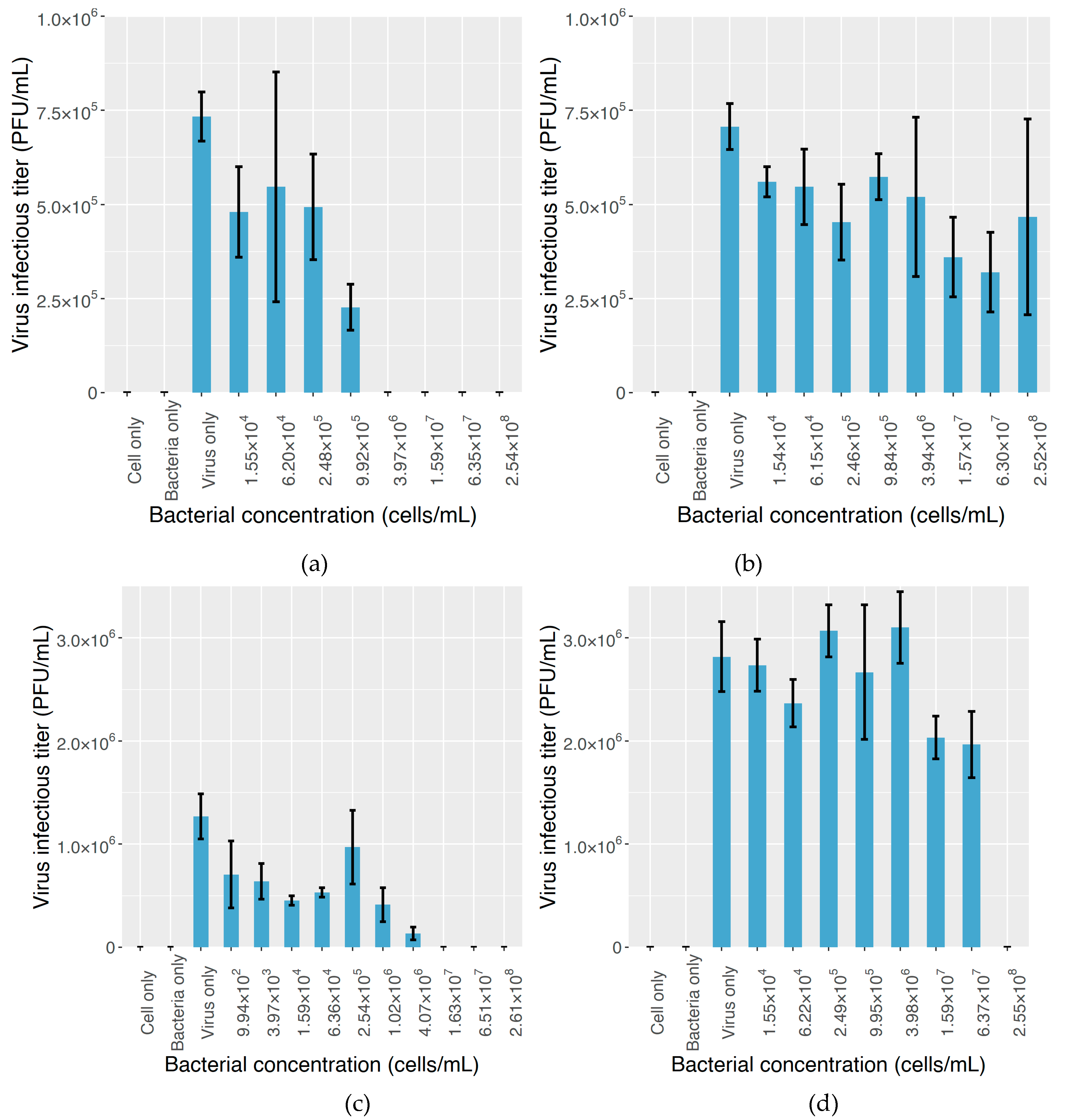
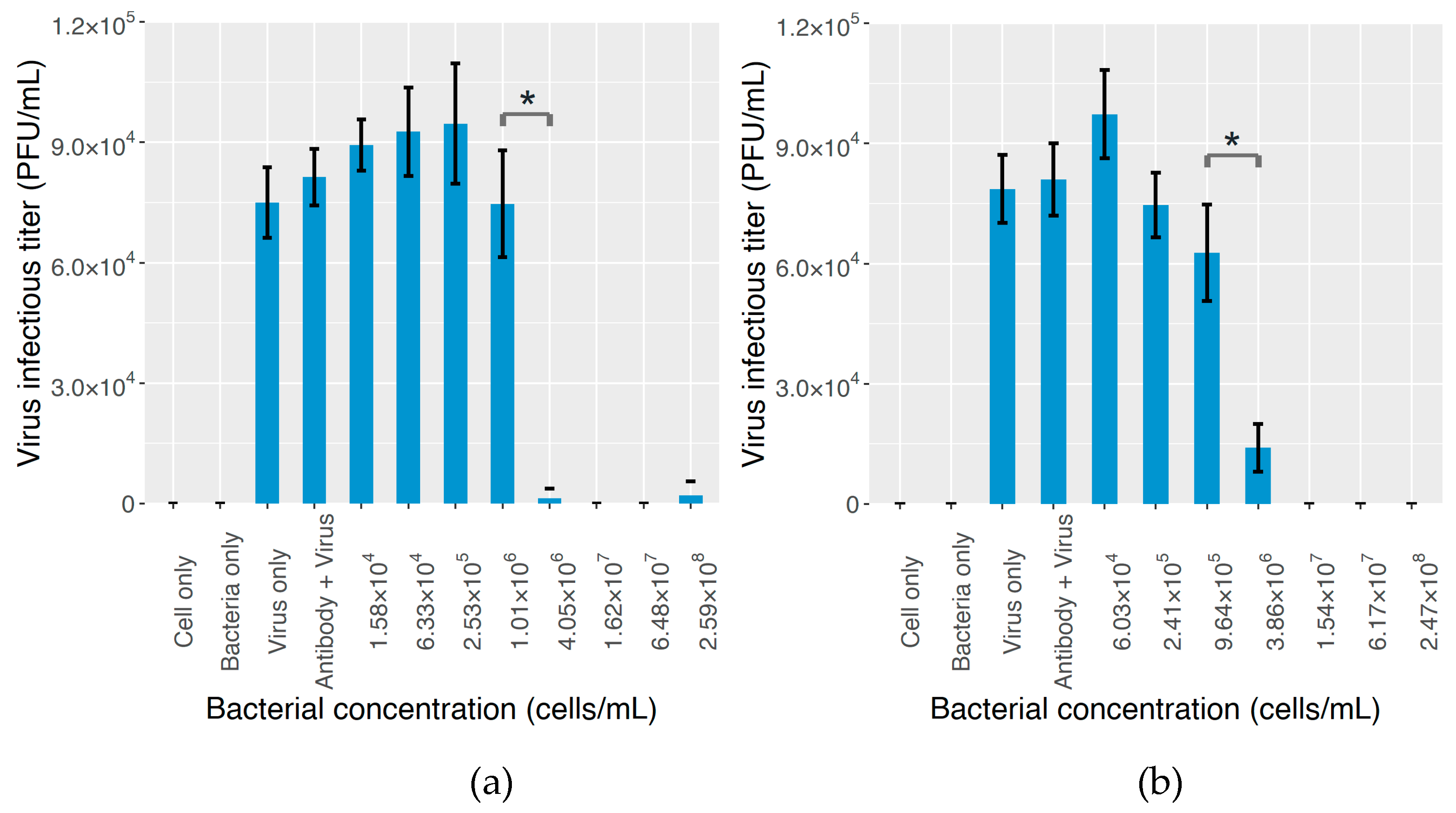
2.3. The Effect on MNV Infectivity
3. Discussion
4. Materials and Methods
4.1. Screening Out GD1a-Positive Bacterial Strains from Sewage Sample
4.2. Preparation of Media, Bacterial Suspension, and Virus Stock
4.3. ELISA for MNV–Bacterium Binding Assessment
4.4. Plaque Assay in the Presence of Bacterial Components
4.5. Viral Infectivity Assessment
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Forouzanfar, M.H.; Afshin, A.; Alexander, L.T.; Anderson, H.R.; Bhutta, Z.A.; Biryukov, S.; Brauer, M.; Burnett, R.; Cercy, K.; Charlson, F.J. Global, regional, and national comparative risk assessment of 79 behavioural, environmental and occupational, and metabolic risks or clusters of risks, 1990–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet 2016, 388, 1659–1724. [Google Scholar] [CrossRef]
- Gibson, K.E. Viral pathogens in water: Occurrence, public health impact, and available control strategies. Curr. Opin. Virol. 2014, 4, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Kocwa-Haluch, R. Waterborne enteroviruses as a hazard for human health. Pol. J. Environ. Stud. 2001, 10, 485–487. [Google Scholar]
- Gibney, K.B.; O’Toole, J.; Sinclair, M.; Leder, K. Burden of disease attributed to waterborne transmission of selected enteric pathogens, Australia, 2010. Am. J. Trop. Med. Hyg. 2017, 96, 1400–1403. [Google Scholar] [CrossRef] [PubMed]
- Mawatari, M.; Kato, Y. Norovirus gastroenteritis. Emerg. Infect. Dis. Clin. Case Stud. 2014, 203–212. [Google Scholar] [CrossRef]
- Atmar, R.L. Noroviruses: State of the art. Food Environ. Virol. 2010, 2, 117–126. [Google Scholar] [CrossRef]
- Lopman, B.; Gastañaduy, P.; Park, G.W.; Hall, A.J.; Parashar, U.D.; Vinjé, J. Environmental transmission of norovirus gastroenteritis. Curr. Opin. Virol. 2012, 2, 96–102. [Google Scholar] [CrossRef]
- Lanata, C.F.; Fischer-Walker, C.L.; Olascoaga, A.C.; Torres, C.X.; Aryee, M.J.; Black, R.E. Global causes of diarrheal disease mortality in children <5 years of age: A systematic review. PLoS ONE 2013, 8, e72788. [Google Scholar] [CrossRef]
- Patel, M.M.; Widdowson, M.A.; Glass, R.I.; Akazawa, K.; Vinjé, J.; Parashar, U.D. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg. Infect. Dis. 2008, 14, 1224–1231. [Google Scholar] [CrossRef]
