Towards the Inhibition of Protein–Protein Interactions (PPIs) in STAT3: Insights into a New Class of Benzothiadiazole Derivatives
Abstract
1. Introduction
2. Results and Discussion
2.1. Molecular Modeling—Design and Chemical Space Exploration
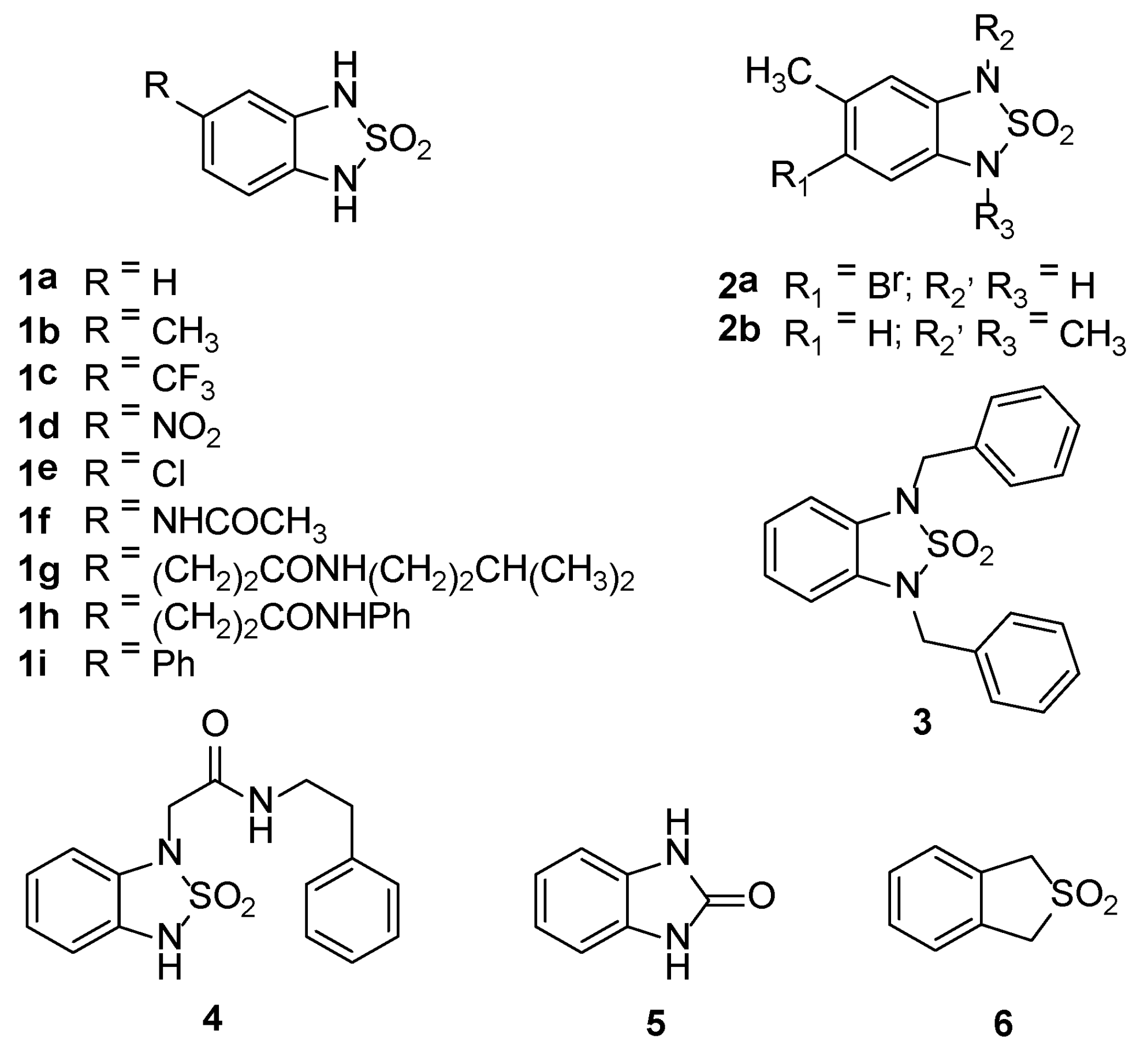
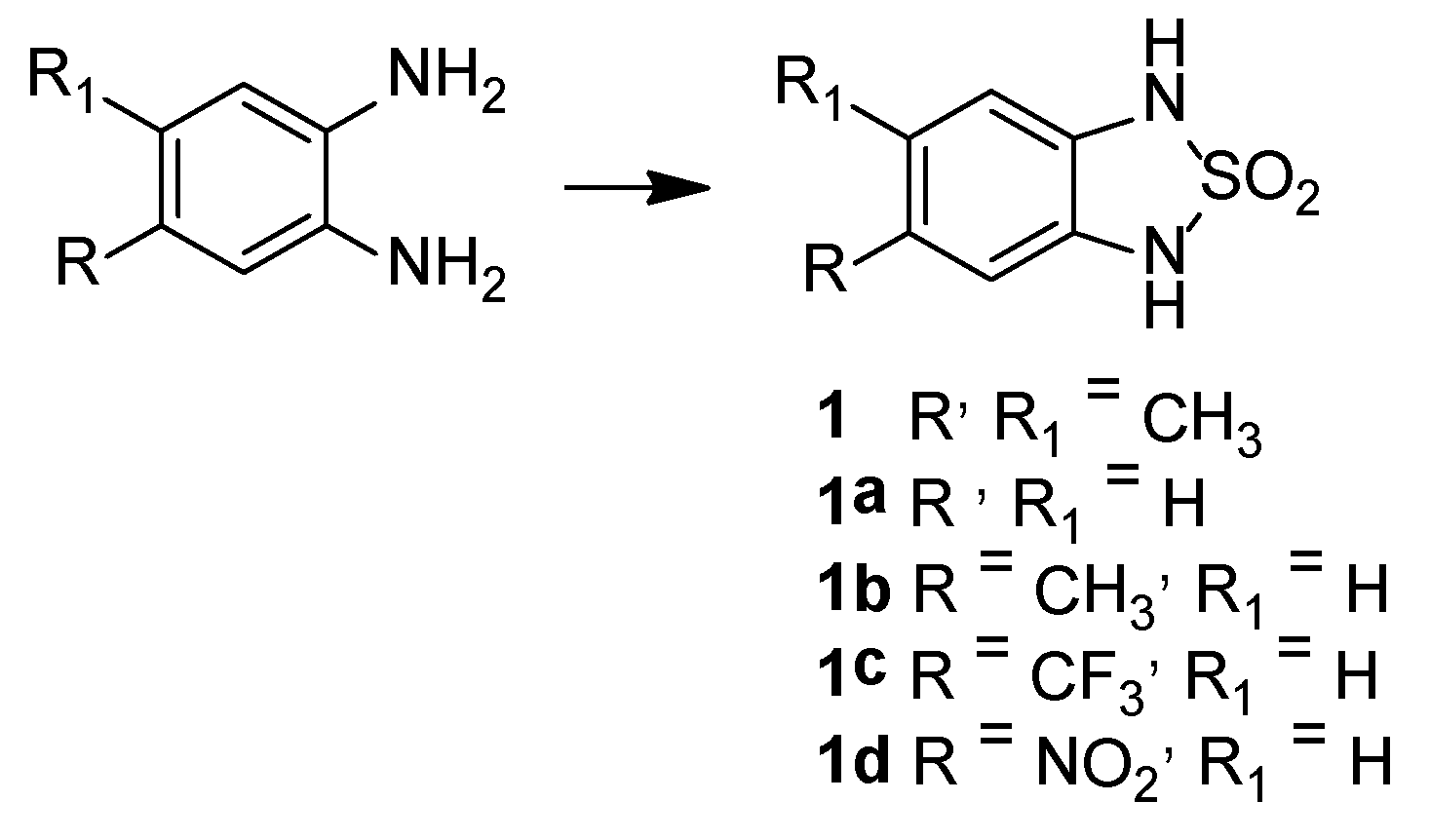
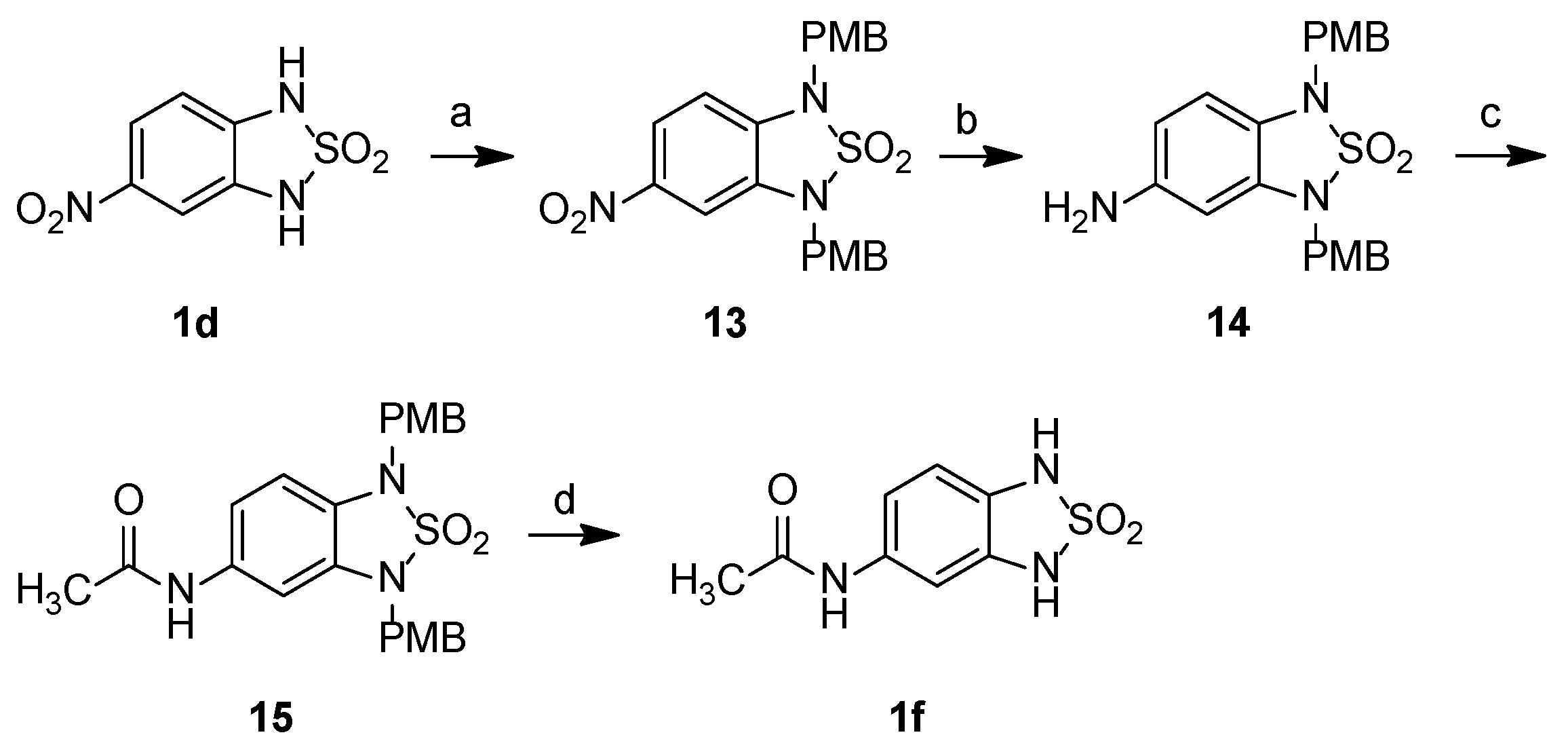
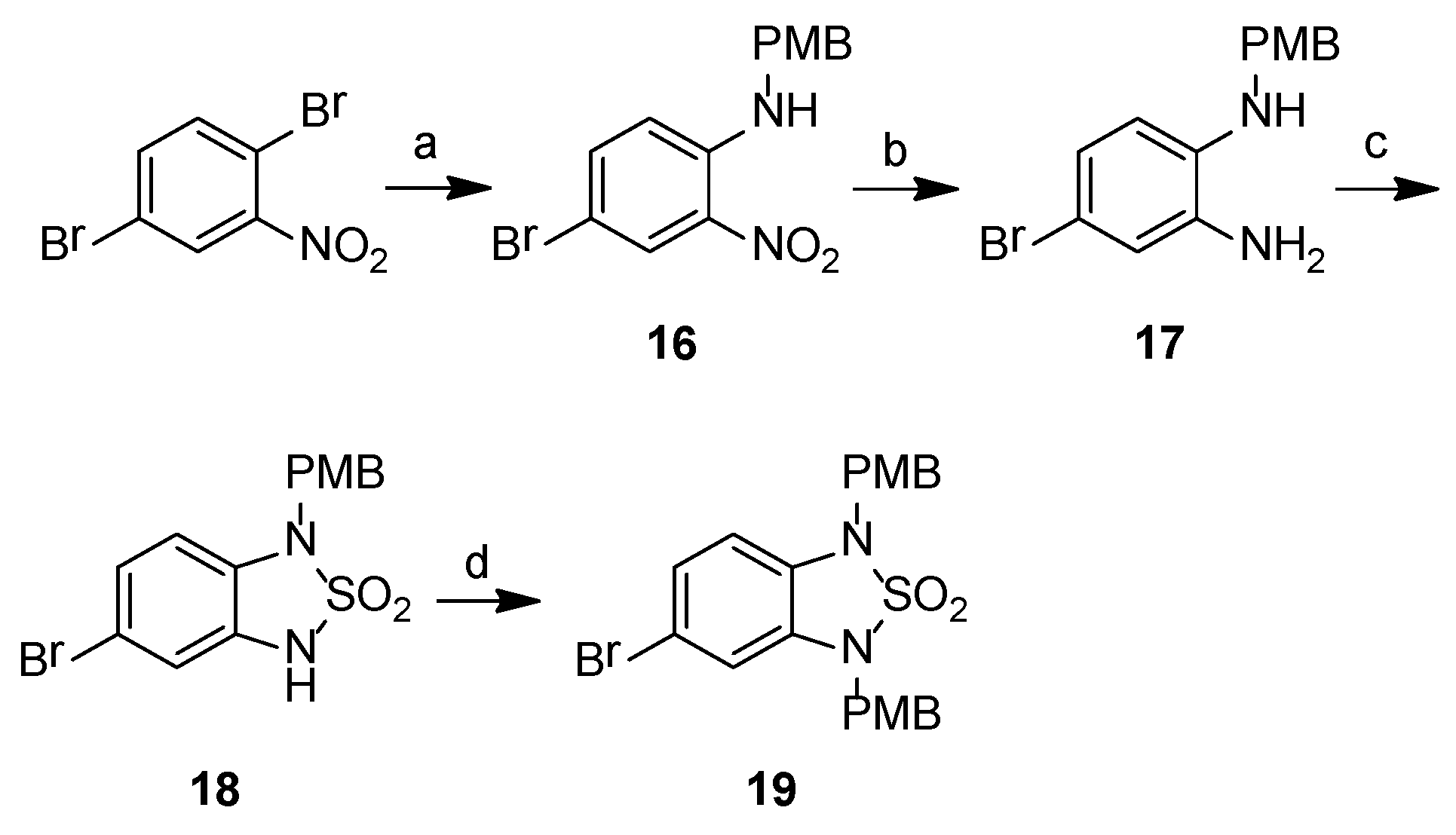
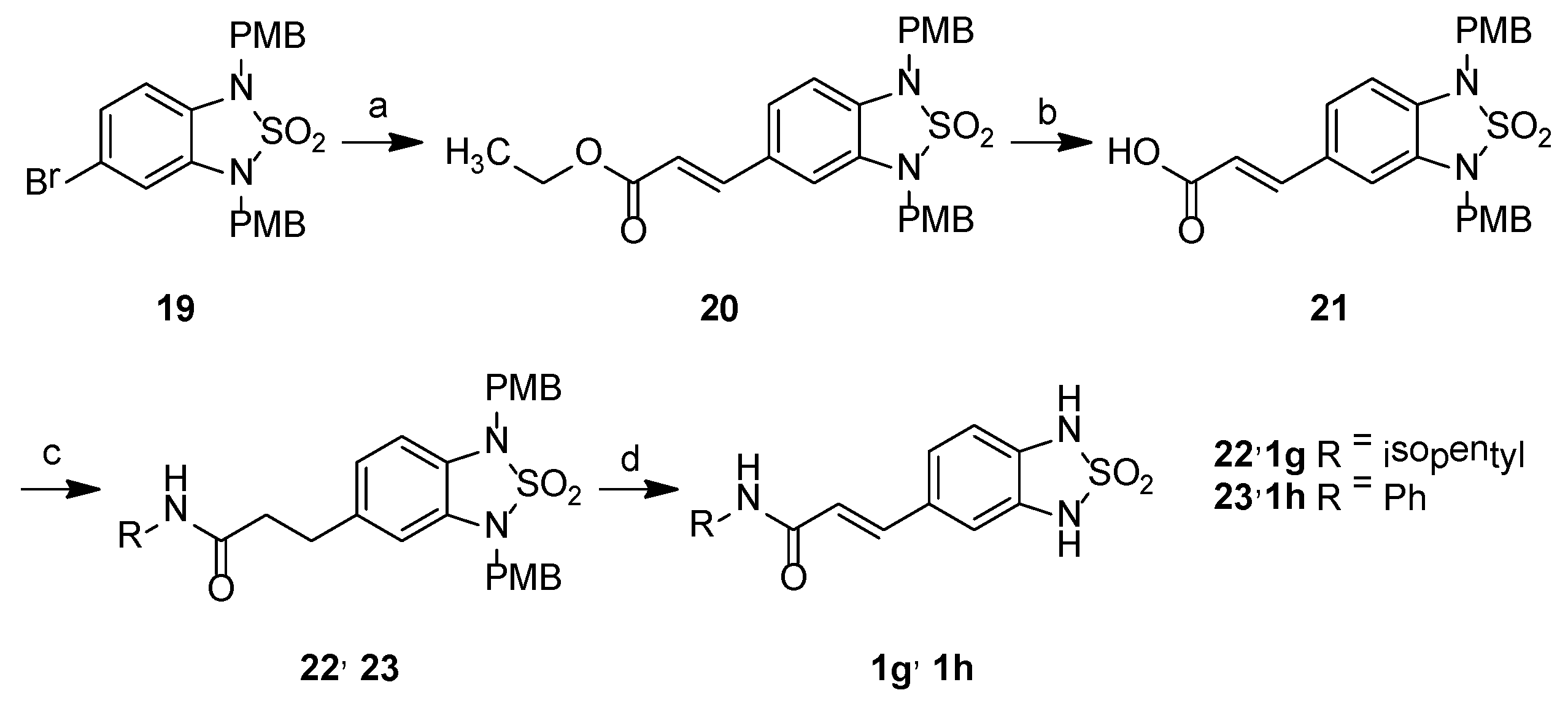
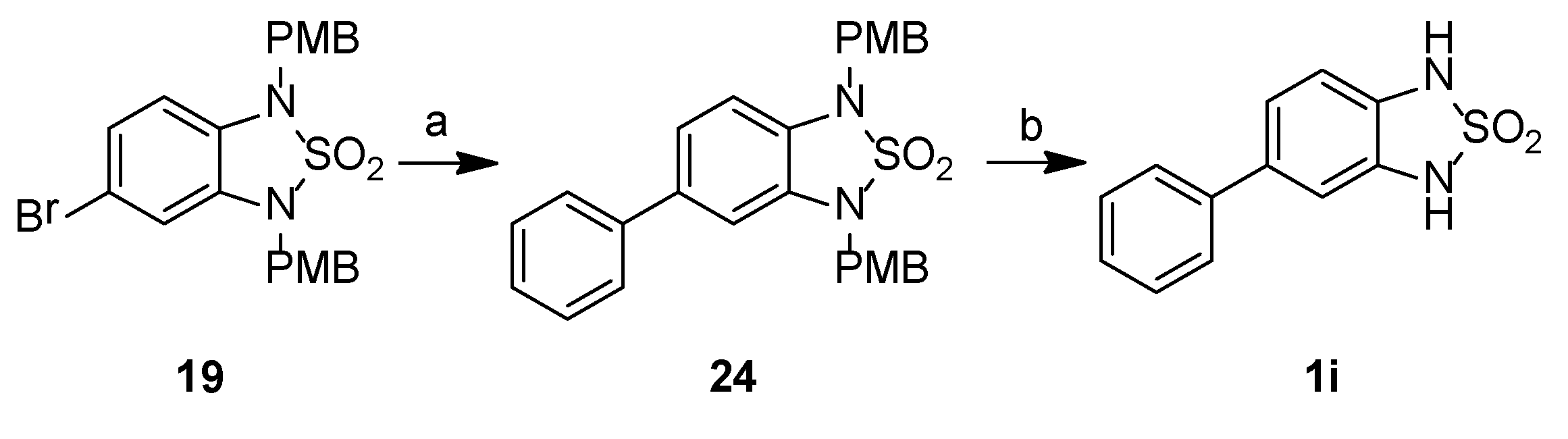
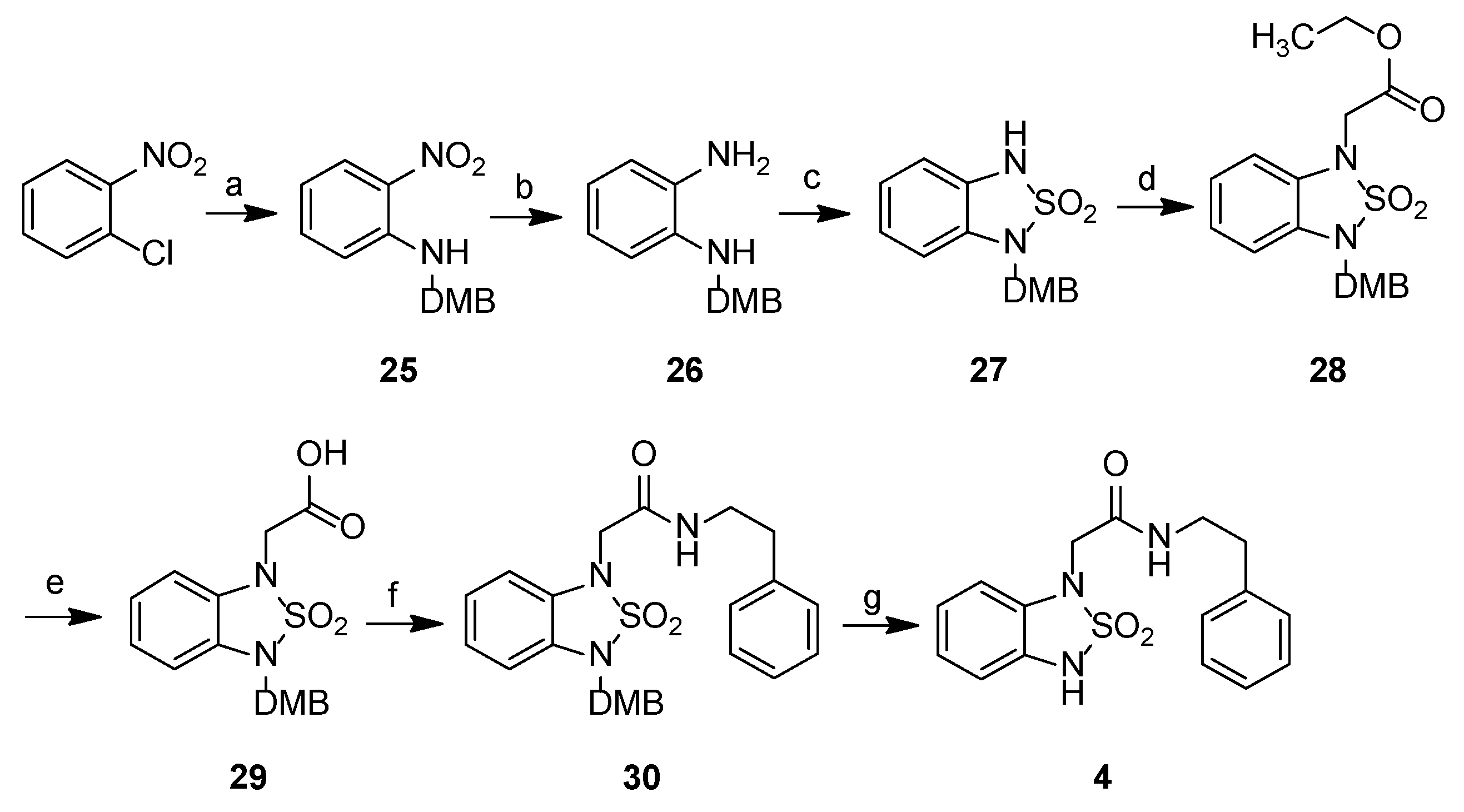
2.2. Chemistry
2.3. Biological Evaluation
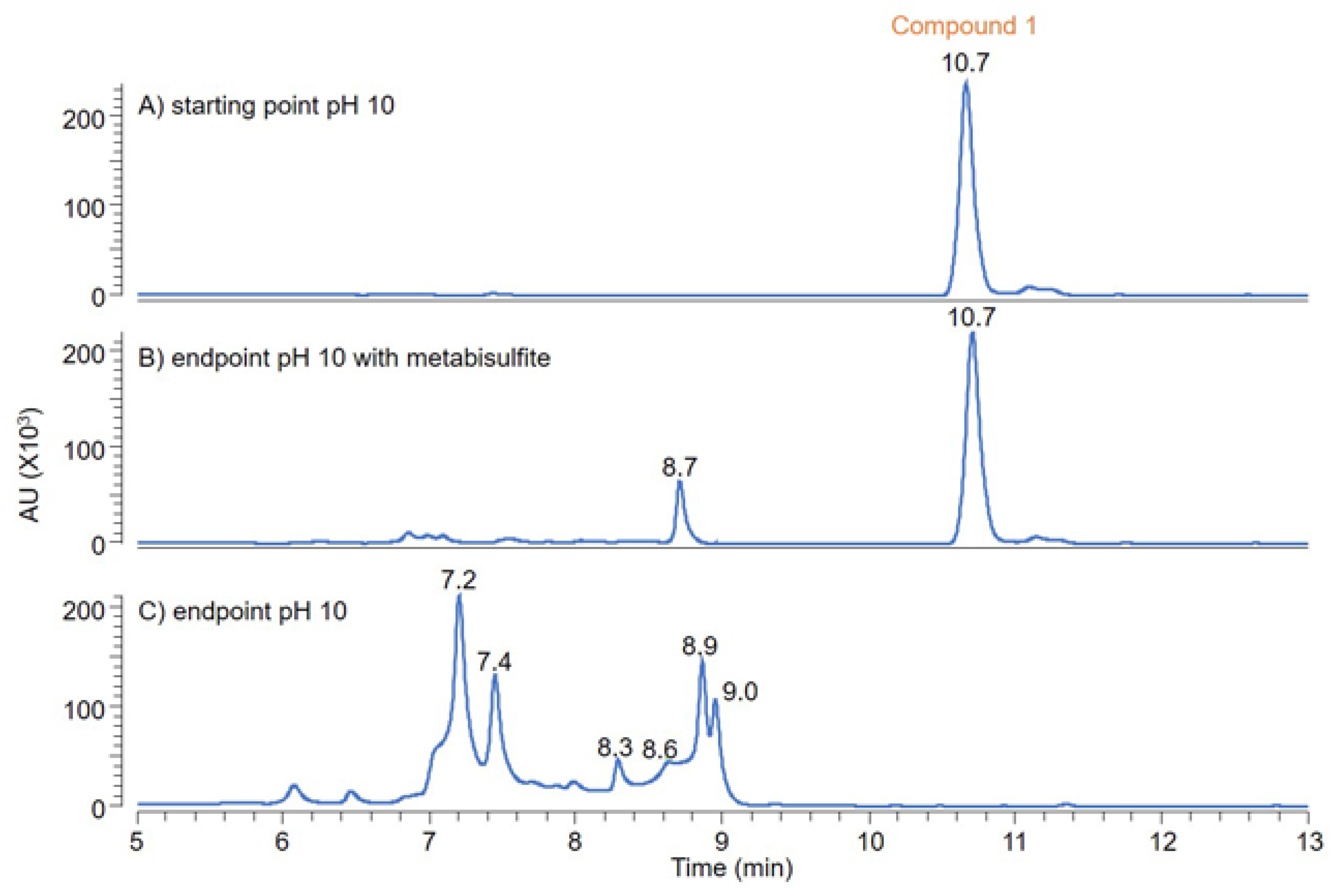
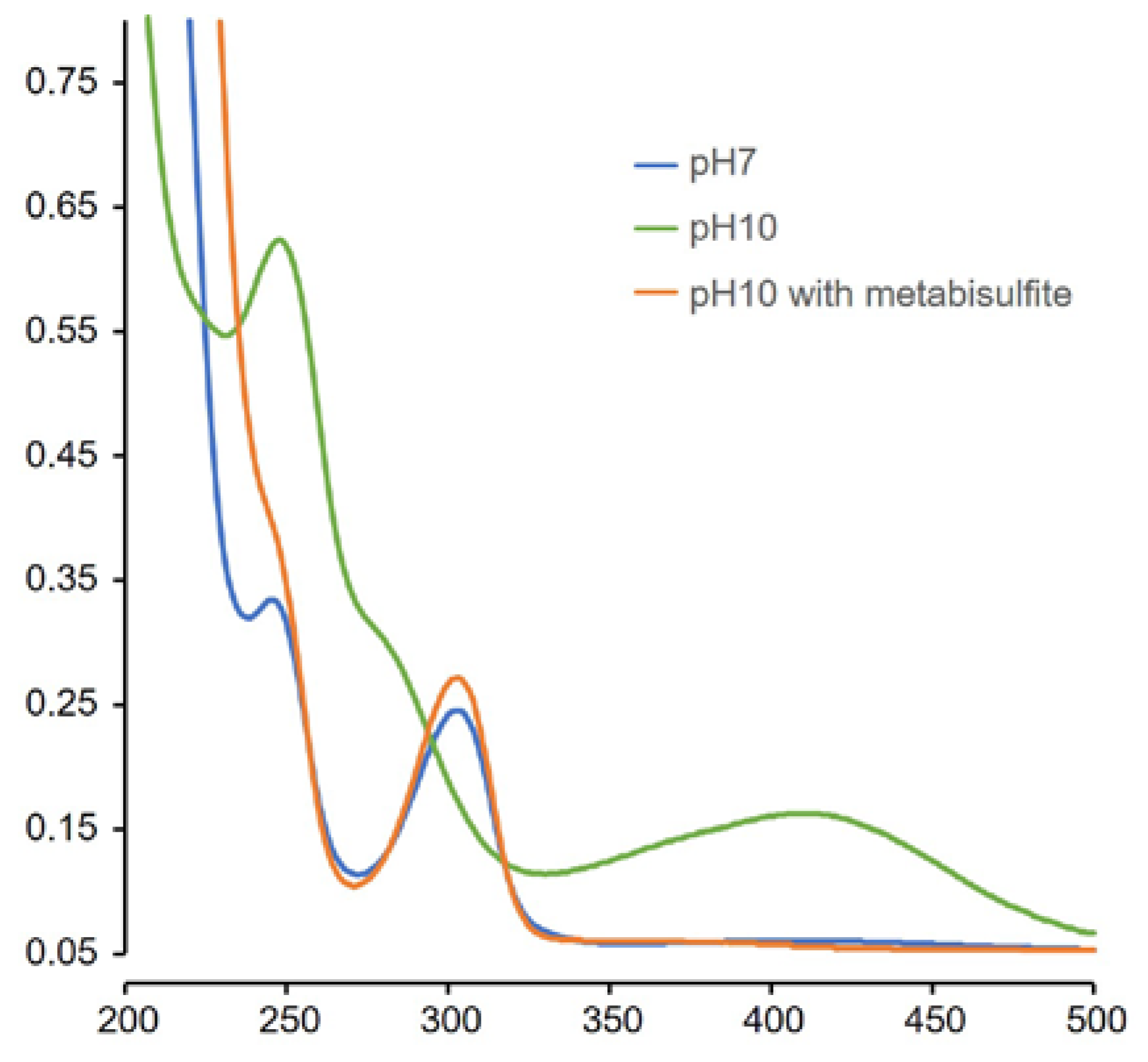
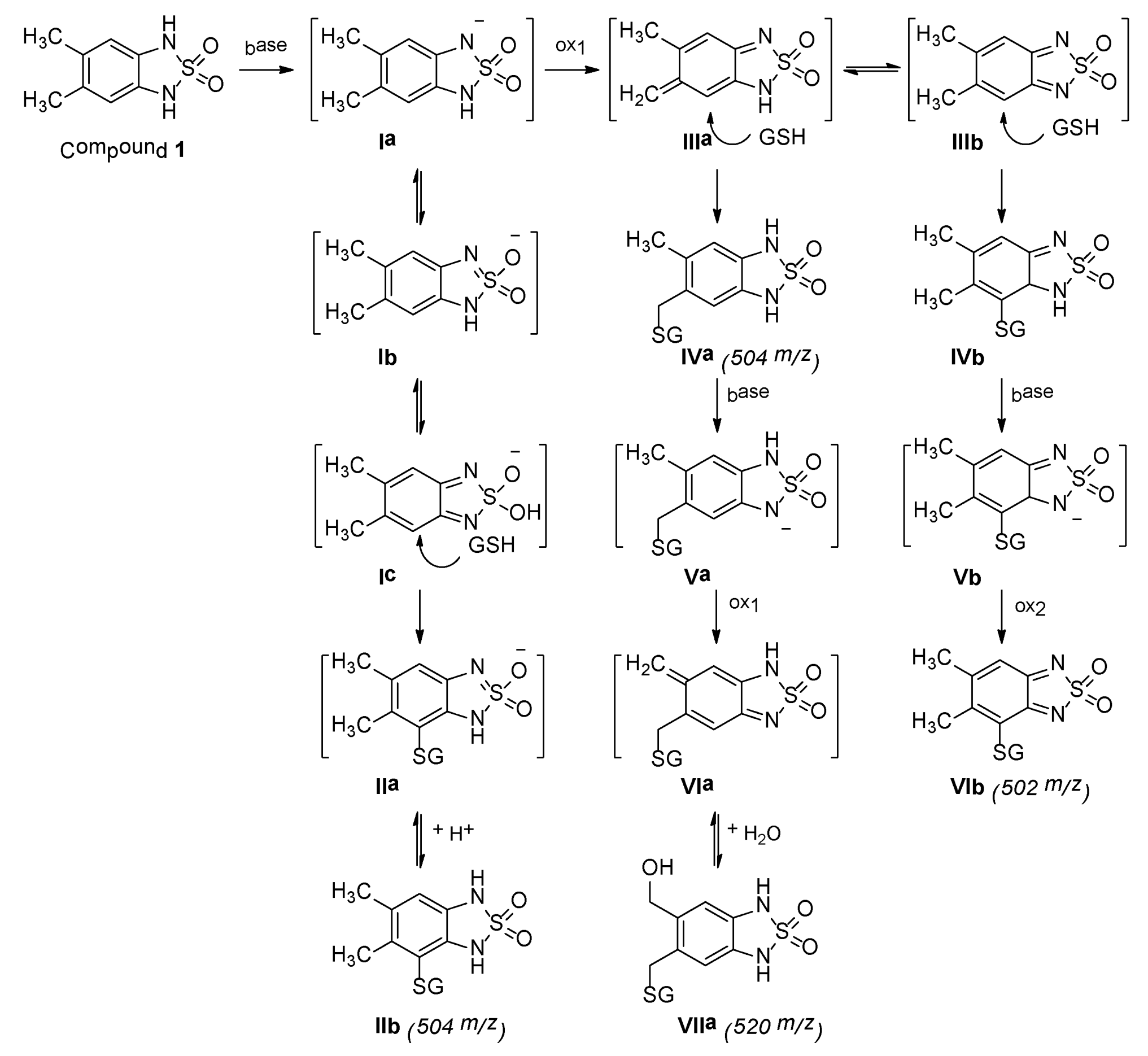
2.4. Analytical Studies
2.4.1. MS, UV, and LC Studies
2.4.2. 1H-NMR Analysis
3. Experimental Section
3.1. Computational Methods
3.2. Chemistry
3.2.1. Materials and Methods
3.2.2. Synthetic Procedures
5,6-Dimethyl-1H,3H-2,1,3-benzothiadiazole-2,2-dioxide, 1
1H,3H-2,1,3-Benzothiadiazole-2,2-dioxide, 1a
5-Methyl-1H,3H-2,1,3-benzothiadiazole-2,2-dioxide, 1b
5-(Trifluoromethyl)-1H,3H-2,1,3-benzothiadiazole-2,2-dioxide, 1c
5-Nitro-1H,3H-2,1,3-benzothiadiazole-2,2-dioxide, 1d
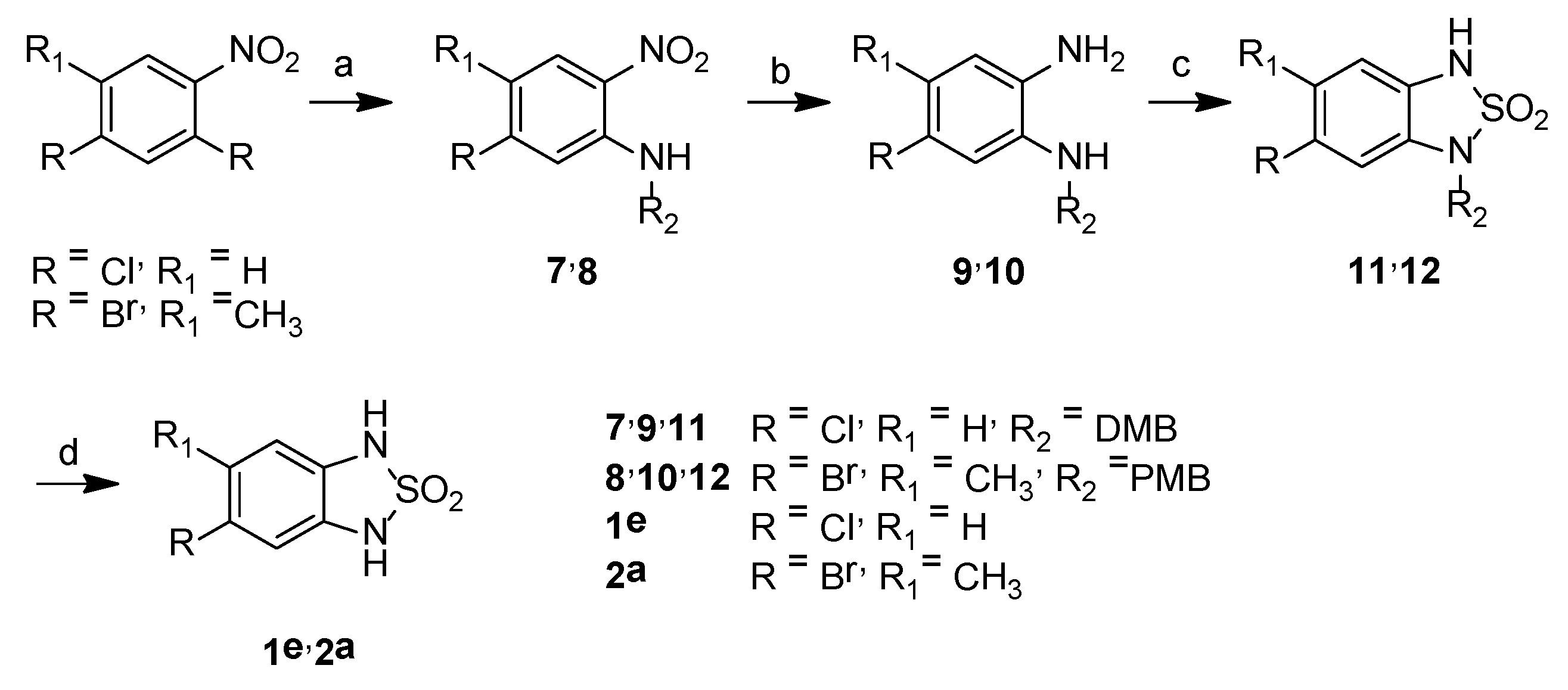
5-Chloro-N-[(2,4-dimethoxyphenyl)methyl]-2-nitroaniline, 7
5-Bromo-N-[(4-methoxyphenyl)methyl]-4-methyl-2-nitroaniline, 8
5-Chloro-N1-[(2,4-dimethoxyphenyl)methyl]benzene-1,2-diamine, 9
5-Bromo-N1-[(4-methoxyphenyl)methyl]-4-methylbenzene-1,2-diamine, 10
6-Chloro-1-[(2,4-dimethoxyphenyl)methyl]-1H,3H-2,1,3-benzothiadiazole-2,2-dioxide, 11
