Abstract
Schistosomiasis, a neglected tropical disease of medical and veterinary importance, transmitted through specific freshwater snail intermediate hosts, is targeted for elimination in several endemic regions in sub-Saharan Africa. Multi-disciplinary methods are required for both human and environmental diagnostics to certify schistosomiasis elimination when eventually reached. Molecular xenomonitoring protocols, a DNA-based detection method for screening disease vectors, have been developed and trialed for parasites transmitted by hematophagous insects, such as filarial worms and trypanosomes, yet few have been extensively trialed or proven reliable for the intermediate host snails transmitting schistosomes. Here, previously published universal and Schistosoma-specific internal transcribed spacer (ITS) rDNA primers were adapted into a triplex PCR primer assay that allowed for simple, robust, and rapid detection of Schistosoma haematobium and Schistosoma bovis in Bulinus snails. We showed this two-step protocol could sensitively detect DNA of a single larval schistosome from experimentally infected snails and demonstrate its functionality for detecting S. haematobium infections in wild-caught snails from Zanzibar. Such surveillance tools are a necessity for succeeding in and certifying the 2030 control and elimination goals set by the World Health Organization.
Keywords:
bovine; control; elimination; schistosomiasis; urogenital; surveillance; disease; parasite 1. Introduction
Schistosomiasis is a disease affecting an estimated 229 million people worldwide caused by infection with parasitic worms of the genus Schistosoma, leading to severe morbidity and mortality due to the associated complications of worm presence [1]. Schistosoma spp. in Africa are transmitted through specific freshwater snail intermediate hosts of the Bulinus and Biomphalaria genera [2]. Infections occur when humans or animals come into contact with freshwater containing infectious larval stages (cercariae) shed from the infected snails. Human schistosomiasis in Africa, where at least ~90% of the people requiring treatment live [3], consists of two forms of the disease—urogenital and intestinal schistosomiasis, caused predominantly by Schistosoma haematobium and Schistosoma mansoni, respectively [1]. Bovine, ovine, and caprine schistosomiasis is also of significant veterinary and economic importance across sub-Saharan Africa [4,5] and is caused by infection of cattle, sheep, and goats with species closely related to S. haematobium (termed S. haematobium group species), primarily Schistosoma bovis, Schistosoma curassoni, and Schistosoma mattheei. Overlapping geographical distribution of multiple schistosome and intermediate snail host species strains complicates disease transmission surveillance in (co)endemic zones [2,6,7].
The World Health Organization (WHO) aims for the elimination of human schistosomiasis as a public health problem, defined as >1% of the population with heavy intensity infections (≥50 schistosome eggs per 10 mL of urine, or ≥400 schistosome eggs per gram of feces [8]), in all endemic countries by 2030 [9]. Despite great advances in schistosomiasis control mainly via preventative chemotherapy (praziquantel), the lack of protection against rapid re-infection together with prolific asexual replication of schistosomes within their intermediate snail host presents substantial hurdles to achieving the targeted elimination of schistosomiasis. Very quickly, snails can become infected by eggs emanating from untreated humans, leading to a rapid resurgence of transmission [10]. Therefore, adaptive treatment strategies that take into account the transmission dynamics of Schistosoma spp. with their snail hosts are required to control and eliminate the disease [11].
To better understand the local transmission dynamics of different Schistosoma species, allowing both human and bovine schistosomiasis to be monitored, a need exists for methodologies that detect schistosome infections in the intermediate host snails. These tools for assessing Schistosoma transmission could eventually be used during elimination programs to identify focal areas of persisting transmission or certify elimination and/or transmission interruption [12,13,14]. Defining ongoing transmission in snail populations through traditional methods of observing cercariae shed from snails is particularly challenging in an elimination setting, such as the Zanzibar Archipelago, where few snails (0.5–2.3%) are observed shedding cercariae [6,15]. Snails with non-patent (including pre-patent) infections are missed using these approaches. Additionally, larval schistosomes are not easily identifiable to a species level using morphological characteristics (although the relative position of sensory receptors is of some value [16,17]).
Molecular xenomonitoring is a DNA-based method that has been developed to monitor the transmission of several vector-borne diseases, including trypanosomiasis [18,19], filariasis and malaria [20], helminthiases [21], and fascioliasis [22], including to some extent schistosomiasis [23,24,25,26,27,28,29]. Screening snails provides evidence on the extent of environmental contamination (i.e., schistosome miracidia penetrating snails), as well as environmental infection risk (i.e., schistosome sporocysts and cercariae developing inside the (pre-patent) snails, eventually emerging from the (patent) snail. Most of the available snail-schistosome xenomonitoring assays do not include internal controls [23,28,30], an important feature in any diagnostic tool that helps prevent false-negative results [27]. Many assays will assume that a negative result means non-infection, not necessarily reaction failure.
In the current study, we adapted available universal [31] and Schistosoma-specific [27] internal transcribed spacer (ITS) rDNA primers to design a three primer multiplex assay and tested this as a simple, robust, and rapid xenomonitoring PCR assay to enable the large-scale screening of Bulinus snails for Schistosoma infections (S. haematobium and S. bovis). We used a conventional PCR-based approach focused on simplicity, ease of data interpretation, sensitivity, and specificity, with a primary aim to provide a xenomonitoring tool for monitoring S. haematobium transmission in endemic settings.
2. Results
2.1. In Silico and In Vitro Primer Evaluation
Bulinus globosus and Bulinus nasutus rDNA sequence data showed conserved primer binding sites for the universal primers ETTS2 and ETTS1 [31] at the 3′ end of the 18S and 5′ end of the 28S, the flanking regions of the ITS, respectively. ETTS1 gave a 100% match, and the ETTS2 primer showed just a single base pair mismatch. The resulting snail amplicon size predicted from these alignments was between 1232 and 1263 bp and served as an internal snail control during PCR amplification.
Alignments of the Schistosoma-specific ITS primers (ITS2_Schisto_F and ITS2_Schisto_R [27]) showed 100% and 90% (2 mismatches) homology to S. haematobium and S. bovis, respectively, with no cross-reactivity to the Bulinus reference rDNA data. When paired with their opposing universal primers (ITS2_Schisto_F + ETTS1 or ITS2_Schisto_R + ETTS2), the amplicon sizes of 538 and 835 bp were predicted, respectively, for Schistosoma. With the addition of the other universal primer to each combination (ETTS2 and ETTS1, respectively), the three-primer multiplex ITS xenomonitoring (MIX) reactions were predicted to be able to produce distinct amplicon profiles for non-infected snails (a single snail amplicon) and snails infected with Schistosoma spp. (three-band profile). This was confirmed by in vitro testing of the primer combinations (Figure 1).
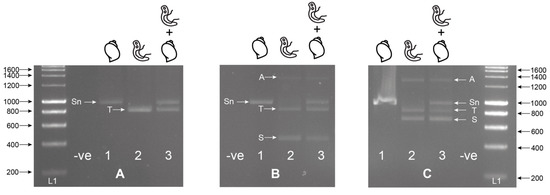
Figure 1.
Singleplex (A); ETTS2 + ETTS1) and multiplex (B); multiplex ETTS2 + ETTS1 + ITS2_Schisto_F, (C); ETTS2 + ETTS1 + ITS2_Schisto_R) PCRs on laboratory-bred Bulinus wrighti (B.w.) and Schistosoma haematobium (S.h.) gDNA separately (1; B.w., 2; S.h.) and combined (3; B.w. + S.h.). When B.w. and S.h. DNA was combined (A3, B3, C3), two amplicons were produced by the ETTS1 + ETTS2 primers, a larger snail amplicon (Sn) (~1200 bp) and a smaller Schistosoma amplicon (T) (~1000), with the additional Schistosoma-specific primers producing either a 538 bp (B3; ITS2_Schisto_F) or 835 bp (C3; ITS2_Schisto_R) amplicon (S). A larger amplicon (A) (~1400–1600 bp) was also observed to be amplified in some reactions, and this was thought to be a PCR artifact or additional primer targets in the Schistosoma gDNA. L1 = HyperLadder I (Bioline, London, UK). -ve = negative, no template control. ITS = internal transcribed spacer.
