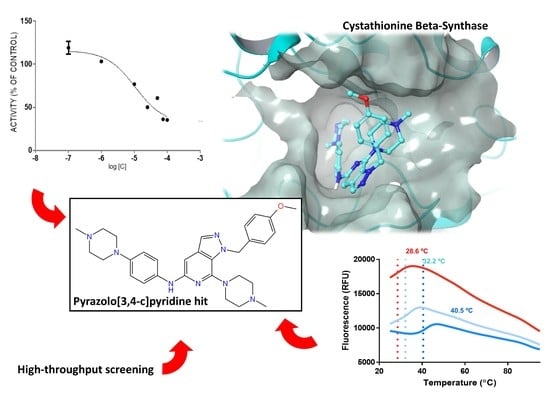
Screening of Heteroaromatic Scaffolds against Cystathionine Beta-Synthase Enables Identification of Substituted Pyrazolo[3,4-c]Pyridines as Potent and Selective Orthosteric Inhibitors
Abstract
1. Introduction
2. Results and Discussion
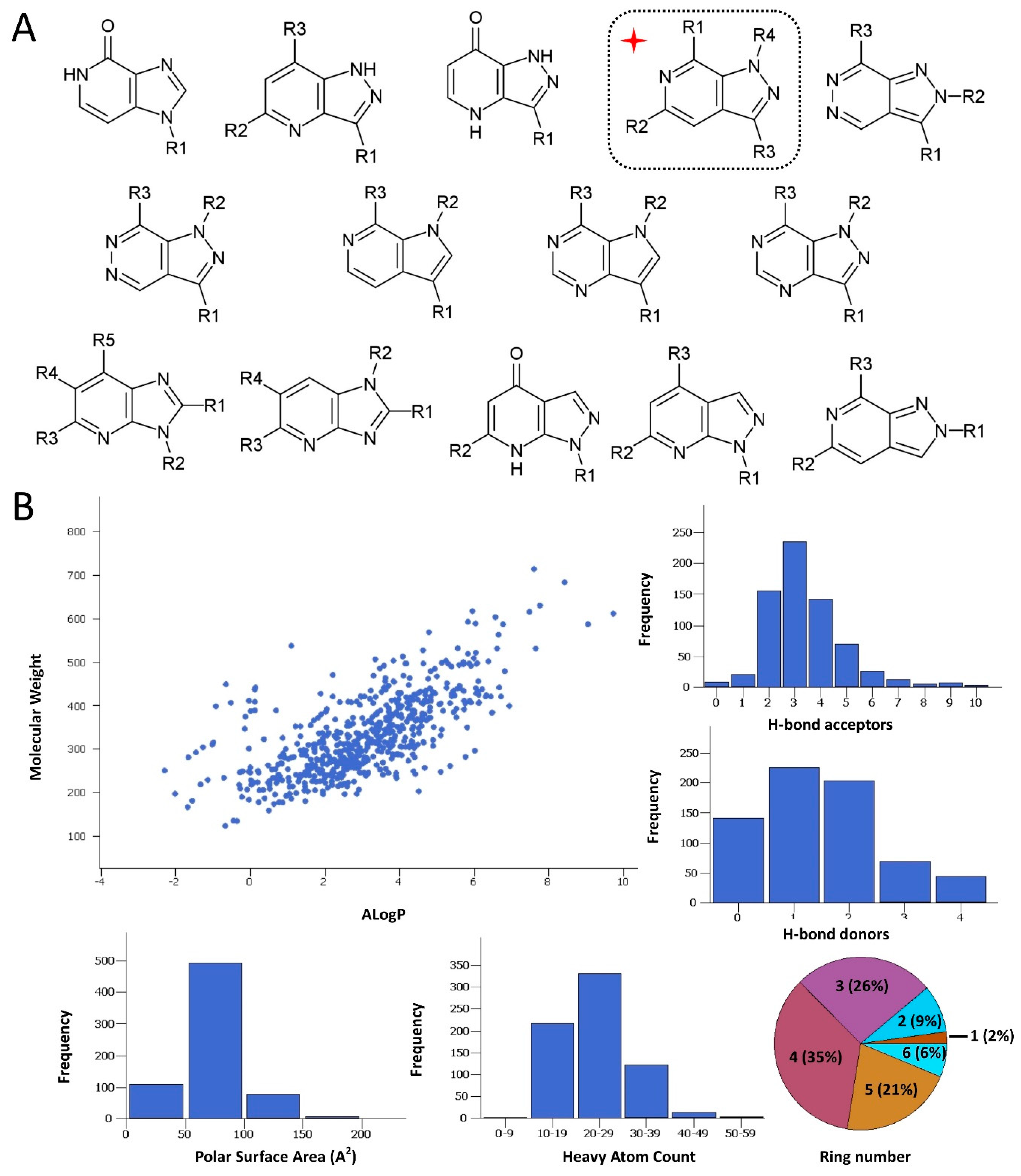
2.1. Rationale and Compound Selection
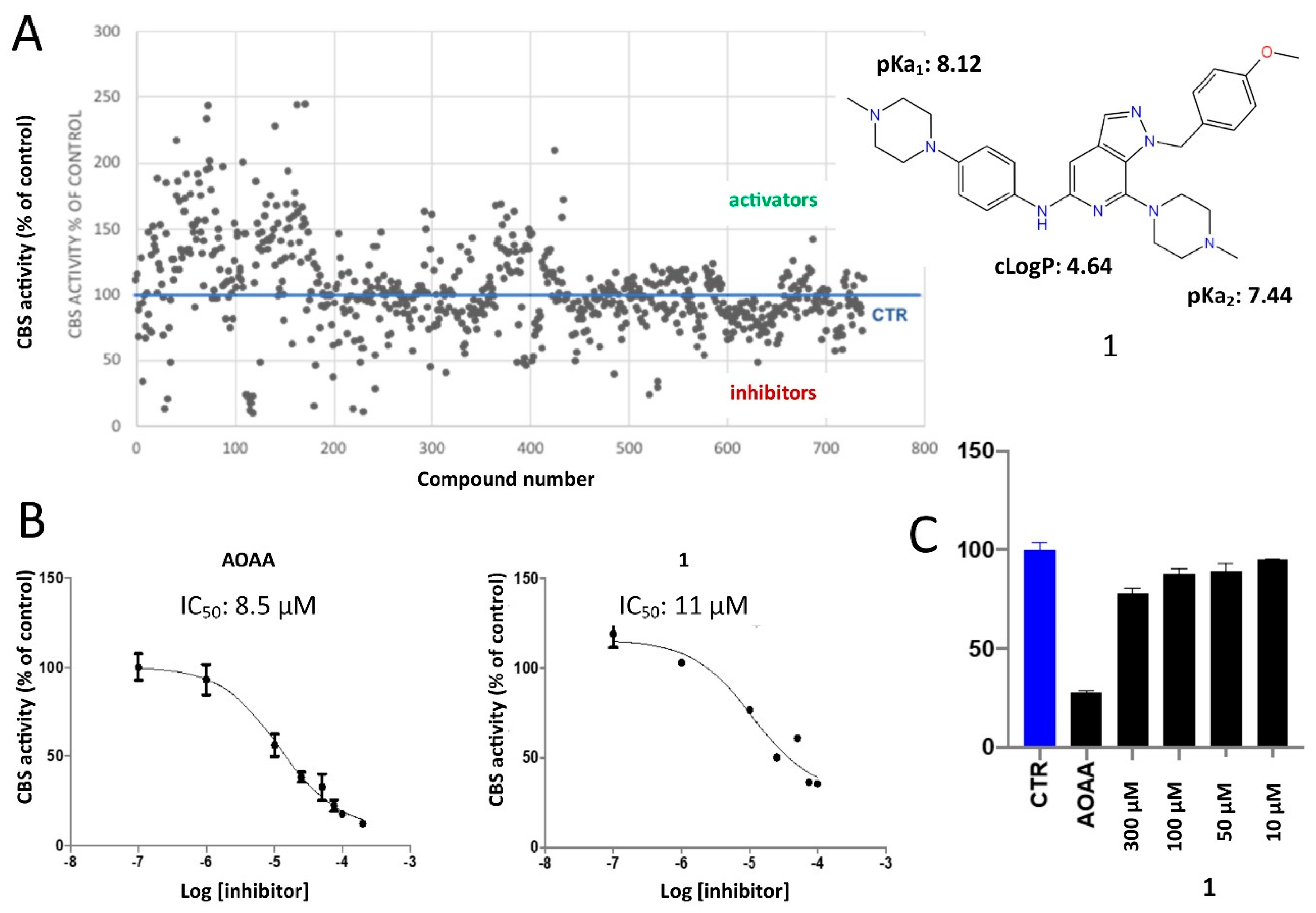
2.2. Protein Expression and In Vitro Evaluations
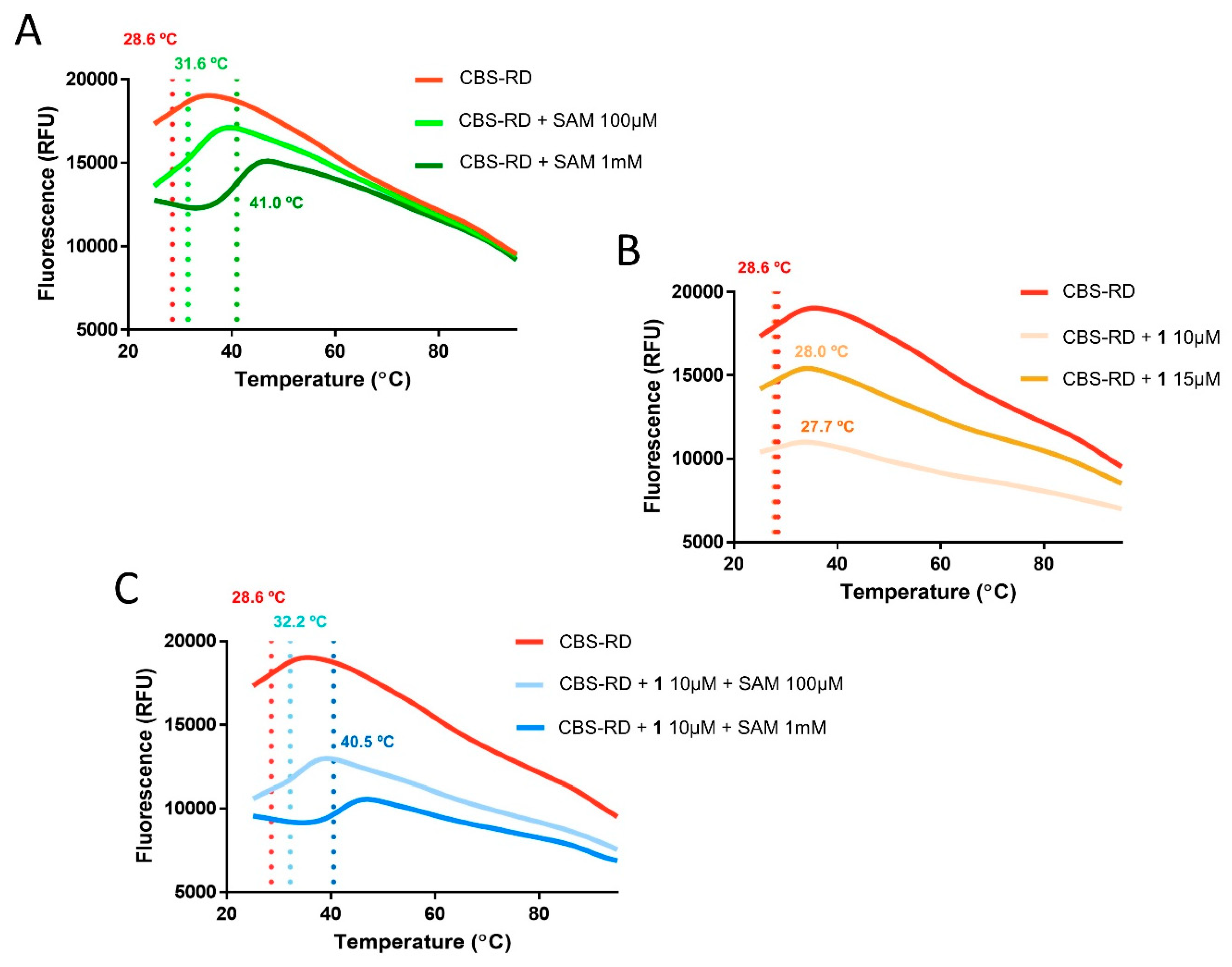
2.3. Differential Scanning Fluorimetry
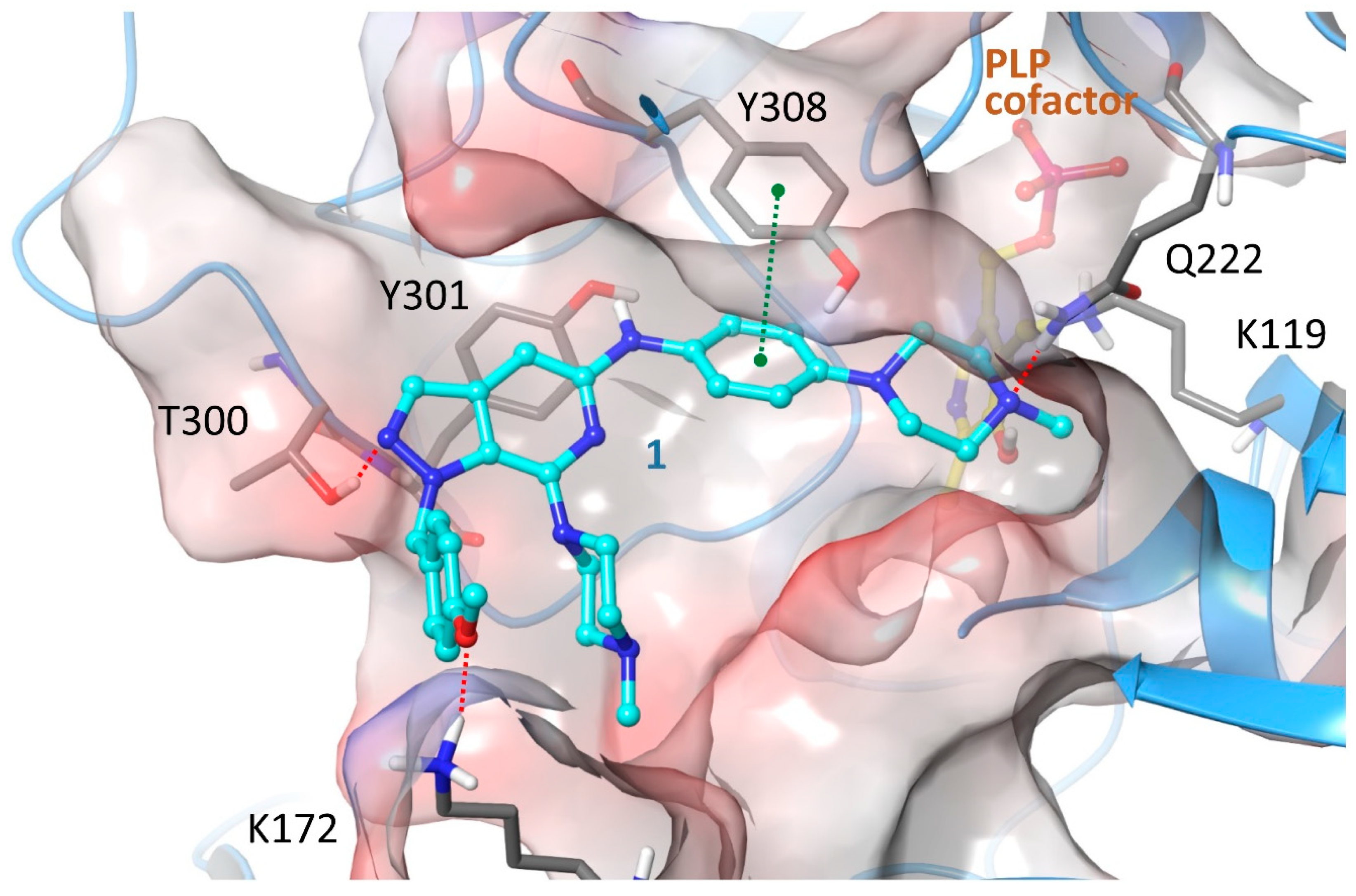
2.4. Structure-Activity Relationships, Neural Networks Modeling and Theoretical Simulations
3. Materials and Methods
3.1. Expression and Purification of Full Length GST-CBS and His Tag-CBS Regulatory Domain
3.2. Sample Preparation and Library Administration
3.3. H2S Detection Using the Methylene Blue Assay
3.4. H2S Detection Using the 7-Azido-4-Methylcoumarin Assay
3.5. Differential Scanning Fluorimetry
3.6. Deep Neural Networks
3.7. Molecular Simulations
3.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Wang, R. Physiological implications of hydrogen sulfide: A whiff exploration that blossomed. Physiol. Rev. 2012, 92, 791–896. [Google Scholar] [CrossRef] [PubMed]
- Szabo, C.; Papapetropoulos, A. International Union of Basic and Clinical Pharmacology. CII: Pharmacological Modulation of H2S Levels: H2S Donors and H2S Biosynthesis Inhibitors. Pharmacol. Rev. 2017, 69, 497–564. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Banerjee, R. PLP-dependent H2S biogenesis. Biochim. Biophys. Acta 2011, 1814, 1518–1527. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, R.; Zou, C.G. Redox regulation and reaction mechanism of human cystathionine-beta-synthase: A PLP-dependent hemesensor protein. Arch. Biochem. Biophys. 2005, 433, 144–156. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Ding, Y.P.; Wang, Z.; Kong, Y.; Gao, R.; Chen, G. Hydrogen sulfide therapy in brain diseases: From bench to bedside. Med. Gas Res. 2017, 7, 113–119. [Google Scholar] [CrossRef]