6-Bromo-1-[(4-methoxyphenyl)methyl]-5-methyl-1H,3H-2,1,3-benzothiadiazole-2,2-dioxide, 12
5-Chloro-1H,3H-2,1,3-benzothiadiazole-2,2-dioxide, 1e
5-Bromo-6-methyl-1H,3H-2,1,3-benzothiadiazole-2,2-dioxide, 2a
1,3-Bis[(4-methoxyphenyl)methyl]-5-nitro-2,1,3-benzothiadiazole-2,2-dioxide, 13
5-Amino-1,3-bis[(4-methoxyphenyl)methyl]-2,1,3-benzothiadiazole-2,2-dioxide, 14
N-{1,3-Bis[(4-methoxyphenyl)methyl]-2,2-dioxo-2,1,3-benzothiadiazol-5-yl}acetamide, 15
N-(2,2-Dioxo-1H,3H-2,1,3-benzothiadiazol-5-yl)acetamide, 1f
4-Bromo-N-[(4-methoxyphenyl)methyl]-2-nitroaniline, 16
4-Bromo-N1-[(4-methoxyphenyl)methyl]benzene-1,2-diamine, 17
5-Bromo-1-[(4-methoxyphenyl)methyl]-3H-2,1,3-benzothiadiazole-2,2-dioxide, 18
5-Bromo-1,3-bis[(4-methoxyphenyl)methyl]-2,1,3-benzothiadiazole-2,2-dioxide, 19
Ethyl (2E)-3-{1,3-bis[(4-methoxyphenyl)methyl]-2,2-dioxo-2,1,3-benzothiadiazol-5-yl} prop-2-enoate, 20
(2E)-3-{1,3-Bis[(4-methoxyphenyl)methyl]-2,2-dioxo-2,1,3-benzothiadiazol-5-yl} prop-2-enoic acid, 21
(2E)-3-{1,3-Bis[(4-methoxyphenyl)methyl]-2,2-dioxo-2,1,3-benzothiadiazol-5-yl}-N-(3-methylbutyl)prop-2-enamide, 22
(2E)-3-{1,3-Bis[(4-methoxyphenyl)methyl]-2,2-dioxo-2,1,3-benzothiadiazol-5-yl}-N-phenylprop-2-enamide, 23
3-(2,2-Dioxo-1H,3H-2,1,3-benzothiadiazol-5-yl)-N-(3-methylbutyl)propanamide, 1g
3-(2,2-Dioxo-1H,3H-2,1,3-benzothiadiazol-5-yl)-N-phenylpropanamide, 1h
1,3-Bis[(4-methoxyphenyl)methyl]-5-phenyl-2,1,3-benzothiadiazole-2,2-dioxide, 24
5-Phenyl-1H,3H-2,1,3-benzothiadiazole-2,2-dioxide, 1i
1,3-Dibenzyl-2,1,3-benzothiadiazole-2,2-dioxide, 3