- Fong, T.-T.; Lipp, E.K. Enteric viruses of humans and animals in aquatic environments: Health risks, detection, and potential water quality assessment tools. Microbiol. Mol. Biol. Rev. 2014, 69, 357–371. [Google Scholar] [CrossRef]
- Matrosovich, M.; Herrler, G.; Klenk, H.D. Sialic acid receptors of viruses. In SialoGlyco Chemistry and Biology II; Gerardy-Schahn, R., Delannoy, P., von Itzstein, M., Eds.; Springer International Publishing: Cham, Switzerland, 2013; pp. 1–28. [Google Scholar]
- Yezli, S.; Otter, J.A. Minimum infective dose of the major human respiratory and enteric viruses transmitted through food and the environment. Food Environ. Virol. 2011, 3, 1–30. [Google Scholar] [CrossRef]
- Bosch, A. Human enteric viruses in the water environment: A minireview. Int. Microbiol. 1998, 1, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Atmar, R.L.; Opekun, A.R.; Gilger, M.A.; Estes, M.K.; Crawford, S.E.; Neill, F.H.; Graham, D.Y. Norwalk virus shedding after experimental human infection. Emerg. Infect. Dis. 2008, 14, 1553–1557. [Google Scholar] [CrossRef] [PubMed]
- Parra, G.I.; Squires, R.B.; Karangwa, C.K.; Johnson, J.A.; Lepore, C.J.; Sosnovtsev, S.V.; Green, K.Y. Static and evolving norovirus genotypes: Implications for epidemiology and immunity. PLoS Pathog. 2017, 13, e1006136. [Google Scholar] [CrossRef] [PubMed]
- Teunis, P.F.M.; Sukhrie, F.H.A.; Vennema, H.; Bogerman, J.; Beersma, M.F.C.; Koopmans, M.P.G. Shedding of norovirus in symptomatic and asymptomatic infections. Epidemiol. Infect. 2015, 143, 1710–1717. [Google Scholar] [CrossRef]
- Belliot, G.; Lopman, B.A.; Ambert-Balay, K.; Pothier, P. The burden of norovirus gastroenteritis: An important foodborne and healthcare-related infection. Clin. Microbiol. Infect. 2014, 20, 724–730. [Google Scholar] [CrossRef]
- Hellmér, M.; Paxéus, N.; Magnius, L.; Enache, L.; Arnholm, B.; Johansson, A.; Bergström, T.; Norder, H. Detection of pathogenic viruses in sewage provided early warnings of hepatitis A virus and norovirus outbreaks. Appl. Environ. Microbiol. 2014, 80, 6771–6781. [Google Scholar] [CrossRef]
- Seitz, S.R.; Leon, J.S.; Schwab, K.J.; Lyon, G.M.; Dowd, M.; McDaniels, M.; Abdulhafid, G.; Fernandez, M.L.; Lindesmith, L.C.; Baric, R.S.; et al. Norovirus infectivity in humans and persistence in water. Appl. Environ. Microbiol. 2011, 77, 6884–6888. [Google Scholar] [CrossRef]