- Keller, R.; Chrastina, P.; Pavlíková, M.; Gouveia, S.; Ribes, A.; Kölker, S.; Blom, H.J.; Baumgartner, M.R.; Bártl, J.; Dionisi-Vici, C.; et al. Newborn screening for homocystinurias: Recent recommendations versus current practice. J. Inherit. Metab. Dis. 2019, 42, 128–139. [Google Scholar] [CrossRef]
- Wen, Y.D.; Wang, H.; Zhu, Y.Z. The drug developments of hydrogen sulfide on cardiovascular disease. Oxidative Med. Cell. Longev. 2018, 2018, 4010395. [Google Scholar] [CrossRef]
- Wu, D.; Si, W.; Wang, M.; Lv, S.; Ji, A.; Li, Y. Hydrogen sulfide in cancer: Friend or foe? Nitric Oxide Biol. Chem. 2015, 50, 38–45. [Google Scholar] [CrossRef]
- Asimakopoulou, A.; Panopoulos, P.; Chasapis, C.T.; Coletta, C.; Zhou, Z.; Cirino, G.; Giannis, A.; Szabo, C.; Spyroulias, G.A.; Papapetropoulos, A. Selectivity of commonly used pharmacological inhibitors for cystathionine beta synthase (CBS) and cystathionine gamma lyase (CSE). Br. J. Pharmacol. 2013, 169, 922–932. [Google Scholar] [CrossRef]
- Druzhyna, N.; Szczesny, B.; Olah, G.; Módis, K.; Asimakopoulou, A.; Pavlidou, A.; Szoleczky, P.; Gerö, D.; Yanagi, K.; Törö, G.; et al. Screening of a composite library of clinically used drugs and well-characterized pharmacological compounds for cystathionine β-synthase inhibition identifies benserazide as a drug potentially suitable for repurposing for the experimental therapy of colon cancer. Pharmacol. Res. 2016, 113, 18–37. [Google Scholar] [CrossRef]
- Hellmich, M.R.; Coletta, C.; Chao, C.; Szabo, C. The therapeutic potential of cystathionine beta-synthetase/hydrogen sulfide inhibition in cancer. Antioxid. Redox Signal. 2015, 22, 424–448. [Google Scholar] [CrossRef] [PubMed]
- Panagaki, T.; Randi, E.B.; Augsburger, F.; Szabo, C. Overproduction of H(2)S, generated by CBS, inhibits mitochondrial Complex IV and suppresses oxidative phosphorylation in Down syndrome. Proc. Natl. Acad. Sci. USA 2019, 116, 18769–18771. [Google Scholar] [CrossRef] [PubMed]
- Marechal, D.; Brault, V.; Leon, A.; Martin, D.; Lopes Pereira, P.; Loaëc, N.; Birling, M.-C.; Friocourt, G.; Blondel, M.; Herault, Y. Cbs overdosage is necessary and sufficient to induce cognitive phenotypes in mouse models of Down syndrome and interacts genetically with Dyrk1α. Hum. Mol. Genet. 2019, 28, 1561–1577. [Google Scholar] [CrossRef] [PubMed]