1,3,5-Trimethyl-2,1,3-benzothiadiazole-2,2-dioxide, 2b
N-[(2,4-Dimethoxyphenyl)methyl]-2-nitroaniline, 25
N1-[(2,4-Dimethoxyphenyl)methyl]benzene-1,2-diamine, 26
1-[(2,4-Dimethoxyphenyl)methyl]-3H-2,1,3-benzothiadiazole-2,2-dioxide, 27
Ethyl 2-{3-[(2,4-dimethoxyphenyl)methyl]-2,2-dioxo-2,1,3-benzothiadiazol-1-yl}acetate, 28
2-{3-[(2,4-Dimethoxyphenyl)methyl]-2,2-dioxo-2,1,3-benzothiadiazol-1-yl} acetic acid, 29
2-{3-[(2,4-Dimethoxyphenyl)methyl]-2,2-dioxo-2,1,3-benzothiadiazol-1-yl}-N-phenethylacetamide, 30
2-(2,2-Dioxo-3H-2,1,3-benzothiadiazol-1-yl)-N-(2-phenylethyl)acetamide, 4
2,3-Dihydro-1H-1,3-benzodiazol-2-one, 5
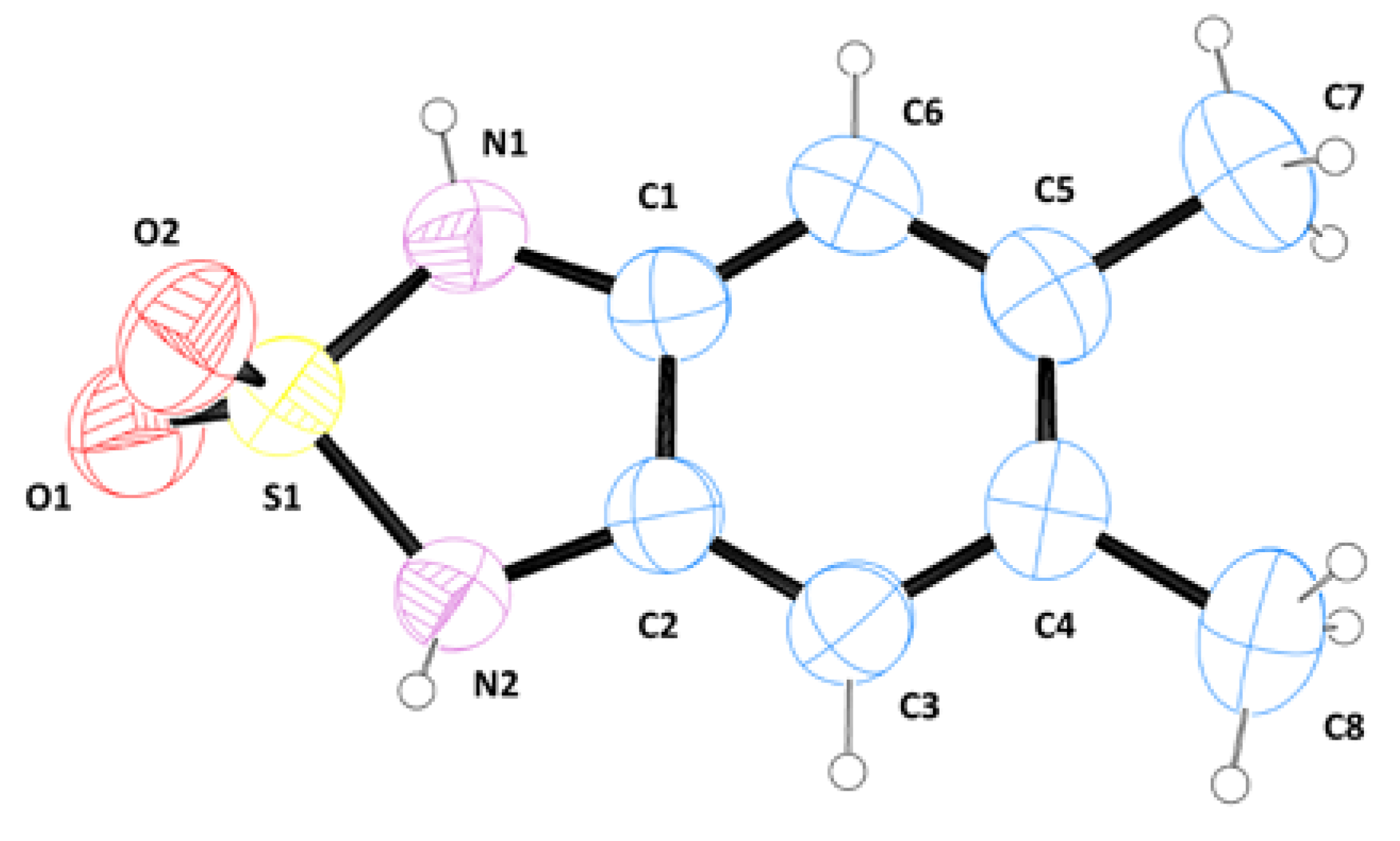
3.3. Single Crystal X-ray Analysis
3.4. Biological Evaluation
3.5. Analytical Studies
3.5.1. MS, UV, and LC Analyses
Sample Preparation
MS Method
UV Method
LC Method
3.5.2. 1H-NMR Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Masciocchi, D.; Gelain, A.; Villa, S.; Meneghetti, F.; Barlocco, D. Signal transducer and activator of transcription 3 (STAT3): A promising target for anticancer therapy. Future Med. Chem. 2011, 3, 567–597. [Google Scholar] [CrossRef] [PubMed]
- Gelain, A.; Mori, M.; Meneghetti, F.; Villa, S. Signal Transducer and Activator of Transcription Protein 3 (STAT3): An Update on its Direct Inhibitors as Promising Anticancer Agents. Curr. Med. Chem. 2018, 26, 5165–5206. [Google Scholar] [CrossRef]
- Furtek, S.L.; Backos, D.S.; Matheson, C.J.; Reigan, P. Strategies and Approaches of Targeting STAT3 for Cancer Treatment. ACS Chem. Biol. 2016, 11, 308–318. [Google Scholar] [CrossRef] [PubMed]