- Kotwal, G.; Cannon, J.L. Environmental persistence and transfer of enteric viruses. Curr. Opin. Virol. 2014, 4, 37–43. [Google Scholar] [CrossRef]
- Pang, X.; Qiu, Y.; Gao, T.; Zurawell, R.; Neumann, N.F.; Craik, S.; Lee, B.E. Prevalence, levels and seasonal variations of human enteric viruses in six major rivers in Alberta, Canada. Water Res. 2019, 153, 349–356. [Google Scholar] [CrossRef]
- Eftim, S.E.; Hong, T.; Soller, J.; Boehm, A.; Warren, I.; Ichida, A.; Nappier, S.P. Occurrence of norovirus in raw sewage—A systematic literature review and meta-analysis. Water Res. 2017, 111, 366–374. [Google Scholar] [CrossRef]
- Gall, A.M.; Mariñas, B.J.; Lu, Y.; Shisler, J.L. Waterborne viruses: A barrier to safe drinking water. PLoS Pathog. 2015, 11, e004867. [Google Scholar] [CrossRef] [PubMed]
- Kukkula, M.; Maunula, L.; Silvennoinen, E.; von Bonsdorff, C. Outbreak of viral gastroenteritis due to drinking water contaminated by Norwalk-like viruses. J. Infect. Dis. 1999, 180, 1771–1776. [Google Scholar] [CrossRef]
- Maunula, L.; Klemola, P.; Kauppinen, A.; Söderberg, K.; Nguyen, T.; Pitkänen, T.; Kaijalainen, S.; Simonen, M.L.; Miettinen, I.T.; Lappalainen, M.; et al. Enteric viruses in a large waterborne outbreak of acute gastroenteritis in Finland. Food Environ. Virol. 2009, 1, 31–36. [Google Scholar] [CrossRef]
- Dickin, S.K.; Schuster-Wallace, C.J.; Qadir, M.; Pizzacalla, K. A review of health risks and pathways for exposure to wastewater use in agriculture. Environ. Health Perspect. 2016, 124, 900–909. [Google Scholar] [CrossRef] [PubMed]
- Okoh, A.I.; Sibanda, T.; Gusha, S.S. Inadequately treated wastewater as a source of human enteric viruses in the environment. Int. J. Environ. Res. Public Health 2010, 7, 2620–2637. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, G.; Bosch, A. Survival of enteric viruses in the environment and food. In Viruses in Foods; Goyal, S.M., Cannon, J.L., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 367–392. ISBN 978-3-319-30723-7. [Google Scholar]
- Armanious, A.; Aeppli, M.; Jacak, R.; Refardt, D.; Sigstam, T.; Kohn, T.; Sander, M. Viruses at solid-water interfaces: A systematic assessment of interactions driving adsorption. Environ. Sci. Technol. 2016, 50, 732–743. [Google Scholar] [CrossRef]