- Gardiner, K.; Davisson, M. The sequence of human chromosome 21 and implications for research into Down syndrome. Genome Biol. 2000, 1, reviews0002.1. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Skovby, F.; Krassikoff, N.; Francke, U. Assignment of the gene for cystathionine β-synthase to human chromosome 21 in somatic cell hybrids. Hum. Genet. 1984, 65, 291–294. [Google Scholar] [CrossRef] [PubMed]
- Kamoun, P.P. Mental retardation in Down syndrome: Two ways to treat. Med. Hypotheses 2019, 131, 109289. [Google Scholar] [CrossRef]
- Szabo, C. The re-emerging pathophysiological role of the cystathionine-beta-synthase-hydrogen sulfide system in Down syndrome. FEBS J. 2020. [Google Scholar] [CrossRef]
- Bhattacharyya, S.; Saha, S.; Giri, K.; Lanza, I.R.; Nair, K.S.; Jennings, N.B.; Rodriguez-Aguayo, C.; Lopez-Berestein, G.; Basal, E.; Weaver, A.L.; et al. Cystathionine beta-synthase (CBS) contributes to advanced ovarian cancer progression and drug resistance. PLoS ONE 2013, 8, e79167. [Google Scholar] [CrossRef]
- Kashfi, K. The dichotomous role of H2S in cancer cell biology? Déjà vu all over again. Biochem. Pharmacol. 2018, 149, 205–223. [Google Scholar] [CrossRef]
- Untereiner, A.A.; Pavlidou, A.; Druzhyna, N.; Papapetropoulos, A.; Hellmich, M.R.; Szabo, C. Drug resistance induces the upregulation of H2S-producing enzymes in HCT116 colon cancer cells. Biochem. Pharmacol. 2018, 149, 174–185. [Google Scholar] [CrossRef]
- Modis, K.; Coletta, C.; Asimakopoulou, A.; Szczesny, B.; Chao, C.; Papapetropoulos, A.; Hellmich, M.R.; Szabo, C. Effect of S-adenosyl-l-methionine (SAM), an allosteric activator of cystathionine-beta-synthase (CBS) on colorectal cancer cell proliferation and bioenergetics in vitro. Nitric Oxide Biol. Chem. 2014, 41, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Giuffrè, A.; Tomé, C.S.; Fernandes, D.G.F.; Zuhra, K.; Vicente, J.B. Hydrogen sulfide metabolism and signaling in the tumor microenvironment. Adv. Exp. Med. Biol. 2020, 1219, 335–353. [Google Scholar] [CrossRef] [PubMed]
- Zuhra, K.; Augsburger, F.; Majtan, T.; Szabo, C. Cystathionine-β-synthase: Molecular regulation and pharmacological inhibition. Biomolecules 2020, 10, 697. [Google Scholar] [CrossRef] [PubMed]