- Lis, C.; Rubner, S.; Roatsch, M.; Berg, A.; Gilcrest, T.; Fu, D.; Nguyen, E.; Schmidt, A.M.; Krautscheid, H.; Meiler, J.; et al. Development of Erasin: A chromone-based STAT3 inhibitor which induces apoptosis in Erlotinib-resistant lung cancer cells. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.Y.; Lee, S.H.; Han, S.W.; Kim, M.J.; Kim, T.M.; Kim, T.Y.; Heo, D.S.; Yuasa, M.; Yanagihara, Y.; Bang, Y.J. Phase I study of OPB-31121, an oral STAT3 inhibitor, in patients with advanced solid tumors. Cancer Res. Treat. 2015, 47, 607–615. [Google Scholar] [CrossRef]
- Keskin, O.; Gursoy, A.; Ma, B.; Nussinov, R. Principles of protein-protein interactions: What are the preferred ways for proteins to interact? Chem. Rev. 2008, 108, 1225–1244. [Google Scholar] [CrossRef]
- Wells, J.A.; McClendon, C.L. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature 2007, 450, 1001–1009. [Google Scholar] [CrossRef]
- Shin, D.S.; Masciocchi, D.; Gelain, A.; Villa, S.; Barlocco, D.; Meneghetti, F.; Pedretti, A.; Han, Y.M.; Han, D.C.; Kwon, B.M.; et al. Synthesis, modeling, and crystallographic study of 3,4-disubstituted-1,2,5-oxadiazoles and evaluation of their ability to decrease STAT3 activity. Medchemcomm 2010, 1, 156–164. [Google Scholar] [CrossRef]
- Masciocchi, D.; Villa, S.; Meneghetti, F.; Pedretti, A.; Barlocco, D.; Legnani, L.; Toma, L.; Kwon, B.M.; Nakano, S.; Asai, A.; et al. Biological and computational evaluation of an oxadiazole derivative (MD77) as a new lead for direct STAT3 inhibitors. Medchemcomm 2012, 3, 592–599. [Google Scholar] [CrossRef]
