Identification of Potential Chemical Substrates as Fuel for Hypoxic Tumors That May Be Linked to Invadopodium Formation in Hypoxia-Induced MDA-MB-231 Breast-Cancer Cell Line
Abstract
1. Introduction
2. Results
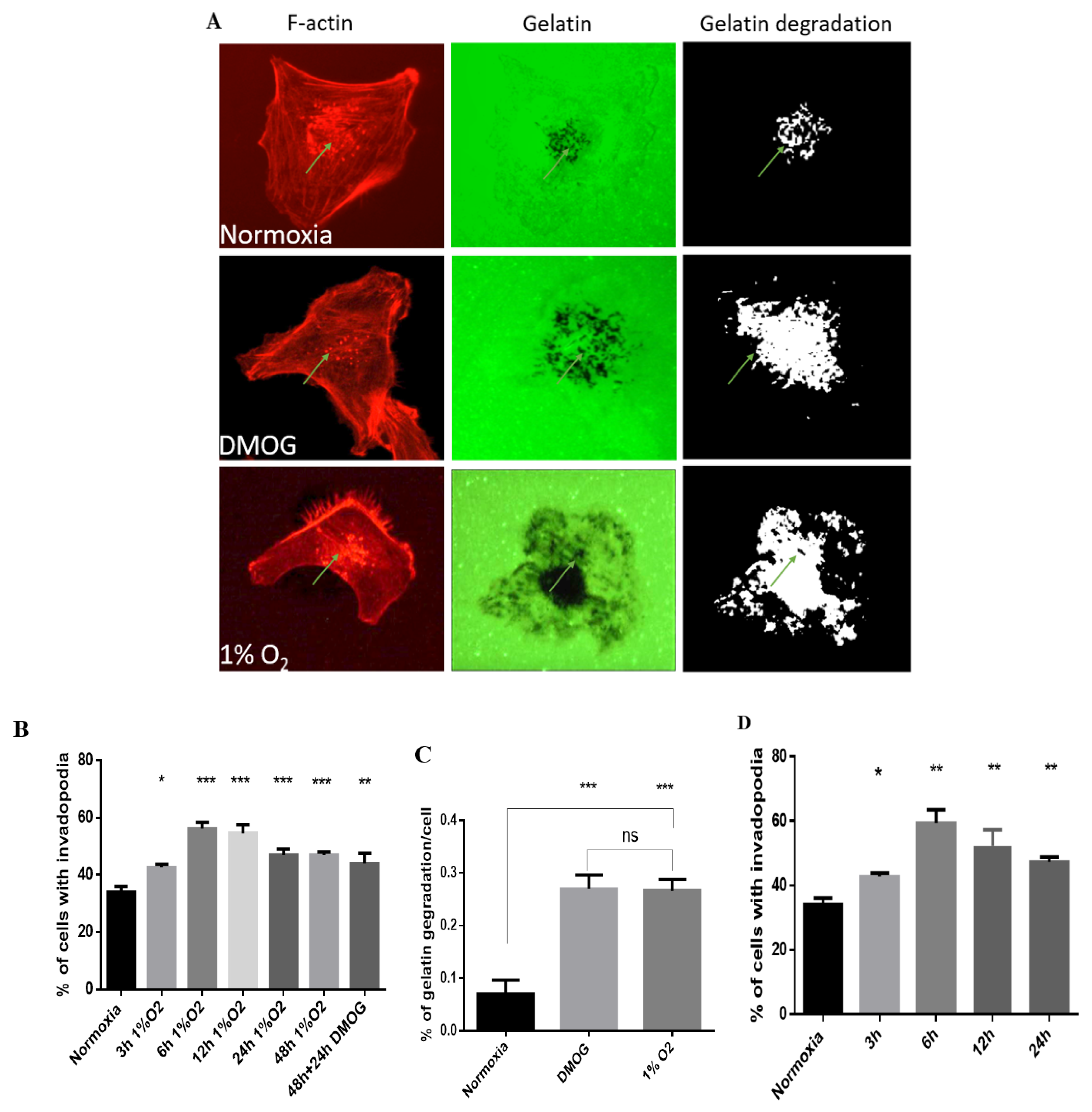
2.1. Hypoxia Induction Using Hypoxia Chamber and DMOG Treatment to Visualize and Quantify Invadopodia Formation
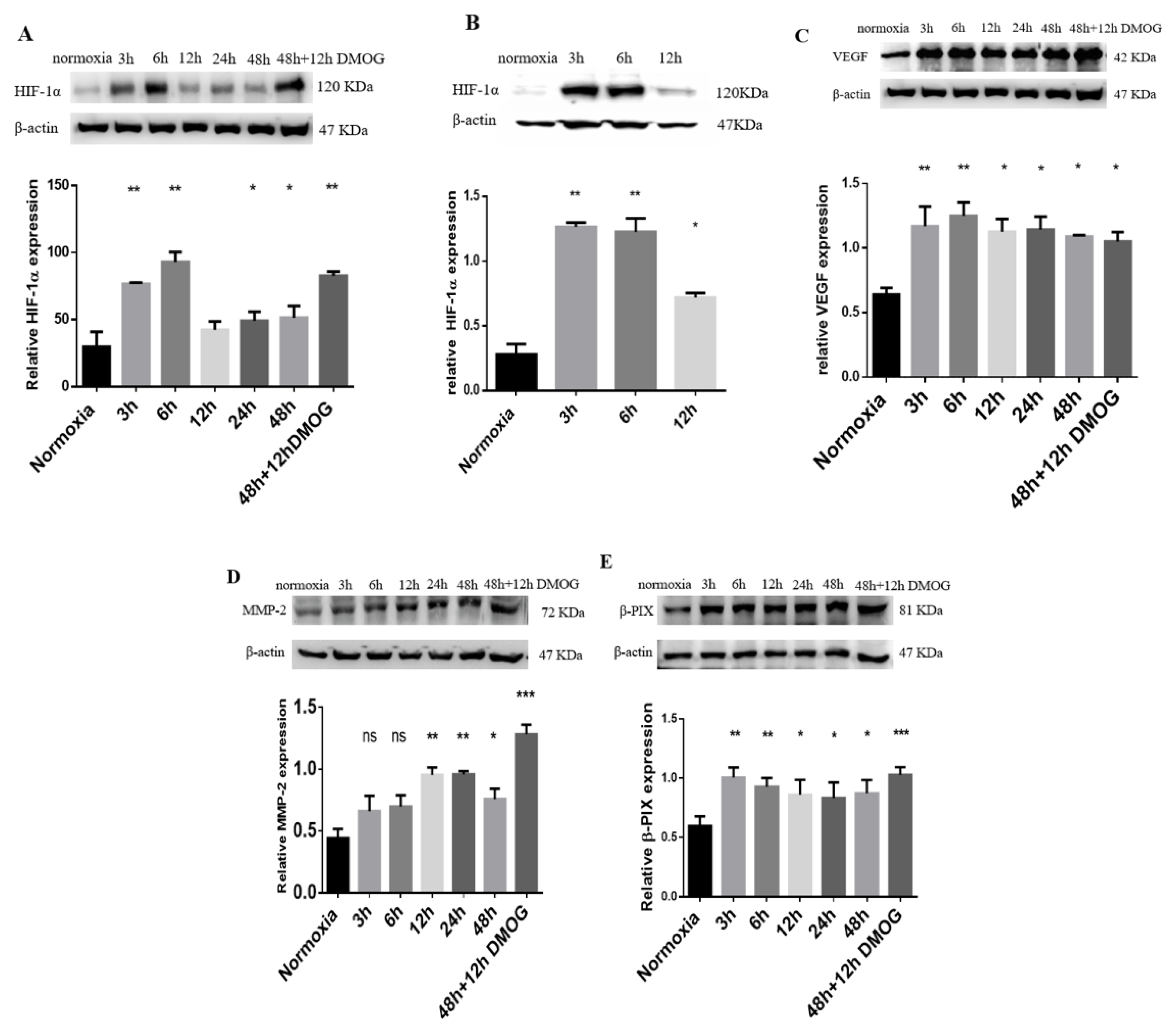
2.2. Expression of HIF-1α and VEGF in MDA-MB-231 Cells
2.3. MMP-2 and β-PIX Expression Levels in Hypoxia-Induced MDA-MB-231 Cells
2.4. Phenotypic Characterization of Hypoxia-Induced MDA-MB-231 Cells
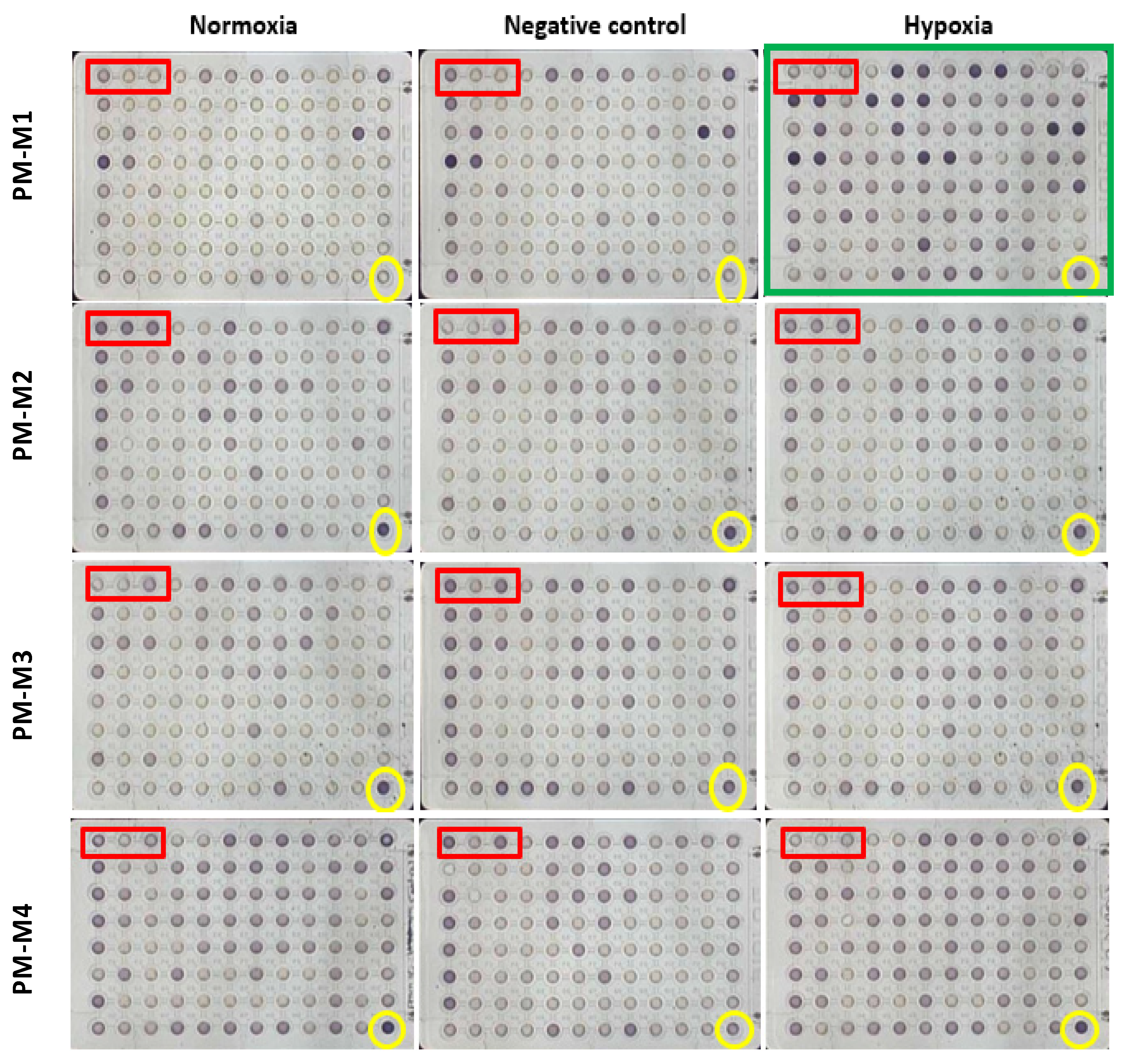
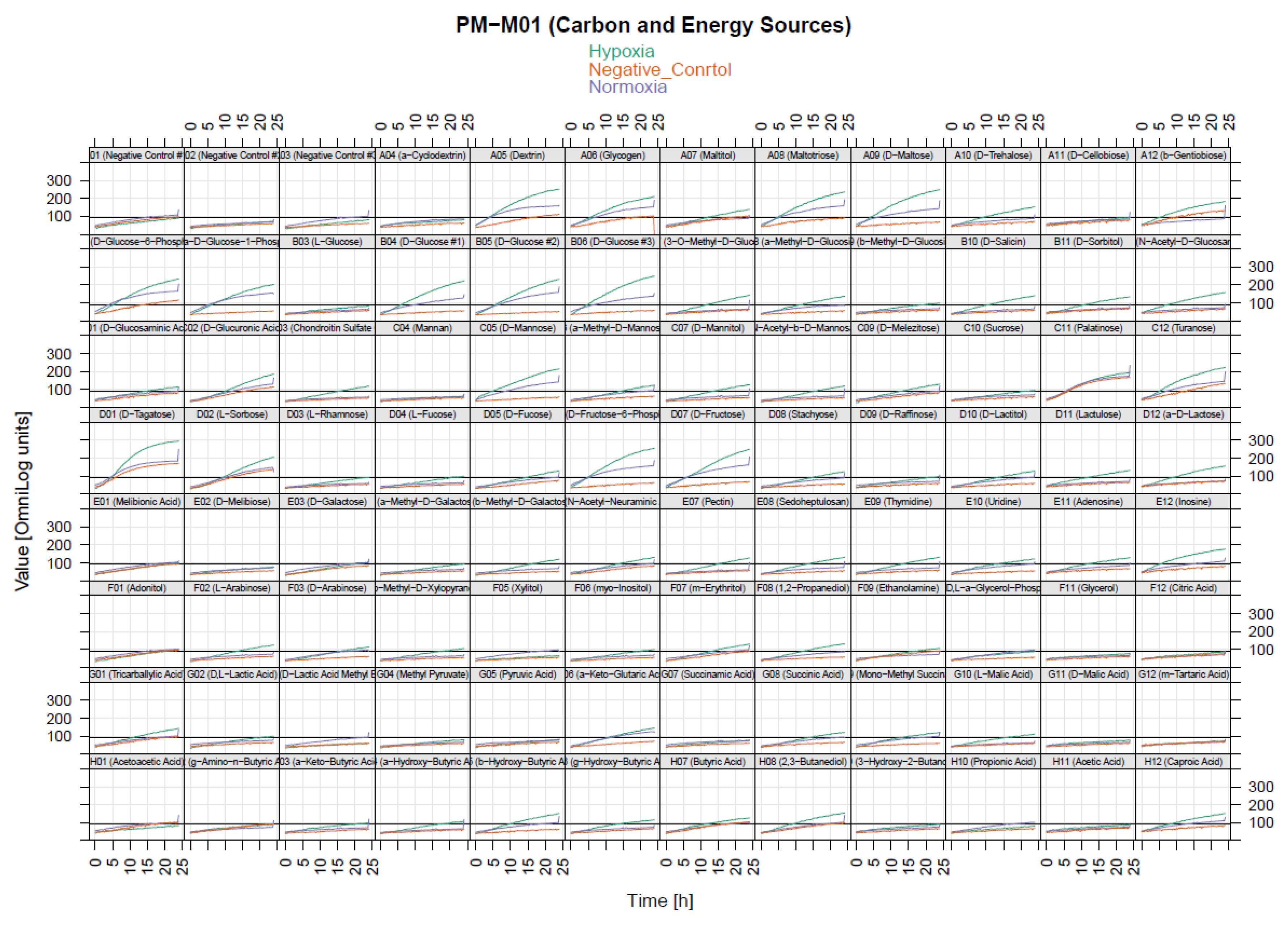
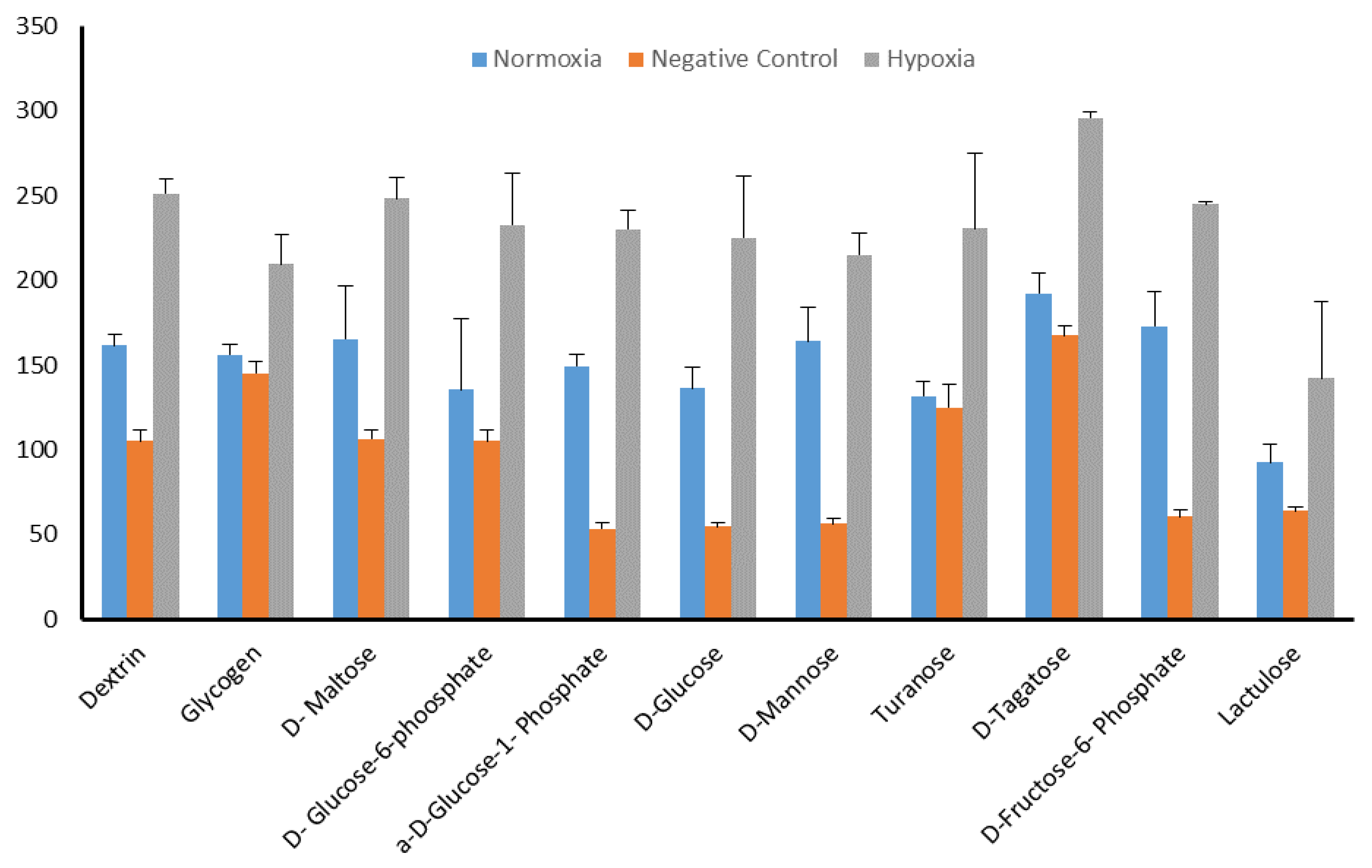
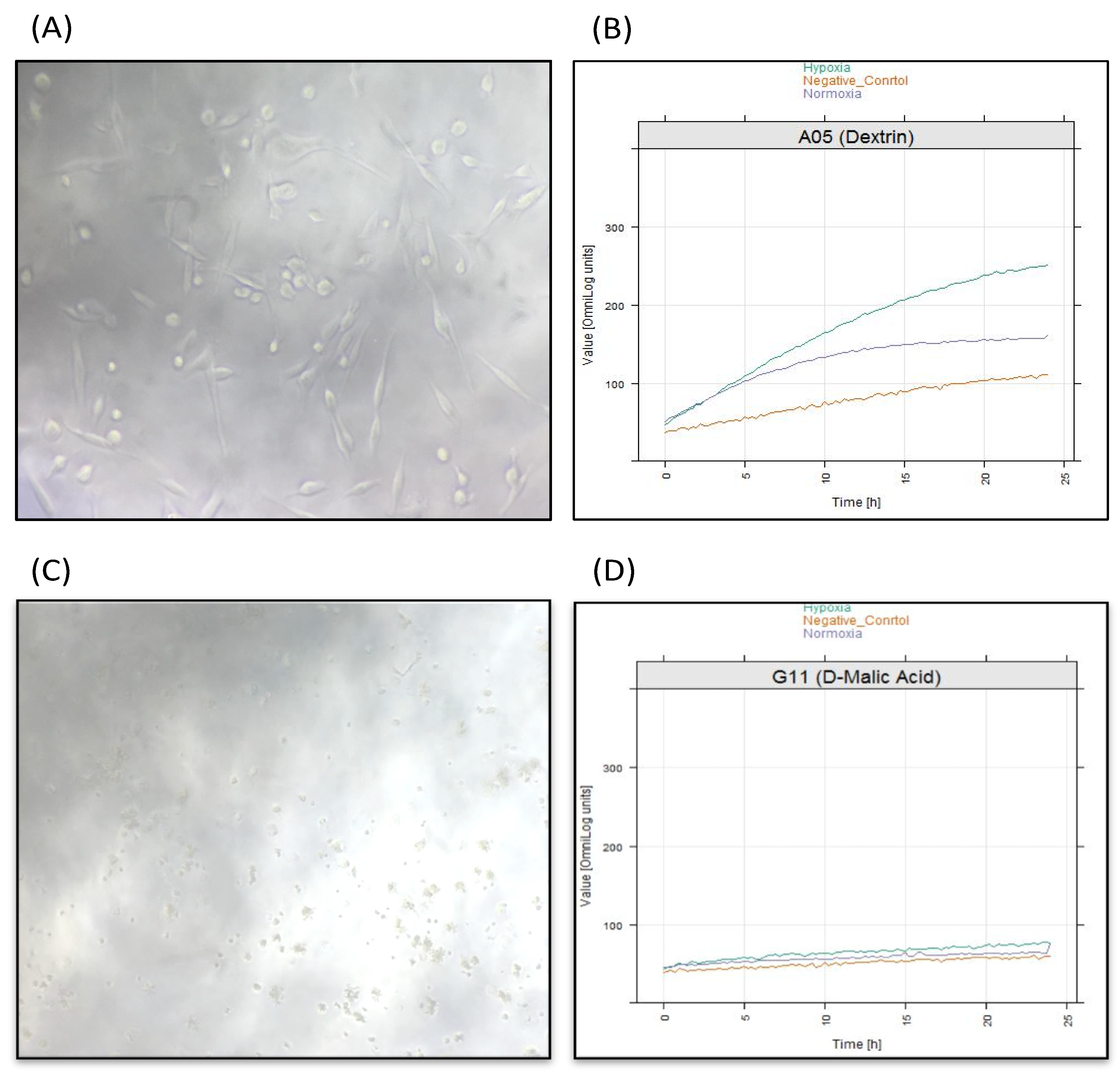
2.5. Identification of Significant Chemical Substrates in PM-M1 Plate
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Preparation of Gelatin-Coated Coverslips for Invadopodium Assay
4.3. Protein Extraction and Western Blot Analysis
4.4. Phenotype Microarray (PM) for Mammalian Cells
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Muz, B.; de la Puente, P.; Azab, F.; Azab, A.K. The Role of Hypoxia in Cancer Progression, Angiogenesis, Metastasis, and Resistance to Therapy. Hypoxia 2015, 3, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Vaupel, P.; Höckel, M. Tumor Hypoxia: Definitions and Current Clinical, Biologic, and Molecular Aspects. J. Natl. Cancer Inst. 2001, 93, 266–276. [Google Scholar]