- Meier, M.; Janosik, M.; Kery, V.; Kraus, J.P.; Burkhard, P. Structure of human cystathionine beta-synthase: A unique pyridoxal 5’-phosphate-dependent heme protein. EMBO J. 2001, 20, 3910–3916. [Google Scholar] [CrossRef] [PubMed]
- Ereno-Orbea, J.; Majtan, T.; Oyenarte, I.; Kraus, J.P.; Martinez-Cruz, L.A. Structural insight into the molecular mechanism of allosteric activation of human cystathionine beta-synthase by S-adenosylmethionine. Proc. Natl. Acad. Sci. USA 2014, 111, E3845–E3852. [Google Scholar] [CrossRef]
- Pey, A.L.; Martinez-Cruz, L.A.; Kraus, J.P.; Majtan, T. Oligomeric status of human cystathionine beta-synthase modulates AdoMet binding. FEBS Lett. 2016, 590, 4461–4471. [Google Scholar] [CrossRef]
- Ereno-Orbea, J.; Oyenarte, I.; Martinez-Cruz, L.A. CBS domains: Ligand binding sites and conformational variability. Arch. Biochem. Biophys. 2013, 540, 70–81. [Google Scholar] [CrossRef]
- Catazaro, J.; Caprez, A.; Guru, A.; Swanson, D.; Powers, R. Functional evolution of PLP-dependent enzymes based on active-site structural similarities. Proteins Struct. Funct. Bioinform. 2014, 82, 2597–2608. [Google Scholar] [CrossRef]
- Koutmos, M.; Kabil, O.; Smith, J.L.; Banerjee, R. Structural basis for substrate activation and regulation by cystathionine beta-synthase (CBS) domains in cystathionine {beta}-synthase. Proc. Natl. Acad. Sci. USA 2010, 107, 20958–20963. [Google Scholar] [CrossRef]
- McCorvie, T.J.; Kopec, J.; Hyung, S.J.; Fitzpatrick, F.; Feng, X.; Termine, D.; Strain-Damerell, C.; Vollmar, M.; Fleming, J.; Janz, J.M.; et al. Inter-domain communication of human cystathionine beta-synthase: Structural basis of S-adenosyl-L-methionine activation. J. Biol. Chem. 2014, 289, 36018–36030. [Google Scholar] [CrossRef]
- Hnizda, A.; Spiwok, V.; Jurga, V.; Kozich, V.; Kodicek, M.; Kraus, J.P. Cross-talk between the catalytic core and the regulatory domain in cystathionine beta-synthase: Study by differential covalent labeling and computational modeling. Biochemistry 2010, 49, 10526–10534. [Google Scholar] [CrossRef] [PubMed]