To maximize amplification efficiency/sensitivity and to provide good amplicon size differentiation, the multiplex PCR incorporating the internal ITS2_Schisto_F (Figure 1B) was selected for further development and testing. This primer combination was also selected as it targeted the ITS2 region for Schistosoma containing four species-specific SNPs, enabling species identification (Table 1).

Table 1.
Schistosoma species-specific SNP positions (including base position) in the internal transcribed spacer (ITS)2 region.
The MIX assay proved robust at varying annealing temperatures (55 °C, 60 °C Figure 2A, 58 °C Figure 2B), with 58 °C proving to be the most efficient, maximizing specificity without decreasing sensitivity. Each of the three amplicons was extracted from the gel and sequenced, confirming the band identity and specificity of the primers to their target gDNA amplicon. These three bands have been described as the snail (Sn) (1232–1263 bp), trematode (T) (~1000 bp), and Schistosoma (S) (538 bp) bands going forward. The secondary Schistosoma ITS xenomonitoring (SIX) PCR, solely targeting the Schistosoma amplicon, proved robust, enabling single amplicon generation and sequencing (Figure 3). This provided a two-step PCR methodology with the MIX PCR for the initial high-throughput screening of the samples and the secondary SIX PCR to target specific samples for further infection clarification by Schistosoma species identification through DNA sequencing.
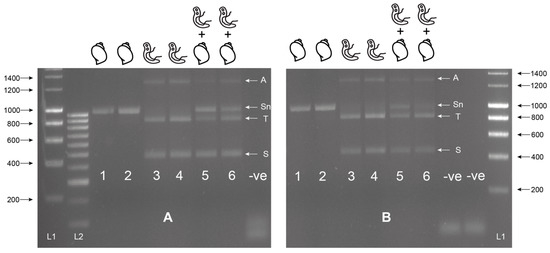
Figure 2.
Multiplex ITS xenomonitoring assay trial at 55 °C (A) and 60 °C (B). Includes gDNA of Bulinus wrighti of both BioSprint (Lane 1 and 5) and DNeasy extractions (Lane 2 and 6) and gDNA of Schistosoma haematobium (Lane 3 and 5) and S. bovis (Lane 4 and 6). Combinations of B. wrighti and S. haematobium (Lane 5) or S. bovis (Lane 6) gDNA shown. Sn = snail amplicon, T = trematode amplicon, S = Schistosoma amplicon, and A = non-specific amplicon or artifact. L1 = HyperLadder I. L2 = HyperLadder IV (Bioline, London, UK). -ve = negative, no template control.
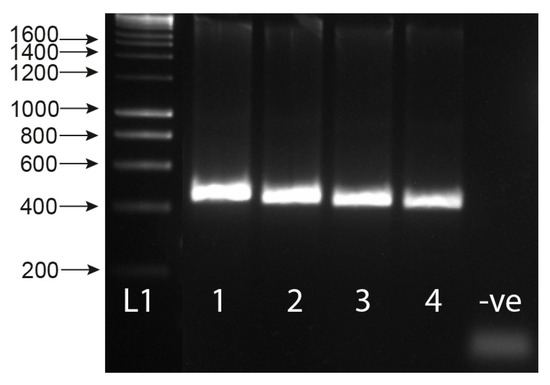
Figure 3.
Gel showing the secondary singleplex ITS xenomonitoring (SIX) PCR for 1) Schistosoma haematobium gDNA; 2) S. bovis gDNA; 3) S. haematobium + B. wrighti gDNA; 4) S. bovis + B. wrighti gDNA. -ve = non-template negative control. L1 = HyperLadder I (Bioline, London, UK).
2.2. Analytical Sensitivity
The assay proved highly sensitive with a limit-of-detection (LoD) of 0.02 ng and 0.002 ng of gDNA for S. bovis and S. haematobium, respectively (Figure 4). Sensitivity appeared higher for S. haematobium (Figure 4), but in both the cases, the assay’s sensitivity was above that necessary to detect gDNA from a single miracidium, which ranges from 1.6–3.65 ng/µL [32]. At lower Schistosoma DNA concentrations, the 1005 bp trematode band (T) lost sensitivity compared with the smaller Schistosoma-specific band.
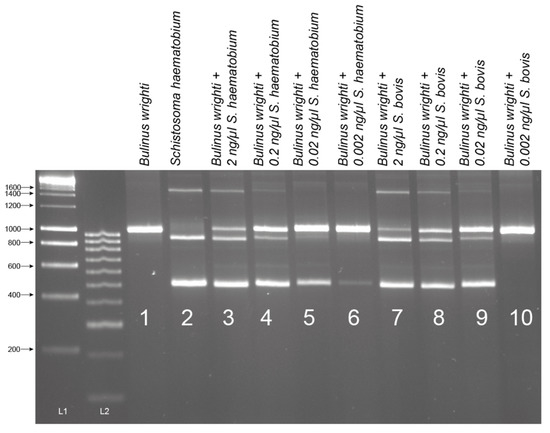
Figure 4.
Sensitivity tests of ITS1-2-F PCR performed with serial dilutions of Schistosoma haematobium and S. bovis gDNA in the presence of Bulinus wrighti gDNA. L1 = HyperLadder I. L2 = HyperLadder IV (Bioline, London, UK).
2.3. Experimental Snail Infections
For the non-patent infections of Bulinus truncatus with S. haematobium, preserved 24 h after exposure, 61.1% (11 out of 18) of the snails were observed to be penetrated by the S. haematobium miracidia, presenting the Schistosoma-specific ITS2 band (Figure 5). Infections were detected in snails exposed to 1, 2, and 7 miracidia. Two of the five (40%) B. truncatus, exposed to one or two miracidia and left for 11 weeks, did not reach patency but were also confirmed to be penetrated by S. haematobium miracidia (Figure 5: Lanes 20 and 21). The secondary SIX PCR was performed on all 13 non-patent infected snails, and the single amplicons were sequenced and confirmed as S. haematobium. Out of all the snails infected that survived until the end of the experiment (11 weeks), 15% (nine out of 62) reached patency, of which two had been infected with two miracidia, and seven with seven miracidia. One of these samples, infected with two miracidia, was analyzed using the MIX PCR, giving the expected triple banding pattern (snail, trematode, and Schistosoma) (Figure 5: Lane 24). All three amplicons from this sample (Figure 5: Lane 24) were gel extracted and sequenced, confirming their identity. Interestingly, in all the non-patent infections, the large trematode amplicons (ETTS2-ETTS2) did not amplify (Figure 5) due to the low level of Schistosoma DNA present in the snails that did not reach patency.
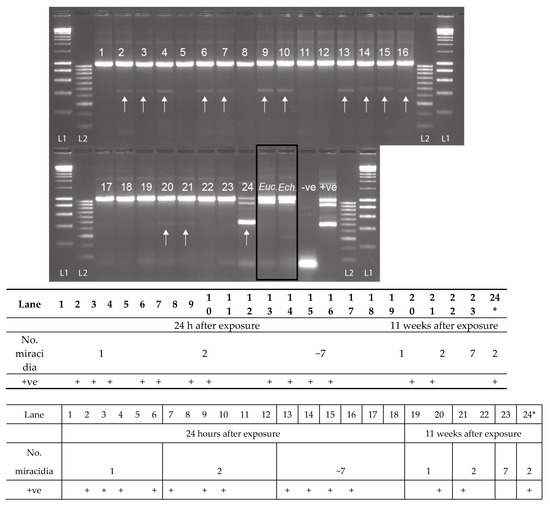
Figure 5.
Experimental infections of Bulinus truncatus with Schistosoma haematobium (1–24), field-collected B. globosus infected with Euclinostomum sp. (Euc.) and field-collected B. nasutus shedding Echinostoma sp. cercariae (Ech.). The S. haematobium DNA amplicon was present (+ve) in 13 of the 23 non-patent snails (11 at 24 h post-exposure, and two at 11 weeks post-exposure), highlighted by the arrow. Lane 24 = B. truncatus sample that was shedding S. haematobium cercariae 11 weeks after exposure. The positive control (+ve) is a mix of B. wrighti and S. haematobium control gDNA. L1: HyperLadder I, L2: HyperLadder IV (Bioline, London, UK).