- Wang, W.; He, Y.F.; Sun, Q.K.; Wang, Y.; Han, X.H.; Peng, D.F.; Yao, Y.W.; Ji, C.S.; Hu, B. Hypoxia-Inducible Factor 1α in Breast Cancer Prognosis. Clin. Chim. Acta 2014, 428, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Harper, K.; Lavoie, R.R.; Charbonneau, M.; Brochu-Gaudreau, K.; Dubois, C.M. The Hypoxic Tumor Microenvironment Promotes Invadopodia Formation and Metastasis through LPA1 Receptor and EGFR Cooperation. Mol. Cancer Res. 2018, 16, 1601–1613. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, C.; Mao, X.; Brown-Clay, J.; Moreau, F.; Al Absi, A.; Wurzer, H.; Sousa, B.; Schmitt, F.; Berchem, G.; Janji, B.; et al. Hypoxia Promotes Breast Cancer Cell Invasion through HIF-1α-Mediated up-Regulation of the Invadopodial Actin Bundling Protein CSRP2. Sci. Rep. 2018, 8, 1–14. [Google Scholar] [CrossRef]
- Leal-Esteban, L.C.; Fajas, L. Cell cycle regulators in cancer cell metabolism. Biochim. Biophys. Acta 2020, 1866, 165715. [Google Scholar] [CrossRef]
- Doktorova, H.; Hrabeta, J.; Khalil, M.A.; Eckschlager, T. Hypoxia-Induced Chemoresistance in Cancer Cells: The Role of Not Only HIF-1. Biomed. Pap. 2015, 159, 166–177. [Google Scholar] [CrossRef]
- Kim, J.Y.; Lee, J.Y. Targeting Tumor Adaption to Chronic Hypoxia: Implications for Drug Resistance, and How It Can Be Overcome. Int. J. Mol. Sci. 2017, 18, 1854. [Google Scholar]
- Lv, Y.; Zhao, S.; Han, J.; Zheng, L.; Yang, Z.; Zhao, L. Hypoxia-Inducible Factor-1α Induces Multidrug Resistance Protein in Colon Cancer. Onco Targets 2015, 8, 1941–1948. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- García, E.; Ragazzini, C.; Yu, X.; Cuesta-García, E.; Bernardino De La Serna, J.; Zech, T.; Sarrió, D.; Machesky, L.M.; Antón, I.M. WIP and WICH/WIRE Co-Ordinately Control Invadopodium Formation and Maturation in Human Breast Cancer Cell Invasion. Sci. Rep. 2016, 6, 23590. [Google Scholar] [CrossRef] [PubMed]