- Chaudhry, R.M.; Holloway, R.W.; Cath, T.Y.; Nelson, K.L. Impact of virus surface characteristics on removal mechanisms within membrane bioreactors. Water Res. 2015, 84, 144–152. [Google Scholar] [CrossRef]
- Matsushita, T.; Suzuki, H.; Shirasaki, N.; Matsui, Y.; Ohno, K. Adsorptive virus removal with super-powdered activated carbon. Sep. Purif. Technol. 2013, 107, 79–84. [Google Scholar] [CrossRef]
- Ströh, L.J.; Stehle, T. Glycan engagement by viruses: Receptor switches and specificity. Annu. Rev. Virol. 2014, 1, 285–306. [Google Scholar] [CrossRef]
- Shirato, H. Norovirus and histo-blood group antigens. Jpn. J. Infect. Dis. 2011, 64, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Venkataram Prasad, B.V.; Shanker, S.; Hu, L.; Choi, J.M.; Crawford, S.E.; Ramani, S.; Czako, R.; Atmar, R.L.; Estes, M.K. Structural basis of glycan interaction in gastroenteric viral pathogens. Curr. Opin. Virol. 2014, 7, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Amarasiri, M.; Sano, D. Specific interactions between human norovirus and environmental matrices: Effects on the virus ecology. Viruses 2019, 11, 224. [Google Scholar] [CrossRef] [PubMed]
- Hirneisen, K.A.; Kniel, K.E. Norovirus attachment: Implications for food safety. Food Prot. Trends 2013, 33, 290–299. [Google Scholar]
- Stencel-Baerenwald, J.E.; Reiss, K.; Reiter, D.M.; Stehle, T.; Dermody, T.S. The sweet spot: Defining virus-sialic acid interactions. Nat. Rev. Microbiol. 2014, 12, 739–749. [Google Scholar] [CrossRef]
- Taube, S.; Perry, J.W.; McGreevy, E.; Yetming, K.; Perkins, C.; Henderson, K.; Wobus, C.E. Murine noroviruses bind glycolipid and glycoprotein attachment receptors in a strain-dependent manner. J. Virol. 2012, 86, 5584–5593. [Google Scholar] [CrossRef]
- Thompson, A.J.; de Vries, R.P.; Paulson, J.C. Virus recognition of glycan receptors. Curr. Opin. Virol. 2019, 34, 117–129. [Google Scholar] [CrossRef]
- Rydell, G.E.; Kindberg, E.; Larson, G.; Svensson, L. Susceptibility to winter vomiting disease: A sweet matter. Rev. Med. Virol. 2011, 21, 370–382. [Google Scholar] [CrossRef]
- Murakami, K.; Kurihara, C.; Oka, T.; Shimoike, T.; Fujii, Y.; Takai-Todaka, R.; Park, Y.B.; Wakita, T.; Matsuda, T.; Hokari, R.; et al. Norovirus binding to intestinal epithelial cells is independent of histo-blood group antigens. PLoS ONE 2013, 8, e66534. [Google Scholar] [CrossRef]