2.4. Specificity Testing
The B. globosus with patent S. haematobium (n = 2) and S. bovis (n = 5) infections showed the expected triple banding pattern (snail, trematode, and Schistosoma amplicons, results not shown), and following gel extraction and sequencing the data matched those from the cercariae collected from these samples (GenBank Accessions: MH014047 and MH014044, see [6]).
When the MIX PCR was tested on snails confirmed to be infected with other commonly found trematode species (B. globosus infected with Euclinostomum sp., and the B. nasutus infected with Echinostoma sp. (Figure 5: Lanes Euc. and Ech.)), no Schistosoma amplicon was observed. However, there was strong amplification of the trematode band together with the snail band. These trematode amplicons were gel extracted, sequenced, and the infections were confirmed as Euclinostomum sp. and Echinostoma sp., matching data from the cercariae originally collected from each snail.
2.5. Testing on Field Samples
From the 94 field-collected B. globosus, 33 were shown to be infected with Schistosoma spp. with amplification of the Schistosoma-specific band (Figure 6). Among them, eight also presented the trematode band. The internal snail control was amplified in all samples apart from one. The one that failed was predicted to be due to poor sample preservation, gDNA extraction, or PCR error, and so was disregarded (Figure 6). Of all the samples that gave the Schistosoma-specific band, the secondary SIX PCR (ITS2_Schisto_F + ETTS2) was conducted, and all amplicons were Sanger sequenced. Two failed to amplify, but the remaining 31 produced the Schistosoma amplicon that all sequenced as S. haematobium. One sample also gave the trematode band without the Schistosoma band, indicating a non-Schistosoma trematode infection.
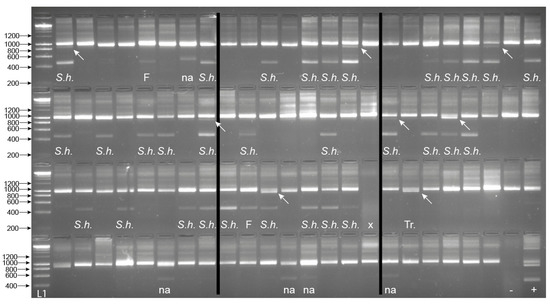
Figure 6.
Gel images for the multiplex ITS xenomonitoring (MIX) PCR amplicons for 94 non-patent Bulinus globosus collected from Wambaa, Pemba, United Republic of Tanzania. The text under each amplicon denotes the outcome of the Schistosoma sp. targeted sequencing where relevant (i.e., presence of 538 bp amplicon), which resulted in either S. haematobium (S.h.) or sequencing failure (F). The presence of a trematode band without the presence of the Schistosoma band indicated a non-Schistosoma trematode infection (Tr.). Other non-specific bands, in this case, larger bands (NA), were also observed in these snail populations, which did not amplify with the secondary SIX PCR. x = sample failure with no control amplicon. Arrows highlight the presence of the ~1000 bp trematode band when present (n = 8). B. globosus with a patent S. haematobium infection (Cham10.1 see [6]) was run as a positive control (+ve) and also represented the amplicon profile obtained for the seven patent B. globosus snails (five and two with S. bovis and S. haematobium infections, respectively (see Section 2.4). -ve = the non-template negative control. L1—HyperLadder IV (Bioline, London, UK).
2.6. Schistosoma spp. cox1 RD-PCR
Despite trying different annealing temperatures and gDNA template amounts used, the cox1 RD-PCR, developed by Webster et al. (2010) [33], tested on the patent S. haematobium and S. bovis-infected B. globosus, only generated the species-specific amplicon for S. bovis-infected snails. PCRs for the S. haematobium-infected snails repeatedly failed to produce a clear amplicon. The cox1 amplicons produced for the S. bovis-infected snails were sequenced, and the data matched that obtained from the cercariae collected and analyzed from the snails (see [6]).
3. Discussion
Pre-patent and non-patent snail screening methods for schistosomes, such as molecular xenomonitoring, offer a higher sensitivity over traditional snail shedding methods that can only detect patent infections by the observation of schistosome cercariae. Molecular xenomonitoring better helps to assess the impact of schistosomiasis control interventions in local communities, particularly where local elimination is being achieved, and certification of the absence of transmission is required at specific foci. However, the diversity of schistosomes circulating in co-endemic areas means that species-specific methods are needed to prevent false-positive data due to non-target species cross-reactivity.
Here, we describe the development and application of a molecular xenomonitoring pipeline for the detection and differentiation of S. haematobium and S. bovis patent and non-patent infections in Bulinus freshwater snails, using three previously developed primers [27,31]. The MIX assay screens for Schistosoma and other trematode species, while also incorporating an internal control, in this case, gastropod DNA, an important feature for any molecular diagnostic assay. The MIX PCR generates clearly identifiable amplicons, of different sizes, for each target (snail, trematode, Schistosoma), which are visible by simple agarose gel electrophoresis. However, the trematode target lacks sensitivity at low DNA concentrations, probably due to its large size and PCR biases for small amplicons at reduced gDNA concentrations. Interestingly, a PCR artifact (~1400–1600 bp) is also observed when using the MIX assay in the presence of Schistosoma DNA, suggesting that the primers may have a secondary binding site. However, this artifact is clearly identifiable from the main target amplicons and does not mislead the interpretation of the results.
3.1. The Sensitivity of MIX PCR Assay
Our in silico and in vitro testing of the MIX assay shows that the presence of S. haematobium and S. bovis DNA can be routinely detected at low concentrations and also is able to identify non-patent Schistosoma infections in snails where the level of DNA varies depending on the development of the infection. The LoD for Schistosoma DNA is ≤0.02 ng/µL, which is 80-fold lower than the minimum amount of gDNA usually observed from a single miracidium [32]. This is also demonstrated by the assay’s ability to detect pre-patent snail infections 24 h after exposure to a single miracidium. This provides sufficient sensitivity for the LoD needed to detect any stage of snail infection, from initial miracidia penetration of a single miracidium to full patency, in natural settings. The fact that not all the snails tested from the experimental snail infections give positive results is corroborative with observations that, even in the experimental systems, many snails avoid penetration or destroy the miracidia rapidly upon invasion. The MIX and SIX methodologies also prove robust when used to screen ‘wild-caught’ snails from Pemba, with uninfected, pre-patent S. haematobium-infected snails, and non-Schistosoma trematode infections are clearly identified.
3.2. Benefits of an Updated Molecular Xenomonitoring Protocol for Schistosomiasis Surveillance
The molecular xenomonitoring protocol requires few consumables and no cold chain, and results can be interpreted using basic molecular laboratory equipment (thermocycler and gel electrophoresis), making the molecular assay accessible in lower resource settings, such as schistosomiasis endemic regions. The molecular xenomonitoring approach described here, therefore, provides a useful tool for monitoring schistosomiasis transmission, as has been outlined as a necessary method for leading toward the WHO 2030 goals for schistosomiasis control and elimination [9].
Molecular xenomonitoring surveillance techniques are often associated with parasites transmitted by hematophagous insects, such as lymphatic filaria in mosquito vectors [20,34,35,36] and trypanosomes in tsetse flies [18,19]. However, several assays have been developed for detecting trematode species in freshwater snails, including Fasciola spp. [22,37,38,39,40,41,42,43,44], other wildlife trematode species [45], and medically important schistosome species—S. japonicum [46,47], S. mansoni [24,27,28,48,49,50,51,52,53,54], and S. haematobium [23,26,27,30,52,54,55]. The first developed assay for the molecular detection of S. haematobium DNA in Bulinus employs the highly repetitive Dra1 target, and this has been the marker of choice for studies investigating S. haematobium infections in snails due to its high sensitivity [55]. However, the specificity of the Dra1 and the interpretation of results can be problematic due to the frequent false-positive and -negative results, lack of internal control, and difficulties in interpreting the amplicon patterns. Furthermore, this marker does not allow for species identification. Kane et al. (2013) [54] employed the use of another repetitive marker, intergenic spacer (IGS), for the detection of snail infections, and a post-amplification restriction digest allowed for the downstream species identification of S. haematobium and S. bovis. However, the method lacks internal controls. In addition, many of these assays use quantitative-PCR (qPCR), rather than conventional PCR/gel electrophoresis. Although able to quantify levels of DNA within a sample, qPCR is more arduous to carry out and lesser suited for use in endemic settings. However, recent technological advances in sample preparation and DNA extraction methods have demonstrated robust field setting methodologies to conduct qPCR analysis capable of detecting avian trematodes and host species in Canadian lakes [56,57,58], which could potentially be modified to suit the detection of human and bovine schistosomes in sub-Saharan Africa, although cost and throughput would need to be considered.