- Bravo-Cordero, J.J.; Hodgson, L.; Condeelis, J. Directed Cell Invasion and Migration during Metastasis. Curr. Opin. Cell Biol. 2012, 24, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.S.; Kim, S.; Kim, B.G.; Won, C.; Nam, S.H.; Kang, S.; Kim, H.J.; Kang, M.; Ryu, J.; Song, H.E.; et al. Snail1 Induced in Breast Cancer Cells in 3D Collagen I Gel Environment Suppresses Cortactin and Impairs Effective Invadopodia Formation. Biochim. Biophys. Acta Mol. Cell Res. 2014, 1843, 2037–2054. [Google Scholar] [CrossRef] [PubMed]
- Weaver, A.M. Invadopodia: Specialized Cell Structures for Cancer Invasion. Clin. Exp. Metastasis 2006, 23, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Meirson, T.; Genna, A.; Lukic, N.; Makhnii, T.; Alter, J.; Sharma, V.P.; Wang, Y.; Samson, A.O.; Condeelis, J.S.; Gil-Henn, H.; et al. Targeting Invadopodia-Mediated Breast Cancer Metastasis by Using ABL Kinase Inhibitors. Oncotarget 2018, 9, 22158–22183. [Google Scholar] [CrossRef]
- Kang, Y.; Siegel, P.M.; Shu, W.; Drobnjak, M.; Kakonen, S.M.; Cordón-Cardo, C.; Guise, T.A.; Massagué, J. A Multigenic Program Mediating Breast Cancer Metastasis to Bone. Cancer Cell 2003, 3, 537–549. [Google Scholar] [CrossRef]
- Wang, Z.; Liang, X.; Cai, M.; Du, G. Analysis of Invadopodia Formation in Breast Cancer Cells. In Breast Cancer; Humana Press: New York, NY, USA, 2016; pp. 203–210. [Google Scholar]