- Masciocchi, D.; Gelain, A.; Porta, F.; Meneghetti, F.; Pedretti, A.; Celentano, G.; Barlocco, D.; Legnani, L.; Toma, L.; Kwon, B.M.; et al. Synthesis, structure-activity relationships and stereochemical investigations of new tricyclic pyridazinone derivatives as potential STAT3 inhibitors. Medchemcomm 2013, 4, 1181–1188. [Google Scholar] [CrossRef]
- Meneghetti, F.; Villa, S.; Masciocchi, D.; Barlocco, D.; Toma, L.; Han, D.C.; Kwon, B.M.; Ogo, N.; Asai, A.; Legnani, L.; et al. Ureido-Pyridazinone Derivatives: Insights into the Structural and Conformational Properties for STAT3 Inhibition. Eur. J. Org. Chem. 2015, 2015, 4907–4912. [Google Scholar] [CrossRef]
- Gabriele, E.; Porta, F.; Facchetti, G.; Galli, C.; Gelain, A.; Meneghetti, F.; Rimoldi, I.; Romeo, S.; Villa, S.; Ricci, C.; et al. Synthesis of new dithiolethione and methanethiosulfonate systems endowed with pharmaceutical interest. Arkivoc 2016, 2017, 235–250. [Google Scholar] [CrossRef]
- Porta, F.; Facchetti, G.; Ferri, N.; Gelain, A.; Meneghetti, F.; Villa, S.; Barlocco, D.; Masciocchi, D.; Asai, A.; Miyoshi, N.; et al. An in vivo active 1,2,5-oxadiazole Pt(II) complex: A promising anticancer agent endowed with STAT3 inhibitory properties. Eur. J. Med. Chem. 2017, 131, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Colyer, D.E.; Nortcliffe, A.; Wheeler, S. A general synthetic strategy for 1,3-dihydro-2,1,3-benzothiadiazole 2,2-dioxides (benzosulfamides). Tetrahedron Lett. 2010, 51, 5306–5308. [Google Scholar] [CrossRef]