- Janošík, M.; Kery, V.; Gaustadnes, M.; Maclean, K.N.; Kraus, J.P. Regulation of human cystathionine β-synthase by S-adenosyl-l-methionine: evidence for two catalytically active conformations involving an autoinhibitory domain in the C-terminal region. Biochemistry 2001, 40, 10625–10633. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Cai, H.; Hu, Y.; Liu, F.; Huang, S.; Zhou, Y.; Yu, J.; Xu, J.; Wu, F. A pharmacological probe identifies cystathionine β-synthase as a new negative regulator for ferroptosis. Cell Death Dis. 2018, 9, 1005. [Google Scholar] [CrossRef] [PubMed]
- Niu, W.; Chen, F.; Wang, J.; Qian, J.; Yan, S. Antitumor effect of sikokianin C, a selective cystathionine β-synthase inhibitor, against human colon cancer in vitro and in vivo. MedChemComm 2018, 9, 113–120. [Google Scholar] [CrossRef]
- Niu, W.; Wu, P.; Chen, F.; Wang, J.; Shang, X.; Xu, C. Discovery of selective cystathionine β-synthase inhibitors by high-throughput screening with a fluorescent thiol probe. MedChemComm 2017, 8, 198–201. [Google Scholar] [CrossRef]
- Zuhra, K.; Sousa, P.M.F.; Paulini, G.; Lemos, A.R.; Kalme, Z.; Bisenieks, I.; Bisenieks, E.; Vigante, B.; Duburs, G.; Bandeiras, T.M.; et al. Screening pyridine derivatives against human hydrogen sulfide-synthesizing enzymes by orthogonal methods. Sci. Rep. 2019, 9, 684. [Google Scholar] [CrossRef]
- Nakai, T.; Nakagawa, N.; Maoka, N.; Masui, R.; Kuramitsu, S.; Kamiya, N. Structure of P-protein of the glycine cleavage system: Implications for nonketotic hyperglycinemia. EMBO J. 2005, 24, 1523–1536. [Google Scholar] [CrossRef]
- Macarron, R.; Banks, M.N.; Bojanic, D.; Burns, D.J.; Cirovic, D.A.; Garyantes, T.; Green, D.V.; Hertzberg, R.P.; Janzen, W.P.; Paslay, J.W.; et al. Impact of high-throughput screening in biomedical research. Nat. Rev. Drug Discov. 2011, 10, 188–195. [Google Scholar] [CrossRef]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra precision Glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef]