- Zhang, Y.; Nolan, M.; Yamada, H.; Watanabe, M.; Nasu, Y.; Takei, K.; Takeda, T. Dynamin2 GTPase Contributes to Invadopodia Formation in Invasive Bladder Cancer Cells. Biochem. Biophys. Res. Commun. 2016, 480, 409–414. [Google Scholar] [CrossRef]
- Yamaguchi, H. Pathological Roles of Invadopodia in Cancer Invasion and Metastasis. Eur. J. Cell Biol. 2012, 91, 902–907. [Google Scholar] [CrossRef]
- Revach, O.Y.; Geiger, B. The Interplay between the Proteolytic, Invasive, and Adhesive Domains of Invadopodia and Their Roles in Cancer Invasion. Cell Adhes. Migr. 2014, 8, 215–225. [Google Scholar] [CrossRef]
- Leong, H.S.; Robertson, A.E.; Stoletov, K.; Leith, S.J.; Chin, C.A.; Chien, A.E.; Hague, M.N.; Ablack, A.; Carmine-Simmen, K.; McPherson, V.A.; et al. Invadopodia Are Required for Cancer Cell Extravasation and Are a Therapeutic Target for Metastasis. Cell Rep. 2014, 8, 1558–1570. [Google Scholar] [CrossRef]
- Mayers, J.R.; Vander Heiden, M.G. Nature and Nurture: What Determines Tumor Metabolic Phenotypes? Cancer Res. 2017, 77, 3131–3134. [Google Scholar] [CrossRef] [PubMed]
- Alexander, N.R.; Estrada, L.; Crooke, C.; Weaver, A.M.; Guelcher, S.A.; Jourquin, J.; Branch, K.M.; Lobdell, N.; Clark, E.S.; Enderling, H.; et al. Dependence of Invadopodia Function on Collagen Fiber Spacing and Cross-Linking: Computational Modeling and Experimental Evidence. Biophys. J. 2008, 95, 2203–2218. [Google Scholar]
- Walk, E.L.; Martin, K.H.; Weed, S.A.; Markwell, S.M.; Ammer, A.G.; Hayes, K.E. Quantitative Measurement of Invadopodia-Mediated Extracellular Matrix Proteolysis in Single and Multicellular Contexts. J. Vis. Exp. 2012, 66, 1–10. [Google Scholar]