- Buettner, R.; Corzano, R.; Rashid, R.; Lin, J.; Senthil, M.; Hedvat, M.; Schroeder, A.; Mao, A.; Herrmann, A.; Yim, J.; et al. Alkylation of cysteine 468 in stat3 defines a novel site for therapeutic development. ACS Chem. Biol. 2011, 6, 432–443. [Google Scholar] [CrossRef]
- Heidelberger, S.; Zinzalla, G.; Antonow, D.; Essex, S.; Piku Basu, B.; Palmer, J.; Husby, J.; Jackson, P.J.M.; Rahman, K.M.; Wilderspin, A.F.; et al. Investigation of the protein alkylation sites of the STAT3:STAT3 inhibitor Stattic by mass spectrometry. Bioorg. Med. Chem. Lett. 2013, 23, 4719–4722. [Google Scholar] [CrossRef]
- Fletcher, S.; Page, B.D.G.; Zhang, X.; Yue, P.; Li, Z.H.; Sharmeen, S.; Singh, J.; Zhao, W.; Schimmer, A.D.; Trudel, S.; et al. Antagonism of the Stat3-Stat3 Protein Dimer with Salicylic Acid Based Small Molecules. Chem. Med. Chem. 2011, 6, 1459–1470. [Google Scholar] [CrossRef]
- Ball, D.P.; Lewis, A.M.; Williams, D.; Resetca, D.; Wilson, D.J.; Gunning, P.T. Signal transducer and activator of transcription 3 (STAT3) inhibitor, S3I-201, acts as a potent and non-selective alkylating agent. Oncotarget 2016, 7, 20669–20679. [Google Scholar] [CrossRef]
- Becker, S.; Groner, B.; Müller, C.W. Three-dimensional structure of the Stat3β homodimer bound to DNA. Nature 1998, 394, 145–151. [Google Scholar] [CrossRef]
- Goodsell, D.S.; Olson, A.J. Automated docking of substrates to proteins by simulated annealing. Proteins Struct. Funct. Bioinform. 1990, 8, 195–202. [Google Scholar] [CrossRef]