- Halgren, T.A.; Murphy, R.B.; Friesner, R.A.; Beard, H.S.; Frye, L.L.; Pollard, W.T.; Banks, J.L. Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 2004, 47, 1750–1759. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Dixon, S.L.; Lowrie, J.F.; Sherman, W. Analysis and comparison of 2D fingerprints: Insights into database screening performance using eight fingerprint methods. J. Mol. Graph. Model. 2010, 29, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Sastry, M.; Lowrie, J.F.; Dixon, S.L.; Sherman, W. Large-scale systematic analysis of 2D fingerprint methods and parameters to improve virtual screening enrichments. J. Chem. Inf. Model. 2010, 50, 771–784. [Google Scholar] [CrossRef] [PubMed]
- Bukovska, G.; Kery, V.; Kraus, J.P. Expression of human cystathionine beta-synthase in Escherichia coli: Purification and characterization. Protein Expr. Purif. 1994, 5, 442–448. [Google Scholar] [CrossRef]
- Frank, N.; Kent, J.O.; Meier, M.; Kraus, J.P. Purification and characterization of the wild type and truncated human cystathionine beta-synthase enzymes expressed in E. coli. Arch. Biochem. Biophys. 2008, 470, 64–72. [Google Scholar] [CrossRef]
- Kraus, J.; Packman, S.; Fowler, B.; Rosenberg, L.E. Purification and properties of cystathionine beta-synthase from human liver. Evidence for identical subunits. J. Biol. Chem. 1978, 253, 6523–6528. [Google Scholar]
- Oyenarte, I.; Majtan, T.; Ereno, J.; Corral-Rodriguez, M.A.; Kraus, J.P.; Martinez-Cruz, L.A. Purification, crystallization and preliminary crystallographic analysis of human cystathionine beta-synthase. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2012, 68, 1318–1322. [Google Scholar] [CrossRef]
- Wingfield, P.T. Overview of the purification of recombinant proteins. Curr. Protoc. Protein Sci. 2015, 80, 6.1.1–6.1.35. [Google Scholar] [CrossRef]
- Moest, R.R. Hydrogen sulfide determination by the methylene blue method. Anal. Chem. 1975, 47, 1204–1205. [Google Scholar] [CrossRef]
- Reese, B.K.; Finneran, D.W.; Mills, H.J.; Zhu, M.-X.; Morse, J.W. Examination and refinement of the determination of aqueous hydrogen sulfide by the methylene blue method. Aquat. Geochem. 2011, 17, 567–582. [Google Scholar] [CrossRef]
- Hartle, M.D.; Pluth, M.D. A practical guide to working with H2S at the interface of chemistry and biology. Chem. Soc. Rev. 2016, 45, 6108–6117. [Google Scholar] [CrossRef] [PubMed]