A recent assay designed by Schols et al. (2019) [27] is a six primer multiplex PCR, which incorporates an internal snail control and offers a xenomonitoring tool for S. haematobium group species that are transmitted by Bulinus snail hosts. Our study simplifies the multiplex process, reducing the primer numbers and mitigating against PCR competition and some of the biases that may occur with multiple primer combinations. It also allows for greater amplicon size differentiation (as amplicon sizes can be more easily distinguished based on size), making results more interpretable. The ITS rDNA is a favorable target within the repeat ribosomal operon of Bulinus and Schistosoma spp., easily detected within small quantities of DNA due to the high copy number of rRNA clusters within eukaryote genomes. Another key feature of the target relates to specificity. The ITS regions of Schistosoma and Bulinus spp. can be routinely amplified using conventional PCR, thanks to its small size (~1000 bp) and highly conserved flanking regions (5′18S and 3′28S), enabling the use of universal primers (ETTS1 + 2) for multiple species [31]. However, interspecies heterogeneity and, to a lesser extent, intraspecies heterogeneity (Pennance et al., unpublished observations) of the ITS regions allow for differentiation between species, such as those of the S. haematobium group [7,33]. The internal Schistosoma-specific primer is situated in a conserved ITS region within the Schistosoma genus, with 100% conservation between several African species, suggesting that it could be utilized for several Schistosoma-snail transmission systems.
3.3. Limitations of Molecular Xenomonitoring Approaches for Schistosomiasis Surveillance
From our study, we have identified two limiting factors for the practical use of this method. First, the laborious nature of testing each individual snail adds time and cost. Further sensitivity testing should be performed to support the development of pooling strategies. This would help to determine whether infections are still detected when the Schistosoma DNA is diluted in the presence of much higher concentrations of snail DNA, which may inhibit the reaction. Pooling strategies have been successful for arthropod xenomonitoring protocols [18] and would allow for higher throughput of samples required for screening large populations of snails, such as those encountered for schistosomiasis.
Second, a limitation does come with the need for the secondary screening (SIX PCR) of the Schistosoma amplicon, via sequencing, to confirm species. Despite best efforts, rapid species diagnostics, such as the rapid diagnostic cox1 RD-PCR developed by Webster et al. (2010) [33] to determine adult worm and larval stage species identity, is not robust when snail DNA is present, particularly for S. haematobium infections. The cox1 RD-PCR was suggested as a secondary screening method by Schols et al. (2019) [27], but it was only theoretically examined as part of that study. Clearly, further ‘wet lab’ testing on infected snails is needed. In regions where Schistosoma hybridization occurs, mitochondrial DNA analysis would be necessary since both nuclear and mitochondrial DNA are required for hybrid identification [33]. Unfortunately, as with most diagnostics, there is a balance between sensitivity and specificity, with sensitivity increasing and specificity decreasing, usually due to the nature of the biomedical targets. Here, rapid screening with high sensitivity is a priority due to the low levels of infections in our study sites, with secondary species-specific screening only required on a subset of samples that are identified as infected. Moreover, Zanzibar previously thought to be a S. haematobium transmission only zone, although, with the recent report of S. bovis transmission [6], the additional species-specific screening is warranted. However, the need for the secondary screening step for Schistosoma species identification does need further exploration, such as trialing more direct methods that negate DNA sequencing, for example, amplicon enzyme restriction digestion demonstrated in Kane et al. (2013) [54]. However, it is also important to gather detailed information, as is obtained through DNA sequencing and analysis [6,7]. It is likely that xenomonitoring methods may need to be adapted to optimize focal surveillance strategies to specific endemic zones due to geographical genetic differences of the target organisms and potential unidentified species.
4. Materials and Methods
4.1. Primer Selection and In Silico Evaluation
The universal primer pair, ETTS2 and ETTS1 (Table 2 and Figure 7), was selected for the development of the internal control for the assay. They anneal to conserved flanking regions either side of the ITS(1 + 2) rDNA region of Schistosoma spp., amplifying the full ITS rDNA regions, resulting in an amplicon of ~1005 bp [6,7,31,33]. These primers have also been demonstrated to amplify the full ITS rDNA region of other organisms, including intermediate gastropod hosts. Primer’s cross-reactivity with the target Bulinus snail hosts was further confirmed through alignments of the ETTS2 and ETTS1 primers with B. globosus and B. nasutus rDNA regions, available from ongoing projects (Briscoe et al. unpublished data, Pennance et al. unpublished data).

Table 2.
Details of the primers selected for the development of the xenomonitoring assay. Universal (U) and specific (S) denote whether the primers universally target both Schistosoma and snail or just specifically target Schistosoma DNA.
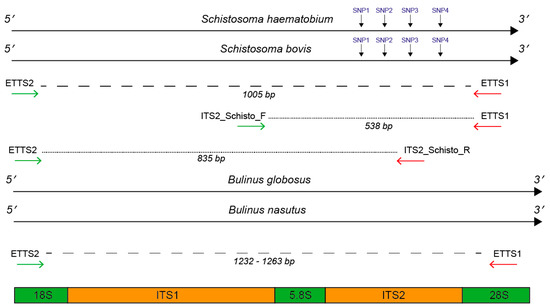
Figure 7.
Primer annealing positions flanking and internal to the ITS1 + 2 rDNA targets. Primer positions were mapped to Schistosoma haematobium and S. bovis ITS1 + 2 reference data [59] and to a Bulinus globosus and B. nasutus DNA reference (Pennance et al., unpublished data). For Schistosoma DNA, the primer combinations produced two fragments; 1) ETTS2–ETTS1 (1005 bp) and either 2) ITS2_Schisto_F-ETTS1 (538 bp) or 3) ITS2_Schisto_R-ETTS2 (835 bp). For Bulinus DNA, the primer combinations produced one fragment, ranging in size between 1232 and 1263 due to interspecies variation. For Schistosoma species identification, four SNPs were present at bp positions 90, 145, 195, and 265 in the ITS2 rDNA region, allowing differentiation of S. haematobium and S. bovis following ITS2 sequencing.
To develop the Schistosoma-specific target, two Schistosoma specific primers (ITS2_Schisto_F and ITS2_Schisto_R), published by Schols et al. (2019) [27], were selected, targeting the internal ITS1 and ITS2 rDNA regions of Schistosoma (Figure 7). These were further tested in silico for specificity by stringently aligning them with rDNA sequence data (Briscoe et al., unpublished data; Pennance et al., unpublished data) of a single B. globosus and B. nasutus from both Unguja and Pemba island (Zanzibar, United Republic of Tanzania) and those previously published for Schistosoma spp. [59,60].
All alignments were performed using Sequencher v5.4.6 (Gene Codes Corporation, Michigan, USA), and the primer positions were used to predict the specific amplicon sizes that would result, following amplification of snail and schistosome DNA using the different primer combinations of ETTS1, ETTS2, ITS2_Schisto_F, and ITS2_Schisto_R.