- Greetham, D. Phenotype Microarray Technology and Its Application in Industrial Biotechnology. Biotechnol. Lett. 2014, 36, 1153–1160. [Google Scholar] [CrossRef] [PubMed]
- Noble, S.; Lei, X.-H.; Siri, M.; Bochner, B.R.; Wagner, B.K.; Clemons, P.A.; Huang, R.H. Assay of the Multiple Energy-Producing Pathways of Mammalian Cells. PLoS ONE 2011, 6, e18147. [Google Scholar]
- Vaas, L.A.I.; Sikorski, J.; Hofner, B.; Fiebig, A.; Buddruhs, N.; Klenk, H.P.; Göker, M. Opm: An R Package for Analysing OmniLog® Phenotype Microarray Data. Bioinformatics 2013, 29, 1823–1824. [Google Scholar] [CrossRef]
- Vehkala, M.; Shubin, M.; Connor, T.R.; Thomson, N.R.; Corander, J. Novel R Pipeline for Analyzing Biolog Phenotypic Microarray Data. PLoS ONE 2015, 10, 1371. [Google Scholar] [CrossRef]
- Jacob, A.; Jing, J.; Lee, J.; Schedin, P.; Gilbert, S.M.; Peden, A.A.; Junutula, J.R.; Prekeris, R. Rab40b Regulates Trafficking of MMP2 and MMP9 during Invadopodia Formation and Invasion of Breast Cancer Cells. J. Cell Sci. 2013, 126, 4647–4658. [Google Scholar] [CrossRef]
- Md Hashim, N.F.; Nicholas, N.S.; Dart, A.E.; Kiriakidis, S.; Paleolog, E.; Wells, C.M. Hypoxia-Induced Invadopodia Formation: A Role for β-PIX. Open Biol. 2013, 3, 120159. [Google Scholar] [CrossRef]
- Hoffmann, C.; Mao, X.; Dieterle, M.; Moreau, F.; Absi, A.A.; Steinmetz, A.; Oudin, A.; Berchem, G.; Janji, B.; Thomas, C.; et al. CRP2, a New Invadopodia Actin Bundling Factor Critically Promotes Breast Cancer Cell Invasion and Metastasis. Oncotarget 2016, 7, 13688–13705. [Google Scholar] [CrossRef]