- Michailidou, M.; Giannouli, V.; Kotsikoris, V.; Papadodima, O.; Kontogianni, G.; Kostakis, I.K.; Lougiakis, N.; Chatziioannou, A.; Kolisis, F.N.; Marakos, P.; et al. Novel pyrazolopyridine derivatives as potential angiogenesis inhibitors: Synthesis, biological evaluation and transcriptome-based mechanistic analysis. Eur. J. Med. Chem. 2016, 121, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Augsburger, F.; Randi, E.B.; Jendly, M.; Ascencao, K.; Dilek, N.; Szabo, C. Role of 3-mercaptopyruvate sulfurtransferase in the regulation of proliferation, migration, and bioenergetics in murine colon cancer cells. Biomolecules 2020, 10, 447. [Google Scholar] [CrossRef] [PubMed]
- Niesen, F.H.; Berglund, H.; Vedadi, M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protocols 2007, 2, 2212–2221. [Google Scholar] [CrossRef]
- Farid, R.; Day, T.; Friesner, R.A.; Pearlstein, R.A. New insights about HERG blockade obtained from protein modeling, potential energy mapping, and docking studies. Bioorganic Med. Chem. 2006, 14, 3160–3173. [Google Scholar] [CrossRef]
- Sherman, W.; Beard, H.S.; Farid, R. Use of an induced fit receptor structure in virtual screening. Chem. Biol. Drug Des. 2006, 67, 83–84. [Google Scholar] [CrossRef]
- Sherman, W.; Day, T.; Jacobson, M.P.; Friesner, R.A.; Farid, R. Novel procedure for modeling ligand/receptor induced fit effects. J. Med. Chem. 2006, 49, 534–553. [Google Scholar] [CrossRef]
- Botou, M.; Yalelis, V.; Lazou, P.; Zantza, I.; Papakostas, K.; Charalambous, V.; Mikros, E.; Flemetakis, E.; Frillingos, S. Specificity profile of NAT/NCS2 purine transporters in Sinorhizobium (Ensifer) meliloti. Mol. Microbiol. 2020. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds 1, 3, 4, 5 and 7 are available (≤1 mg) from the authors. |
| Compound | R1 | R2 | R3 | R4 | Inhibition (%) |
|---|---|---|---|---|---|
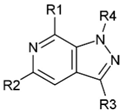
| 1 | 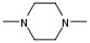 | 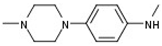 | H | 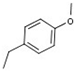 | 70 |
| 2 | 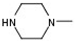 |  | H |  | 30 |