4.2. Bulinus and Schistosoma Genomic DNA Extractions
Whole soft tissue from Bulinus samples (as detailed below) available through the Schistosomiasis Collection at the Natural History Museum (SCAN) [61] and other ongoing projects, including laboratory and field samples, infected/non-infected and patent/non-patent, were used for the assay development and validation. Genomic DNA (gDNA) from all Bulinus samples was extracted using a modified tissue lysis protocol [6]. Two kits were then used to extract total gDNA from the lysed snail tissue—the BioSprint 96 DNA Blood Kit (Qiagen, Manchester, UK) for high-throughput multiple sample processing, and the DNeasy Blood & Tissue Kit (Qiagen, Manchester, UK) for single sample processing. Protocols were carried out according to the manufacturer’s instructions.
Positive control Schistosoma gDNA was obtained from adult worms; S. haematobium (single female worm from Zanzibar) and S. bovis (single male worm from Senegal) were available from SCAN. DNA was extracted following the DNeasy Blood & Tissue Kit protocol according to manufacturer’s instructions (Qiagen, Manchester, UK) [60].
4.3. PCR Conditions, Amplicon Visualization, and Sequencing
All PCR amplifications were performed in 25 µL reactions using illustraTM PuReTaq Ready-To-GoTM PCR Beads (GE Healthcare, UK) with 1 µL of each primer, in their different combinations, as stated in each section, at a concentration of 10 µM. gDNA templates (Schistosoma and/or Bulinus sp.) were added at different volumes and concentrations, as detailed below. The PCR cycling conditions for all multiplex and singleplex reactions were as follows: initial denaturation 5 min at 95 °C, followed by 40 cycles of 30 s at 95 °C, 30 s at 58 °C (unless stated otherwise), 90 s at 72 °C, and a final extension of 10 min at 72 °C. The visualization of all PCR products was performed by running 7.5 µL of each PCR product, mixed with 2 µL of Bioline 5x DNA Loading Buffer Blue (London, UK) and GelRed for visualization under UV light, on a 2% agarose gel for 90 min at 90 V. HyperLadder I and HyperLadder IV were run alongside the PCR amplicons to assess fragment sizes. Gels were visualized using a GBOX-Chemi-XRQ gel documentation system (Syngene, Cambridge, UK).
To validate amplification specificity, selected PCR amplicons from multiplex PCRs, where multiple amplicons were present, were cut from agarose gels and sequenced following purification using the QiaQuick Gel Purification Kit (Qiagen, Manchester, UK) following manufacturer’s instructions. For singleplex reactions, resulting in a single amplicon, PCR products were purified using the QiaQuick PCR Purification Kit (Qiagen, Manchester, UK) following the manufacturer’s instructions. The amplicons were sequenced in both directions using dilutions of the PCR primers. The sequence data were manually edited using Sequencher v5.4.6 (Gene Codes Corporation, Michigan, USA), and the amplicon identification was confirmed by comparison to Schistosoma reference data [59] and by BLAST analysis (BLAST: Basic Local Alignment Search Tool, NCBI).
4.4. In Vitro Primer Testing and Assay Validation
All gDNA extractions from laboratory-bred Bulinus wrighti (not exposed to any trematodes and, therefore, negative for infection) and from the S. haematobium and S. bovis adult worms were quantified using a Qubit® Fluorometer using the dsDNA Broad Range (BR) Assay Kit (Molecular Probes, Life Technologies). The gDNA extracts from the single adult S. haematobium and S. bovis worms were normalized, using nuclease-free water, to 2 ng/µL (± 0.05 ng/µL). The gDNA extract of a B. wrighti snail control was recorded and kept at 31.3 ng/µL. Template gDNA (1 µL) was used in each PCR separately or combined and used to test the different primer combinations (shown in Figure 1). The primers were tested as singleplex PCRs for the internal control (ETTS2 + ETTS1), targeting both snail and Schistosoma gDNA, and then as multiplex PCR’s incorporating each of the internal Schistosoma-specific primers (ETTS2 + ETTS1 + ITS2_Schisto_F or ITS2_Schisto_R). All test PCRs were initially performed at an annealing temperature of 55 °C.
The multiplex primer combination ETTS2 + ITS2_Schisto_F + ETTS1 was selected and taken forward for further development and validation. This is referred to as the multiplex ITS xenomonitoring (MIX) PCR. The MIX PCR was further tested at annealing temperatures of 58 °C and 60 °C to enhance assay specificity, with 58 °C taken forward for further experiments. Additionally, a secondary Schistosoma ITS xenomonitoring (SIX) PCR, incorporating just the Schistosoma-specific primer (ITS2_Schisto_F) and its universal reverse primer (ETTS1), was validated, targeting just the 538 bp Schistosoma DNA amplicon. The SIX PCR was developed to obtain more targeted schistosome species data amplicon sequence analysis, of positive samples, following an initial high-throughput screening of snail populations with the multiplex PCR, which incorporates the internal snail control.
4.5. Sensitivity Testing
The analytical sensitivity and LoD for Schistosoma DNA in the MIX PCR were performed using serial dilutions of S. haematobium and S. bovis gDNA. The S. haematobium and S. bovis gDNA, normalized to 2 ng/µL (± 0.05 ng/µL), was diluted using nuclease-free water by one in ten (0.2 ng/µL), one in one hundred (0.02 ng/µL), and one in one thousand (0.002 ng/µL). 1 µL of each Schistosoma gDNA dilution was used in each multiplex PCR together with 1 µL of the B. wrighti gDNA (31.3 ng/µL).
Sensitivity was tested using controlled laboratory snail infections. Infections were performed by the Schistosomiasis Resource Center (SRC) (Biomedical Research Institute, Maryland, USA [62]) using their B. truncatus/S. haematobium (Egyptian strain) model lifecycle system. Juvenile B. truncatus (2–3 mm, n = 133) was divided into three groups, with individual snails in each group being exposed to either 1, 2, or several (~7) S. haematobium miracidia, respectively (Table 3). Miracidia, hatched in freshwater from eggs collected from S. haematobium-infected male LVG Syrian golden hamsters (see Ethical Statement), were added to individual wells of 24-well ELISA plates containing the B. truncatus snails. A fine-tipped Pasteur pipette was used under a dissection microscope to capture and deliver either an individual miracidium (for 1 and 2 miracidia exposures) or several (~7) miracidia at a time, following the standard operating procedures (SOPs) conducted at SRC (see: https://www.afbr-bri.org/schistosomiasis/standard-operating-procedures/).

Table 3.
Groups of Bulinus truncatus (B.t.) experimentally challenged with either 1, 2, or ~7 S. haematobium (S.h.) miracidia and preserved 24 h post-exposure or checked for patent S.h. infections and preserved 11 weeks (wks) post-exposure.
The snails were kept in their individual wells until no miracidia were observed swimming under a binocular microscope, assumed to have penetrated the snail (~2 h). Following 24 h after initial exposure to the miracidia, half of each infection group was preserved in 100% ethanol for molecular analysis. The remaining exposed B. truncatus were maintained in their separate infection groups for 11 weeks to allow the infections to mature, and since this was the first opportunity to conduct sampling of the infected snails. Snails were maintained according to the SRC’s SOP’s (see above). Snails that died were recorded and promptly removed from the group. At 11 weeks post-exposure, the remaining snails were individually induced to shed cercariae by exposure to freshwater and light. Once it had been determined if the snails were infected and patent, they were washed, to remove any cercariae, and preserved in 100% ethanol for molecular analysis.
The MIX PCR was performed using gDNA (1 µL) extracted from six individual B. truncatus from each group, which were preserved after 24 h—two non-patent snails from group 1 and 2, and one non-patent snail from group 3 (11 weeks post-exposure), and one patent (shedding) snail from group 2 (11 weeks post-exposure) (Table 3). The secondary SIX PCR was performed on selected Schistosoma positive samples, to amplify the 538 bp S. haematobium-specific amplicon for sequence analysis to confirm that the MIX PCR was not a false-positive.