- Jones, M.K.; Watanabe, M.; Zhu, S.; Graves, C.L.; Keyes, L.R.; Grau, K.R.; Gonzalez-Hernandez, M.B.; Iovine, N.M.; Wobus, C.E.; Vinjé, J. Enteric bacteria promote human and mouse norovirus infection of B cells. Science 2014, 346, 755–759. [Google Scholar] [CrossRef]
- Ali, E.S.; Rajapaksha, H.; Lundborg, M.; Carr, J.M.; Petrovsky, N. Norovirus drug candidates that inhibit viral capsid attachment to human histo-blood group antigens. Antiviral Res. 2016, 133, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Miura, T.; Sano, D.; Suenaga, A.; Yoshimura, T.; Fuzawa, M.; Nakagomi, T.; Nakagomi, O.; Okabe, S. Histo-blood group antigen-like substances of human enteric bacteria as specific adsorbents for human noroviruses. J. Virol. 2013, 87, 9441–9451. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Jiang, X. Vaccine against norovirus. Hum. Vaccines Immunother. 2014, 10, 1449–1456. [Google Scholar] [CrossRef] [PubMed]
- Monedero, V.; Buesa, J.; Rodríguez-Díaz, J. The interactions between host glycobiology, bacterial microbiota, and viruses in the gut. Viruses 2018, 10, 96. [Google Scholar] [CrossRef] [PubMed]
- Imai, T.; Sano, D.; Miura, T.; Okabe, S.; Wada, K.; Masago, Y.; Omura, T. Adsorption characteristics of an enteric virus-binding protein to norovirus, rotavirus and poliovirus. BMC Biotechnol. 2011, 11, 123. [Google Scholar] [CrossRef]
- Tamura, M.; Natori, K.; Kobayashi, M.; Miyamura, T.; Takeda, N. Interaction of recombinant Norwalk virus particles with the 105-Kilodalton cellular binding protein, a candidate receptor molecule for virus attachment. J. Virol. 2000, 74, 11589–11597. [Google Scholar] [CrossRef]
- Tamura, M.; Natori, K.; Kobayashi, M.; Miyamura, T.; Takeda, N. Genogroup II noroviruses efficiently bind to heparan sulfate proteoglycan associated with the cellular membrane. J. Virol. 2004, 78, 3817–3826. [Google Scholar] [CrossRef]
- Li, Q.; Wang, D.; Yang, D.; Shan, L.; Tian, P. Binding of Escherichia coli does not protect tulane virus from heat-inactivation regardless the expression of HBGA-like molecules. Front. Microbiol. 2017, 8, 1746. [Google Scholar] [CrossRef]
- Taube, S.; Perry, J.W.; Yetming, K.; Patel, S.P.; Auble, H.; Shu, L.; Nawar, H.F.; Lee, C.H.; Connell, T.D.; Shayman, J.A.; et al. Ganglioside-linked terminal sialic acid moieties on murine macrophages function as attachment receptors for murine noroviruses. J. Virol. 2009, 83, 4092–4101. [Google Scholar] [CrossRef]