- Schmidhammer, H.; Hohenlohe-Oehringen, K. Synthesis of dopa and dopamine analogs with acidic imido-functionalities in heterocyclic ring moieties instead of the phenolic hydroxyl groups. Sci. Pharm. 1983, 51, 8–16. [Google Scholar]
- Burke, P.O.; McDermott, S.D.; Hannigan, T.J.; Spillane, W.J. Basicity of nitrogen-sulphur(VI) compounds. Part 6. Ionization of NN’-di-and N-mono-substituted sulphamides and dihydro-2,1,3-benzothiadiazoline and benzothiadiazine 2,2-dioxides (cyclic sulphamides). J. Chem. Soc. Perkin Trans. 1984, 2, 1851–1854. [Google Scholar] [CrossRef]
- Farrugia, L.J. WinGX suite for small-molecule single-crystal crystallography. J. Appl. Crystallogr. 1999, 32, 837–838. [Google Scholar] [CrossRef]
- Kortylewski, M.; Jove, R.; Yu, H. Targeting STAT3 affects melanoma on multiple fronts. Cancer Metastasis Rev. 2005, 24, 315–327. [Google Scholar] [CrossRef]
- McMurray, J.S.; Klostergaard, J. STAT3 Signaling in Cancer: Small Molecule Intervention as Therapy? In Anti-Angiogenesis Drug Discovery and Development; Elsevier: Amsterdam, The Netherlands, 2014; pp. 216–267. [Google Scholar]
- Dahlin, J.L.; Nissink, J.W.M.; Strasser, J.M.; Francis, S.; Higgins, L.; Zhou, H.; Zhang, Z.; Walters, M.A. PAINS in the assay: Chemical mechanisms of assay interference and promiscuous enzymatic inhibition observed during a sulfhydryl-scavenging HTS. J. Med. Chem. 2015, 58, 2091–2113. [Google Scholar] [CrossRef] [PubMed]
- FAF-Drugs3: A Web Server for Compound Property Calculation and Chemical Library Design—PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/25883137/ (accessed on 13 July 2020).