- Salvi, A.; Thanabalu, T. Expression of N-WASP Is Regulated by HiF1α through the Hypoxia Response Element in the N-WASP Promoter. Biochem. Biophys. Rep. 2017, 9, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Grass, G.D.; Bratoeva, M.; Toole, B.P. Regulation of Invadopodia Formation and Activity by CD147. J. Cell Sci. 2012, 125, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Lynn, K.D.; Roland, C.L.; Brekken, R.A. VEGF and Pleiotrophin Modulate the Immune Profile of Breast Cancer. Cancers 2010, 2, 970–988. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Semenza, G.L.; Zhang, H. Hypoxia-Inducible Factor 1 and Breast Cancer Metastasis. J. Zhejiang Univ. B 2015, 16, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Díaz, B.; Yuen, A.; Iizuka, S.; Higashiyama, S.; Courtneidge, S.A. Notch Increases the Shedding of HB-EGF by ADAM12 to Potentiate Invadopodia Formation in Hypoxia. J. Cell Biol. 2013, 201, 279–292. [Google Scholar] [CrossRef] [PubMed]
- Abe, C.T.S.; Alves Júnior, S.d.M.; Pinheiro, J.d.J.V.; Kataoka, M.S.d.S.; da Costa, N.M.M.; Branco, D.C. HIF-1α, NOTCH1, ADAM12, and HB-EGF Are Overexpressed in Mucoepidermoid Carcinoma. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2018, 127, e8–e17. [Google Scholar]
- Jacob, A.; Prekeris, R. The Regulation of MMP Targeting to Invadopodia during Cancer Metastasis. Front. Cell Dev. Biol. 2015, 3, 1–9. [Google Scholar]
- Linklater, E.; Jacob, A.; Bayless, B.A.; Lyons, T.; Prekeris, R. The Role and Regulation of Rab40b–Tks5 Complex during Invadopodia Formation and Cancer Cell Invasion. J. Cell Sci. 2016, 129, 4341–4353. [Google Scholar]