- Li, D.; Breiman, A.; le Pendu, J.; Uyttendaele, M. Binding to histo-blood group antigen-expressing bacteria protects human norovirus from acute heat stress. Front. Microbiol. 2015, 6, 659. [Google Scholar] [CrossRef]
- Purnell, S.; Ebdon, J.; Buck, A.; Tupper, M.; Taylor, H. Removal of phages and viral pathogens in a full-scale MBR: Implications for wastewater reuse and potable water. Water Res. 2016, 100, 20–27. [Google Scholar] [CrossRef]
- Fox, R.; Stuckey, D. MS-2 and T4 phage removal in an anaerobic membrane bioreactor (AnMBR): Effect of gas sparging rate. J. Chem. Technol. Biotechnol. 2015, 90, 384–390. [Google Scholar] [CrossRef] [PubMed]
- Hai, F.I.; Riley, T.; Shawkat, S.; Magram, S.F.; Yamamoto, K. Removal of pathogens by membrane bioreactors: A review of the mechanisms, influencing factors and reduction in chemical disinfectant dosing. Water 2014, 6, 3603–3630. [Google Scholar] [CrossRef]
- Lv, W.; Zheng, X.; Yang, M.; Zhang, Y.; Liu, Y.; Liu, J. Virus removal performance and mechanism of a submerged membrane bioreactor. Process Biochem. 2006, 41, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, K.O.; Furukawa, K. Biosynthesis and functions of gangliosides: Recent advances. Glycoconj. J. 1998, 15, 627–636. [Google Scholar] [CrossRef]
- Schnaar, R.L. Gangliosides of the vertebrate nervous system. J. Mol. Biol. 2016, 428, 3325–3336. [Google Scholar] [CrossRef] [PubMed]
- Amarasiri, M.; Hashiba, S.; Miura, T.; Nakagomi, T.; Nakagomi, O.; Ishii, S.; Okabe, S.; Sano, D. Bacterial histo-blood group antigens contributing to genotype-dependent removal of human noroviruses with a microfiltration membrane. Water Res. 2016, 95, 383–391. [Google Scholar] [CrossRef]
- Macpherson, A.J.; Harris, N.L. Interactions between commensal intestinal bacteria and the immune system. Nat. Rev. Immunol. 2004, 4, 478–485. [Google Scholar] [CrossRef]
- Maalouf, H.; Zakhour, M.; Pendu, J.L.; Le Saux, J.C.; Atmar, R.L.; Le Guyader, F.S. Distribution in tissue and seasonal variation of norovirus genogroup I and II ligands in oysters. Appl. Environ. Microbiol. 2010, 76, 5621–5630. [Google Scholar] [CrossRef]