4.6. Specificity Testing and Validation on Field Samples
As part of a longitudinal xenomonitoring project on Pemba in relation to urogenital schistosomiasis transmission [6], the ‘wild-caught’ B. globosus and B. nasutus field isolates were available for further validation of the MIX assay. Individual snails had been collected during malacological surveys, individually checked for patent trematode infections by cercarial shedding, and then preserved in 100% ethanol for molecular analysis [6]. Cercariae from infected B. globosus were preserved on Whatman FTA cards and identified using molecular methods as S. haematobium or S. bovis from two and five snails, respectively [6]. In addition, individual B. globosus and B. nasutus (also collected from Pemba), which were shedding two other trematode species, Euclinostomum sp. and Echinostoma sp., respectively (unpublished data), were tested to investigate assay specificity. Additionally, 94 B. globosus snails from Wambaa (Pemba) collected during November 2018, which were not shedding any trematode cercariae, were tested for infections by PCR.
All samples, which gave the 538 bp Schistosoma-specific amplicon (Figure 7), were further subjected to the SIX PCR assay with the resulting amplicons purified and sequenced to confirm the species of the infection. The identity of the S. haematobium and S. bovis species was confirmed by analysis of the four species’ SNPs that exist in the ITS2 region [7] between S. haematobium and S. bovis (Table 1).
4.7. Testing the Schistosoma cox1 Rapid-Diagnostic PCR (RD-PCR) for Secondary Species Identification
The patent B. globosus snails collected from Pemba shedding either S. haematobium (n = 2) or S. bovis (n = 5) (see [6]), as detailed above, were further tested using the published multiplex RD-PCR (see [27,33]) with an aim to provide a secondary species-specific screening method, as described in Schols et al. (2019) [27]. This multiplex RD-PCR, capable of differentiating S. bovis and S. haematobium by species-specific amplicon size (S. haematobium (543 bp), S. bovis (306 bp)), was performed following the published protocol and cycling conditions described by Webster et al. (2010) [33]. Different amount of gDNA (1 µL, 2 µL, and 3 µL) and PCR annealing temperatures (58 °C, 62 °C, and 65 °C) were trialed to investigate sensitivity and specificity. The amplicons were purified, and Sanger sequenced, as described above, using the species-specific reverse primers to confirm species/amplicon identification.
4.8. Ethical Statement
Schistosoma haematobium experimental infections were conducted at the Biomedical Research Institute – Schistosomiasis Resource Center (Rockville, MA, USA) animal facility maintained with AAALAC full accreditation (Site # 000779), operating under the National Institutes of Health’s Office of Laboratory Animal Welfare (OLAW) # A3080-01. S. haematobium parasite material was collected from male LVG Syrian golden hamsters following percutaneous exposure to cercariae. Hamster’s use was approved by the Institutional Animal Care and Use Committee (IACUC) of the Biomedical Research Institute for the Animal Use Protocol, #18-01.
Author Contributions
Conceptualization, T.P., S.M.A. (Said Mohammed Ali), S.M.A. (Shaali Makame Ame), and B.L.W.; Data curation, T.P., F.A., and B.L.W.; Formal analysis, T.P. and B.L.W.; Funding acquisition, B.L.W.; Investigation, T.P., J.A., E.B.L., P.R., F.L., A.K.A., K.R.S., S.L., and B.L.W.; Methodology, T.P., J.A., S.L., J.C., and B.L.W.; Project administration, T.P., S.M.A. (Said Mohammed Ali), S.M.A. (Shaali Makame Ame), and B.L.W.; Resources, T.P.; Supervision, S.M.A. (Said Mohammed Ali), J.C., S.K., S.M.A. (Shaali Makame Ame) and B.L.W.; Validation, T.P., J.A., D.R., F.A. and B.L.W.; Visualization, T.P.; Writing—original draft, T.P.; Writing—review and editing, J.A., E.L., D.R., J.C., S.K., F.A., and B.L.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by a Wellcome Trust Seed Award, grant number 207728 awarded to Dr. Bonnie Lee Webster. The APC was funded by Wellcome Trust Seed Award in line with their open access policies. T.P. was supported by the NERC GW4+ DTP and the Natural Environmental Research Council (NE/L002434/1). F.A. was supported by the Wellcome Trust on the SCAN project (104958/Z/14/Z). SK is funded by a PRIMA award from the Swiss National Science Foundation (PR00P3_179753).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Colley, D.G.; Secor, W.E. Immunology of human schistosomiasis. Parasite Immunol. 2014, 36, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.S. Freshwater Snails of Africa and Their Medical Importance, 1st ed.; Taylor & Francis Ltd.: London, UK, 1994. [Google Scholar]
- WHO Schistosomiasis Fact Sheets. Available online: https://www.who.int/news-room/fact-sheets/detail/schistosomiasis (accessed on 27 April 2020).
- De Bont, J.; Vercruysse, J. Schistosomiasis in Cattle. Adv. Parasitol. 1998, 41, 285–364. [Google Scholar] [CrossRef] [PubMed]
- De Bont, J.; Vercruysse, J. The epidemiology and control of cattle schistosomiasis. Parasitol. Today 1997, 13, 255–262. [Google Scholar] [CrossRef]
- Pennance, T.; Ame, S.M.; Amour, A.K.; Suleiman, K.R.; Allan, F.; Rollinson, D.; Webster, B.L. Occurrence of Schistosoma bovis on Pemba Island, Zanzibar: Implications for urogenital schistosomiasis transmission monitoring. Parasitology 2018, 145, 1727–1731. [Google Scholar] [CrossRef] [PubMed]
- Pennance, T.; Allan, F.; Emery, A.; Rabone, M.; Cable, J.; Garba, A.D.; Hamidou, A.A.; Webster, J.P.; Rollinson, D.; Webster, B.L. Interactions between Schistosoma haematobium group species and their Bulinus spp. intermediate hosts along the Niger River Valley. Parasites Vectors 2020, 13, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Guidelines for the Evaluation of Soil-Transmitted Helminths and Schistosomiasis at Community Level. Available online: https://apps.who.int/iris/handle/10665/63821 (accessed on 23 July 2020).
- Ending the Neglect to Attain the Sustainable Development Goals: A Road Map for Neglected Tropical Diseases 2021–2030. Available online: https://www.who.int/neglected_diseases/Ending-the-neglect-to-attain-the-SDGs--NTD-Roadmap.pdf?ua=1 (accessed on 23 July 2020).