| 3 |  |  | 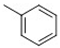 |  | 20 |
| 4 |  | 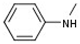 | H |  | 20 |
| 5 |  | N≡C |  |  | No inhibition |
| 6 |  | 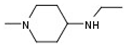 | 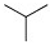 | H | No inhibition |
| 7 |  | N≡C | H | H | No inhibition |
| 8 | 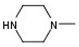 | Cl | H |  | <20 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fantel, A.-M.; Myrianthopoulos, V.; Georgoulis, A.; Lougiakis, N.; Zantza, I.; Lamprinidis, G.; Augsburger, F.; Marakos, P.; Vorgias, C.E.; Szabo, C.; et al. Screening of Heteroaromatic Scaffolds against Cystathionine Beta-Synthase Enables Identification of Substituted Pyrazolo[3,4-c]Pyridines as Potent and Selective Orthosteric Inhibitors. Molecules 2020, 25, 3739. https://doi.org/10.3390/molecules25163739
Fantel A-M, Myrianthopoulos V, Georgoulis A, Lougiakis N, Zantza I, Lamprinidis G, Augsburger F, Marakos P, Vorgias CE, Szabo C, et al. Screening of Heteroaromatic Scaffolds against Cystathionine Beta-Synthase Enables Identification of Substituted Pyrazolo[3,4-c]Pyridines as Potent and Selective Orthosteric Inhibitors. Molecules. 2020; 25(16):3739. https://doi.org/10.3390/molecules25163739
Chicago/Turabian StyleFantel, Anna-Maria, Vassilios Myrianthopoulos, Anastasios Georgoulis, Nikolaos Lougiakis, Iliana Zantza, George Lamprinidis, Fiona Augsburger, Panagiotis Marakos, Constantinos E. Vorgias, Csaba Szabo, and et al. 2020. "Screening of Heteroaromatic Scaffolds against Cystathionine Beta-Synthase Enables Identification of Substituted Pyrazolo[3,4-c]Pyridines as Potent and Selective Orthosteric Inhibitors" Molecules 25, no. 16: 3739. https://doi.org/10.3390/molecules25163739
APA StyleFantel, A.-M., Myrianthopoulos, V., Georgoulis, A., Lougiakis, N., Zantza, I., Lamprinidis, G., Augsburger, F., Marakos, P., Vorgias, C. E., Szabo, C., Pouli, N., Papapetropoulos, A., & Mikros, E. (2020). Screening of Heteroaromatic Scaffolds against Cystathionine Beta-Synthase Enables Identification of Substituted Pyrazolo[3,4-c]Pyridines as Potent and Selective Orthosteric Inhibitors. Molecules, 25(16), 3739. https://doi.org/10.3390/molecules25163739