- Madrigal, J.L.; Bhar, S.; Hackett, S.; Engelken, H.; Joseph, R.; Keyhani, N.O.; Jones, M.K. Attach me if you can: Murine norovirus binds to commensal bacteria and fungi. Viruses 2020, 12, 759. [Google Scholar] [CrossRef]
- Baldridge, M.T.; Nice, T.J.; McCune, B.T.; Yokoyama, C.C.; Kambal, A.; Wheadon, M.; Diamond, M.S.; Ivanova, Y.; Artyomov, M.; Virgin, H.W. Commensal microbes and interferon-λ determine persistence of enteric murine norovirus infection. Science 2015, 347, 266–269. [Google Scholar] [CrossRef]
- Nilsson, E.C.; Storm, R.J.; Bauer, J.; Johansson, S.M.C.; Lookene, A.; Ångström, J.; Hedenström, M.; Eriksson, T.L.; FräCurrency Signngsmyr, L.; Rinaldi, S.; et al. The GD1a glycan is a cellular receptor for adenoviruses causing epidemic keratoconjunctivitis. Nat. Med. 2011, 17, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Looney, W.J.; Narita, M.; Mühlemann, K. Stenotrophomonas maltophilia: An emerging opportunist human pathogen. Lancet Infect. Dis. 2009, 9, 312–323. [Google Scholar] [CrossRef]
- Liu, W.T.; Chan, O.C.; Fang, H.H.P. Characterization of microbial community in granular sludge treating brewery wastewater. Water Res. 2002, 36, 1767–1775. [Google Scholar] [CrossRef]
- Kroon, A.G.M.; Van Ginkel, C.G. Complete mineralization of dodecyldimethylamine using a two-membered bacterial culture. Environ. Microbiol. 2001, 3, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Hoefel, D.; Monis, P.T.; Grooby, W.L.; Andrews, S.; Saint, C.P. Profiling bacterial survival through a water treatment process and subsequent distribution system. J. Appl. Microbiol. 2005, 99, 175–186. [Google Scholar] [CrossRef]
- Crossman, L.C.; Gould, V.C.; Dow, J.M.; Vernikos, G.S.; Okazaki, A.; Sebaihia, M.; Saunders, D.; Arrowsmith, C.; Carver, T.; Peters, N.; et al. The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol. 2008, 9, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Adegoke, A.A.; Stenström, T.A.; Okoh, A.I. Stenotrophomonas maltophilia as an emerging ubiquitous pathogen: Looking beyond contemporary antibiotic therapy. Front. Microbiol. 2017, 8, 2276. [Google Scholar] [CrossRef]
- Mukherjee, P.; Roy, P. Genomic potential of Stenotrophomonas maltophilia in bioremediation with an assessment of its multifaceted role in our environment. Front. Microbiol. 2016, 7, 967. [Google Scholar] [CrossRef]