- Mutsaka-Makuvaza, M.J.; Zingoni, Z.M.; Tshuma, C.; Ray, S.; Zhou, X.-N.; Webster, B.L.; Midzi, N. Reinfection of urogenital schistosomiasis in pre-school children in a highly endemic district in Northern Zimbabwe: A 12 months compliance study. Infect. Dis. Poverty 2018, 7, 102. [Google Scholar] [CrossRef]
- Tchuente, L.-A.T.; Rollinson, D.; Stothard, J.R.; Molyneux, D.H. Moving from control to elimination of schistosomiasis in sub-Saharan Africa: Time to change and adapt strategies. Infect. Dis. Poverty 2017, 6, 42. [Google Scholar] [CrossRef]
- King, C.H.; Sturrock, R.F.; Kariuki, H.C.; Hamburger, J. Transmission control for schistosomiasis—Why it matters now. Trends Parasitol. 2006, 22, 575–582. [Google Scholar] [CrossRef]
- Rollinson, D.; Knopp, S.; Levitz, S.; Stothard, J.R.; Tchuenté, L.-A.T.; Garba, A.; Mohammed, K.A.; Schur, N.; Person, B.; Colley, D.G.; et al. Time to set the agenda for schistosomiasis elimination. Acta Trop. 2013, 128, 423–440. [Google Scholar] [CrossRef]
- Secor, W.E.; Colley, D.G. When Should the Emphasis on Schistosomiasis Control Move to Elimination? Trop. Med. Infect. Dis. 2018, 3, 85. [Google Scholar] [CrossRef]
- Pennance, T.; Person, B.; Muhsin, M.A.; Khamis, A.N.; Muhsin, J.; Khamis, I.S.; Mohammed, K.A.; Kabole, F.; Rollinson, D.; Knopp, S. Urogenital schistosomiasis transmission on Unguja Island, Zanzibar: Characterisation of persistent hot-spots. Parasites Vectors 2016, 9, 646. [Google Scholar] [CrossRef] [PubMed]
- Bayssade-Dufour, C. Chétotaxies cercariennes comparées de dix espèces de Schistosomes. Ann. Parasitol. Hmm. Comp. 1982, 57, 467–485. [Google Scholar] [CrossRef]
- Bayssade-Dufour, C.; Cabaret, J.; Ngendahayo, L.; Albaret, J.-L.; Carrat, C.; Chabaud, A. Identification of schistosoma haematobium, S. bovis and S. curassoni by multivariate analysis of cercarial papillae indices. Int. J. Parasitol. 1989, 19, 839–846. [Google Scholar] [CrossRef]
- Cunningham, L.J.; Lingley, J.K.; Haines, L.R.; Ndung’U, J.M.; Torr, S.J.; Adams, E.R. Illuminating the Prevalence of Trypanosoma brucei s.l. in Glossina Using LAMP as a Tool for Xenomonitoring. PLoS Negl. Trop. Dis. 2016, 10, e0004441. [Google Scholar] [CrossRef] [PubMed]
- Garrod, G.; Adams, E.R.; Lingley, J.K.; Saldanha, I.; Torr, S.J.; Cunningham, L.J. Identification of Trypanosoma brucei gambiense and T. b. rhodesiense in vectors using multiplexed high-resolution melt analysis. bioRxiv 2020. Available online: https://www.biorxiv.org/content/10.1101/2020.04.21.052928v1 (accessed on 22 April 2020). [CrossRef]
- Cook, D.A.; Pilotte, N.; Minetti, C.; Williams, S.A.; Reimer, L. A superhydrophobic cone to facilitate the xenomonitoring of filarial parasites, malaria, and trypanosomes using mosquito excreta/feces. Gates Open Res. 2018, 1, 1. [Google Scholar] [CrossRef]
- Minetti, C.; LaCourse, E.J.; Reimer, L.; Stothard, J.R. Focusing nucleic acid-based molecular diagnostics and xenomonitoring approaches for human helminthiases amenable to preventive chemotherapy. Parasitology 2016, 2, 16. [Google Scholar] [CrossRef]
- Rathinasamy, V.; Hosking, C.; Tran, L.; Kelley, J.; Williamson, G.; Swan, J.; Elliott, T.; Rawlin, G.; Beddoe, T.; Spithill, T.W. Development of a multiplex quantitative PCR assay for detection and quantification of DNA from Fasciola hepatica and the intermediate snail host, Austropeplea tomentosa, in water samples. Vet. Parasitol. 2018, 259, 17–24. [Google Scholar] [CrossRef]
- Hamburger, J.; Hoffman, O.; Kariuki, H.C.; Muchiri, E.M.; King, C.H.; Ouma, J.H.; Koech, D.K.; Sturrock, R.F. Large-Scale, Polymerase Chain Reaction–Based Surveillance Of Schistosoma Haematobium DNA In Snails from Transmission Sites in Coastal Kenya: A New Tool for Studying the Dynamics of Snail Infection. Am. J. Trop. Med. Hyg. 2004, 71, 765–773. [Google Scholar] [CrossRef]
- Lu, L.; Zhang, S.-M.; Mutuku, M.W.; Mkoji, G.M.; Loker, E.S. Relative compatibility of Schistosoma mansoni with Biomphalaria sudanica and B. pfeifferi from Kenya as assessed by PCR amplification of the S. mansoni ND5 gene in conjunction with traditional methods. Parasites Vectors 2016, 9, 166. [Google Scholar] [CrossRef]
- Allan, F.; Rollinson, D.; Smith, J.; Dunn, A.M. Host choice and penetration bySchistosoma haematobiummiracidia. J. Helminthol. 2009, 83, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Allan, F.; Dunn, A.M.; Emery, A.M.; Stothard, J.R.; Johnston, D.A.; Kane, R.A.; Khamis, A.N.; Mohammed, K.A.; Rollinson, D. Use of sentinel snails for the detection of Schistosoma haematobium transmission on Zanzibar and observations on transmission patterns. Acta Trop. 2013, 128, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Schols, R.; Carolus, H.; Hammoud, C.; Mulero, S.; Mudavanhu, A.; Huyse, T. A rapid diagnostic multiplex PCR approach for xenomonitoring of human and animal schistosomiasis in a ‘One Health’ context. Trans. R. Soc. Trop. Med. Hyg. 2019, 113, 722–729. [Google Scholar] [CrossRef]
- Casotti, M.O.; Gryschek, R.C.B.; De Paula, F.M.; Gomes-Gouvêa, M.; Pinho, J.R.R.; Tuan, R.; Dias-Neto, E.; Luna, E.J.D.A.; Espírito-Santo, M.C.C.D. Molecular detection of prepatent Schistosoma mansoni infection in Biomphalaria glabrata snail vectors. Rev. Inst. Med. Trop. Sao Paulo 2020, 62, e17. [Google Scholar] [CrossRef] [PubMed]
- Kaiglová, A.; Changoma, M.J.S.; Špajdelová, J.; Jakubcová, D.; Bírová, K. Urinary schistosomosis in patients of rural medical health centers in Kwale county, Kenya. J. Helminthol. 2020, 57, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Abbasi, I.; Webster, B.L.; King, C.H.; Rollinson, D.; Hamburger, J. The substructure of three repetitive DNA regions of Schistosoma haematobium group species as a potential marker for species recognition and interbreeding detection. Parasites Vectors 2017, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kane, R.A.; Rollinson, D. Repetitive sequences in the ribosomal DNA internal transcribed spacer of Schistosoma haematobium, Schistosoma intercalatum and Schistosoma mattheei. Mol. Biochem. Parasitol. 1994, 63, 153–156. [Google Scholar] [CrossRef]
- Webster, B.L. Isolation and preservation of schistosome eggs and larvae in RNAlater® facilitates genetic profiling of individuals. Parasites Vectors 2009, 2, 50. [Google Scholar] [CrossRef]
- Webster, B.L.; Rollinson, D.; Stothard, J.R.; Huyse, T. Rapid diagnostic multiplex PCR (RD-PCR) to discriminate Schistosoma haematobium and S. bovis. J. Helminthol. 2009, 84, 107–114. [Google Scholar] [CrossRef]
- Schmaedick, M.A.; Koppel, A.L.; Pilotte, N.; Torres, M.; Williams, S.A.; Dobson, S.L.; Lammie, P.J.; Won, K.Y. Molecular Xenomonitoring Using Mosquitoes to Map Lymphatic Filariasis after Mass Drug Administration in American Samoa. PLoS Neglected Trop. Dis. 2014, 8, e3087. [Google Scholar] [CrossRef]
- Pilotte, N.; Unnasch, T.R.; Williams, S.A. The Current Status of Molecular Xenomonitoring for Lymphatic Filariasis and Onchocerciasis. Trends Parasitol. 2017, 33, 788–798. [Google Scholar] [CrossRef] [PubMed]
- Pilotte, N.; Zaky, W.I.; Abrams, B.P.; Chadee, D.D.; Williams, S.A. A Novel Xenomonitoring Technique Using Mosquito Excreta/Feces for the Detection of Filarial Parasites and Malaria. PLoS Neglected Trop. Dis. 2016, 10, e0004641. [Google Scholar] [CrossRef] [PubMed]