- Gonzalez-Avila, G.; Sommer, B.; Mendoza-Posada, D.A.; Ramos, C.; Garcia-Hernandez, A.A.; Falfan-Valencia, R. Matrix Metalloproteinases Participation in the Metastatic Process and Their Diagnostic and Therapeutic Applications in Cancer. Crit. Rev. Oncol. Hematol. 2019, 137, 57–83. [Google Scholar] [CrossRef]
- Laderoute, K.R.; Calaoagan, J.M.; Knapp, M.; Johnson, R.S. Glucose Utilization Is Essential for Hypoxia-Inducible Factor 1 -Dependent Phosphorylation of c-Jun. Mol. Cell. Biol. 2004, 24, 4128–4137. [Google Scholar] [CrossRef]
- Condeelis, J.; Wang, Y.; Li, J.; Entenberg, D.; Wang, W.; Wang, H.; Xue, A. Direct Visualization of the Phenotype of Hypoxic Tumor Cells at Single Cell Resolution in Vivo Using a New Hypoxia Probe. IntraVital 2016, 5, e1187803. [Google Scholar]
- Van Horssen, R.; Buccione, R.; Willemse, M.; Cingir, S.; Wieringa, B.; Attanasio, F. Cancer Cell Metabolism Regulates Extracellular Matrix Degradation by Invadopodia. Eur. J. Cell Biol. 2013, 92, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Manchun, S.; Dass, C.R.; Sriamornsak, P. Designing Nanoemulsion Templates for Fabrication of Dextrin Nanoparticles via Emulsion Cross-Linking Technique. Carbohydr. Polym. 2014, 101, 650–655. [Google Scholar] [CrossRef] [PubMed]
- Patra, P.; Das, D.; Dhara, S.; Rameshbabu, A.P.; Pal, S.; Ghosh, P. Biocompatible Nanogel Derived from Functionalized Dextrin for Targeted Delivery of Doxorubicin Hydrochloride to MG 63 Cancer Cells. Carbohydr. Polym. 2017, 171, 27–38. [Google Scholar]