- Purnell, S.; Ebdon, J.; Buck, A.; Tupper, M.; Taylor, H. Bacteriophage removal in a full-scale membrane bioreactor (MBR)-Implications for wastewater reuse. Water Res. 2015, 73, 109–117. [Google Scholar] [CrossRef]
- Miura, T.; Schaeffer, J.; Le Saux, J.C.; Le Mehaute, P.; Le Guyader, F.S. Virus type-specific removal in a full-scale membrane bioreactor treatment process. Food Environ. Virol. 2018, 10, 176–186. [Google Scholar] [CrossRef] [PubMed]
- Shang, C.; Hiu, M.W.; Chen, G. Bacteriophage MS-2 removal by submerged membrane bioreactor. Water Res. 2005, 39, 4211–4219. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Li, H.; Huang, X. Indigenous somatic coliphage removal from a real municipal wastewater by a submerged membrane bioreactor. Water Res. 2010, 44, 1853–1862. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Son, K.; Koo, K.; Kim, J.; Alfajaro, M.; Park, J.; Hosmillo, M.; Soliman, M.; Baek, Y.; Cho, E.; et al. Porcine sapelovirus uses α2,3-linked sialic acid on GD1a ganglioside as a receptor. J. Virol. 2016, 90, 4067–4077. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, J.; Benjamin, T. Uptake pathway of polyomavirus via ganglioside GD1a. J. Virol. 2004, 78, 12259–12267. [Google Scholar] [CrossRef]
Sample Availability: The bacterial strains are available from the authors upon request. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Y.; Kawai, H.; Hashiba, S.; Amarasiri, M.; Kitajima, M.; Okabe, S.; Sano, D. The Effect of GD1a Ganglioside-Expressing Bacterial Strains on Murine Norovirus Infectivity. Molecules 2020, 25, 4084. https://doi.org/10.3390/molecules25184084
Zhu Y, Kawai H, Hashiba S, Amarasiri M, Kitajima M, Okabe S, Sano D. The Effect of GD1a Ganglioside-Expressing Bacterial Strains on Murine Norovirus Infectivity. Molecules. 2020; 25(18):4084. https://doi.org/10.3390/molecules25184084
Chicago/Turabian StyleZhu, Yifan, Hiroki Kawai, Satoshi Hashiba, Mohan Amarasiri, Masaaki Kitajima, Satoshi Okabe, and Daisuke Sano. 2020. "The Effect of GD1a Ganglioside-Expressing Bacterial Strains on Murine Norovirus Infectivity" Molecules 25, no. 18: 4084. https://doi.org/10.3390/molecules25184084
APA StyleZhu, Y., Kawai, H., Hashiba, S., Amarasiri, M., Kitajima, M., Okabe, S., & Sano, D. (2020). The Effect of GD1a Ganglioside-Expressing Bacterial Strains on Murine Norovirus Infectivity. Molecules, 25(18), 4084. https://doi.org/10.3390/molecules25184084