- Caron, Y.; Righi, S.; Lempereur, L.; Saegerman, C.; Losson, B. An optimized DNA extraction and multiplex PCR for the detection of Fasciola sp. in lymnaeid snails. Vet. Parasitol. 2011, 178, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Cucher, M.A.; Carnevale, S.; Prepelitchi, L.; Labbé, J.H.; Wisnivesky-Colli, C. PCR diagnosis of Fasciola hepatica in field-collected Lymnaea columella and Lymnaea viatrix snails. Vet. Parasitol. 2006, 137, 74–82. [Google Scholar] [CrossRef]
- Kozak, M.; Wedrychowicz, H. The performance of a PCR assay for field studies on the prevalence of Fasciola hepatica infection in Galba truncatula intermediate host snails. Vet. Parasitol. 2010, 168, 25–30. [Google Scholar] [CrossRef]
- Magalhães, K.G.; Passos, L.K.J.; Carvalho, O.D.S. Detection of Lymnaea columella infection by Fasciola hepatica through Multiplex-PCR. Mem. Instit. Oswaldo Cruz 2004, 99, 421–424. [Google Scholar] [CrossRef]
- Mostafa, O.M.S.; A Taha, H.; Ramadan, G. Diagnosis of Fasciola gigantica in snail using the polymerase chain reaction (PCR) assay. J. Egypt. Soc. Parasitol. 2003, 33, 733–742. [Google Scholar]
- Velusamy, R.; Singh, B.; Raina, O. Detection of Fasciola gigantica infection in snails by polymerase chain reaction. Vet. Parasitol. 2004, 120, 85–90. [Google Scholar] [CrossRef]
- Kaplan, R.M.; Dame, J.; Reddy, G.; Courtney, C. A repetitive DNA probe for the sensitive detection of Fasciola hepatica infected snails. Int. J. Parasitol. 1995, 25, 601–610. [Google Scholar] [CrossRef]
- Carolus, H.; Muzarabani, K.C.; Hammoud, C.; Schols, R.; Volckaert, F.A.; Barson, M.; Huyse, T. A cascade of biological invasions and parasite spillback in man-made Lake Kariba. Sci. Total Environ. 2018, 659, 1283–1292. [Google Scholar] [CrossRef]
- Born-Torrijos, A.; Poulin, R.; Raga, J.A.; Holzer, A.S. Estimating trematode prevalence in snail hosts using a single-step duplex PCR: How badly does cercarial shedding underestimate infection rates? Parasites Vectors 2014, 7, 243. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, T.; Furushima-Shimogawara, R.; Ohmae, H.; Wang, T.-P.; Lu, S.; Chen, R.; Wen, L.; Ohta, N. Detection of Early and Single Infections of Schistosoma japonicum in the Intermediate Host Snail, Oncomelania hupensis, by PCR and Loop-Mediated Isothermal Amplification (LAMP) Assay. Am. J. Trop. Med. Hyg. 2010, 83, 542–548. [Google Scholar] [CrossRef] [PubMed]
- Tong, Q.; Chen, R.; Zhang, Y.; Yang, G.-J.; Kumagai, T.; Furushima-Shimogawara, R.; Lou, D.; Yang, K.; Wen, L.-Y.; Lu, S.-H.; et al. A new surveillance and response tool: Risk map of infected Oncomelania hupensis detected by Loop-mediated isothermal amplification (LAMP) from pooled samples. Acta Trop. 2015, 141, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Hamburger, J.; Ruppel, A.; Jourdane, J.; Na, H.; Ramzy, R.M.; Xin, X.Y. A polymerase chain reaction assay for detecting snails infected with bilharzia parasites (Schistosoma mansoni) from very early prepatency. Am. J. Trop. Med. Hyg. 1998, 59, 872–876. [Google Scholar] [CrossRef]
- Melo, F.L.; Gomes, A.L.D.V.; Barbosa, C.S.; Werkhäuser, R.P.; Abath, F.G. Development of molecular approaches for the identification of transmission sites of schistosomiasis. Trans. R. Soc. Trop. Med. Hyg. 2006, 100, 1049–1055. [Google Scholar] [CrossRef]
- Fernández-Soto, P.; Arahuetes, J.G.; Hernández, A.S.; López-Abán, J.; Santiago, B.V.; Muro, A. A Loop-Mediated Isothermal Amplification (LAMP) Assay for Early Detection of Schistosoma mansoni in Stool Samples: A Diagnostic Approach in a Murine Model. PLoS Neglected Trop. Dis. 2014, 8, e3126. [Google Scholar] [CrossRef]
- Gandasegui, J.; Fernández-Soto, P.; Hernández-Goenaga, J.; López-Abán, J.; Vicente, B.; Muro, A. Biompha-LAMP: A New Rapid Loop-Mediated Isothermal Amplification Assay for Detecting Schistosoma mansoni in Biomphalaria glabrata Snail Host. PLoS Neglected Trop. Dis. 2016, 10, e0005225. [Google Scholar] [CrossRef]
- Abbasi, I.; King, C.H.; Muchiri, E.M.; Hamburger, J. Detection of Schistosoma mansoni and Schistosoma haematobium DNA by Loop-Mediated Isothermal Amplification: Identification of Infected Snails from Early Prepatency. Am. J. Trop. Med. Hyg. 2010, 83, 427–432. [Google Scholar] [CrossRef]
- Caldeira, R.L.; Jannotti-Passos, L.K.; Carvalho, O.D.S. Use of Molecular Methods for the Rapid Mass Detection of Schistosoma mansoni (Platyhelminthes: Trematoda) in Biomphalariaspp. (Gastropoda: Planorbidae). J. Trop. Med. 2017, 2017, 1–6. [Google Scholar] [CrossRef]
- Kane, R.A.; Stothard, J.R.; Rollinson, D.; Leclipteux, T.; Evraerts, J.; Standley, C.J.; Allan, F.; Betson, M.; Kaba, R.; Mertens, P.; et al. Detection and quantification of schistosome DNA in freshwater snails using either fluorescent probes in real-time PCR or oligochromatographic dipstick assays targeting the ribosomal intergenic spacer. Acta Trop. 2013, 128, 241–249. [Google Scholar] [CrossRef]
- Hamburger, J.; Ruppel, A.; Ramzy, R.M.; Na, H.; Jourdane, J.; Abbasi, I. Polymerase chain reaction assay based on a highly repeated sequence of Schistosoma haematobium: A potential tool for monitoring schistosome-infested water. Am. J. Trop. Med. Hyg. 2001, 65, 907–911. [Google Scholar] [CrossRef] [PubMed]
- Rudko, S.P.; Reimink, R.L.; Froelich, K.; Gordy, M.A.; Blankespoor, C.L.; Hanington, P.C. Use of qPCR-Based Cercariometry to Assess Swimmer’s Itch in Recreational Lakes. EcoHealth 2018, 15, 827–839. [Google Scholar] [CrossRef] [PubMed]
- Rudko, S.P.; Reimink, R.L.; Peter, B.; White, J.; Hanington, P.C. Democratizing water monitoring: Implementation of a community-based qPCR monitoring program for recreational water hazards. PLoS ONE 2020, 15, e0229701. [Google Scholar] [CrossRef] [PubMed]
- Rudko, S.P.; Turnbull, A.; Reimink, R.L.; Froelich, K.; Hanington, P.C. Species-specific qPCR assays allow for high-resolution population assessment of four species avian schistosome that cause swimmer’s itch in recreational lakes. Int. J. Parasitol. Parasites Wildl. 2019, 9, 122–129. [Google Scholar] [CrossRef]
- Webster, B.L.; Culverwell, C.L.; Khamis, I.S.; Mohammed, K.A.; Rollinson, D.; Stothard, J.R. DNA barcoding of Schistosoma haematobium on Zanzibar reveals substantial genetic diversity and two major phylogenetic groups. Acta Trop. 2013, 128, 206–217. [Google Scholar] [CrossRef]
- Webster, B.L.; Emery, A.M.; Webster, J.P.; Gouvras, A.N.; Garba, A.; Diaw, O.; Seye, M.M.; Tchuente, L.A.T.; Simoonga, C.; Mwanga, J.; et al. Genetic Diversity within Schistosoma haematobium: DNA Barcoding Reveals Two Distinct Groups. PLoS Neglected Trop. Dis. 2012, 6, e1882. [Google Scholar] [CrossRef]
- Emery, A.M.; Allan, F.; Rabone, M.; Rollinson, D. Schistosomiasis collection at NHM (SCAN). Parasites Vectors 2012, 5, 185. [Google Scholar] [CrossRef]
- Lewis, F.A.; Liang, Y.-S.; Raghavan, N.; Knight, M. The NIH-NIAID Schistosomiasis Resource Center. PLOS Neglected Trop. Dis. 2008, 2, e267. [Google Scholar] [CrossRef]
Sample Availability: Samples (DNA extracts of snails and parasites) are available from the authors upon appropriate request. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).