Sample Availability: Samples of the compounds are available from Dr. Nur Fariesha Md Hashim, faculty of medicine and health sciences, UPM. |

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hamad, H.A.; Enezei, H.H.; Alrawas, A.; Zakuan, N.M.; Abdullah, N.A.; Cheah, Y.K.; Hashim, N.F.M. Identification of Potential Chemical Substrates as Fuel for Hypoxic Tumors That May Be Linked to Invadopodium Formation in Hypoxia-Induced MDA-MB-231 Breast-Cancer Cell Line. Molecules 2020, 25, 3876. https://doi.org/10.3390/molecules25173876
Hamad HA, Enezei HH, Alrawas A, Zakuan NM, Abdullah NA, Cheah YK, Hashim NFM. Identification of Potential Chemical Substrates as Fuel for Hypoxic Tumors That May Be Linked to Invadopodium Formation in Hypoxia-Induced MDA-MB-231 Breast-Cancer Cell Line. Molecules. 2020; 25(17):3876. https://doi.org/10.3390/molecules25173876
Chicago/Turabian StyleHamad, Hamad Ali, Hamid Hammad Enezei, Anmar Alrawas, Noraina Muhamad Zakuan, Nurul Akmaryanti Abdullah, Yoke Kqueen Cheah, and Nur Fariesha Md Hashim. 2020. "Identification of Potential Chemical Substrates as Fuel for Hypoxic Tumors That May Be Linked to Invadopodium Formation in Hypoxia-Induced MDA-MB-231 Breast-Cancer Cell Line" Molecules 25, no. 17: 3876. https://doi.org/10.3390/molecules25173876
APA StyleHamad, H. A., Enezei, H. H., Alrawas, A., Zakuan, N. M., Abdullah, N. A., Cheah, Y. K., & Hashim, N. F. M. (2020). Identification of Potential Chemical Substrates as Fuel for Hypoxic Tumors That May Be Linked to Invadopodium Formation in Hypoxia-Induced MDA-MB-231 Breast-Cancer Cell Line. Molecules, 25(17), 3876. https://doi.org/10.3390/molecules25173876