- Baell, J.B.; Holloway, G.A. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 2010, 53, 2719–2740. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [PubMed]
- Wen, B.; Fitch, W.L. Screening and characterization of reactive metabolites using glutathione ethyl ester in combination with Q-trap mass spectrometry. J. Mass Spectrom. 2009, 44, 90–100. [Google Scholar] [CrossRef]
- Mayer, R.J.; Ofial, A.R. Nucleophilicity of Glutathione: A Link to Michael Acceptor Reactivities. Angew. Chem.-Int. Ed. 2019, 58, 17704–17708. [Google Scholar] [CrossRef]
- Maier, G.P.; Bernt, C.M.; Butler, A. Catechol oxidation: Considerations in the design of wet adhesive materials. Biomater. Sci. 2018, 6, 332–339. [Google Scholar] [CrossRef] [PubMed]
- Asiamah, I.; Krol, E.S. Quadrupole linear ion-trap mass spectrometry studies on glutathione conjugates of nordihydroguaiaretic acid (NDGA) analogues reveals phenol-type analogues are without reactive metabolite-mediated toxic liability. Cogent Chem. 2018, 4, 1562858. [Google Scholar] [CrossRef]
- Iverson, S.L.; Shen, L.; Anlar, N.; Bolton, J.L. Bioactivation of estrone and its catechol metabolites to quinoid-glutathione conjugates in rat liver microsomes. Chem. Res. Toxicol. 1996, 9, 492–499. [Google Scholar] [CrossRef] [PubMed]
- Toteva, M.M.; Richard, J.P. The generation and reactions of quinone methides. Adv. Phys. Org. Chem. 2011, 45, 39–91. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.M.; Gómez-Biagi, R.F.; Rosa, D.A.; Lai, P.-S.; Heaton, W.L.; Park, J.S.; Eiring, A.M.; Vellore, N.A.; de Araujo, E.D.; Ball, D.P.; et al. Disarming an Electrophilic Warhead: Retaining Potency in Tyrosine Kinase Inhibitor (TKI)-Resistant CML Lines While Circumventing Pharmacokinetic Liabilities. ChemMedChem 2016, 11, 850–861. [Google Scholar] [CrossRef]
- Vila-Viçosa, D.; Teixeira, V.H.; Santos, H.A.F.; Machuqueiro, M. Conformational study of GSH and GSSG using constant-pH molecular dynamics simulations. J. Phys. Chem. B 2013, 117, 7507–7517. [Google Scholar] [CrossRef]
- Pedretti, A.; Villa, L.; Vistoli, G. VEGA: A versatile program to convert, handle and visualize molecular structure on Windows-based PCs. J. Mol. Graph. Model. 2002, 21, 47–49. [Google Scholar] [CrossRef]
- Gasteiger, J.; Marsili, M. Iterative partial equalization of orbital electronegativity-a rapid access to atomic charges. Tetrahedron 1980, 36, 3219–3228. [Google Scholar] [CrossRef]
- MacKerell, A.D.; Bashford, D.; Bellott, M.; Dunbrack, R.L.; Evanseck, J.D.; Field, M.J.; Fischer, S.; Gao, J.; Guo, H.; Ha, S.; et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 1998, 102, 3586–3616. [Google Scholar] [CrossRef]
- Foloppe, N.; Alexander, D.; MacKerell, J. All-atom empirical force field for nucleic acids: I. Parameter optimization based on small molecule and condensed phase macromolecular target data. J. Comput. Chem. 2000, 21, 86–104. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2008, 26, 1781–1802. [Google Scholar] [CrossRef]
- Grotthuss, M.; Koczyk, G.; Pas, J.; Wyrwicz, L.; Rychlewski, L. Ligand.Info Small-Molecule Meta-Database. Comb. Chem. High Throughput Screen. 2005, 7, 757–761. [Google Scholar] [CrossRef] [PubMed]
- Pedretti, A.; Mazzolari, A.; Vistoli, G. WarpEngine, a Flexible Platform for Distributed Computing Implemented in the VEGA Program and Specially Targeted for Virtual Screening Studies. J. Chem. Inf. Model. 2018, 58, 1154–1160. [Google Scholar] [CrossRef]
- Krause, L.; Herbst-Irmer, R.; Sheldrick, G.M.; Stalke, D. Comparison of silver and molybdenum microfocus X-ray sources for single-crystal structure determination. J. Appl. Crystallogr. 2015, 48, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Burla, M.C.; Caliandro, R.; Carrozzini, B.; Cascarano, G.L.; Cuocci, C.; Giacovazzo, C.; Mallamo, M.; Mazzone, A.; Polidori, G. Crystal structure determination and refinement via SIR2014. J. Appl. Crystallogr. 2015, 48, 306–309. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Farrugia, L.J. WinGX and ORTEP for Windows: An update. J. Appl. Crystallogr. 2012, 45, 849–854. [Google Scholar] [CrossRef]
- Matsuno, K.; Masuda, Y.; Uehara, Y.; Sato, H.; Muroya, A.; Takahashi, O.; Yokotagawa, T.; Furuya, T.; Okawara, T.; Otsuka, M.; et al. Identification of a new series of STAT3 inhibitors by virtual screening. ACS Med. Chem. Lett. 2010, 1, 371–375. [Google Scholar] [CrossRef]
- Uehara, Y.; Mochizuki, M.; Matsuno, K.; Haino, T.; Asai, A. Novel high-throughput screening system for identifying STAT3-SH2 antagonists. Biochem. Biophys. Res. Commun. 2009, 380, 627–631. [Google Scholar] [CrossRef]
- Asai, A.; Takakuma, K. Expression and Purification of Soluble STAT5b/STAT3 Proteins for SH2 Domain Binding Assay. In SH2 Domains: Methods and Protocols; Machida, K., Liu, B.A., Eds.; Springer: New York, NY, USA, 2017; pp. 163–172. [Google Scholar]
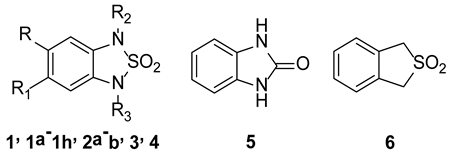
Sample Availability: Samples of the tested compounds (1a–i, 2a–b, 4–6) are available upon request from the authors. |




| Compound | Structure | % Inhibition |
|---|---|---|
| 1 |  | 96.5 |
| I | 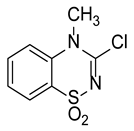 | 79.0 |
| II | 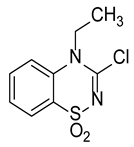 | 43.4 |
| III | 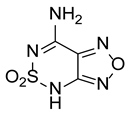 | 85.4 |
 | ||
|---|---|---|
| Compound | Substituents | STAT3 % Inhibition (30 µM) |
| 1 | R, R1 = CH3; R2, R3 = H | 96.5 ± 1.1 [a] |
| 1a | R, R1, R2, R3 = H | 16.4 ± 0.8 [b] |
| 1b | R = H; R1 = CH3; R2, R3 = H | 14.5 ± 0.6 [b] |
| 1c | R = H; R1 = CF3; R2, R3 = H | 8.2 ± 1.6 [b] |
| 1d | R = H; R1 = NO2; R2, R3 = H | 56.9 ± 3.6 [c] |
| 1e | R = H; R1 = Cl; R2, R3 = H | 28.2 ± 1.4 [b] |
| 1f | R = H; R1 = NHCOCH3; R2, R3 = H | 18.9 ± 2.3 [b] |
| 1g | R = H; R1 = (CH2)2CONH(CH2)2CH3; R2, R3 = H | 11 ± 2.1 [b] |
| 1h | R = H; R1 = (CH2)2CONHPh; R2, R3 = H | n.a. |
| 1i | R = H; R1 = Ph; R2, R3 = H | 6.1 ± 0.9 [b] |
| 2a | R = CH3; R1 = Br; R2, R3 = H | 25.6 ± 1.2 [b] |
| 2b | R = H; R1, R2, R3 = CH3 | 8.9 ± 1.1 [b] |
| 3 | R, R1 = H; R2, R3 = CH2Ph | n.a. |
| 4 | R, R1, R3 = H; R2 = CH2CONH(CH2)2Ph | 17.3 ± 1.3 [b] |
| 5 | - | 3.5 ± 5.6 [b] |
| 6 | - | 11.5 ± 0.8 [b] |
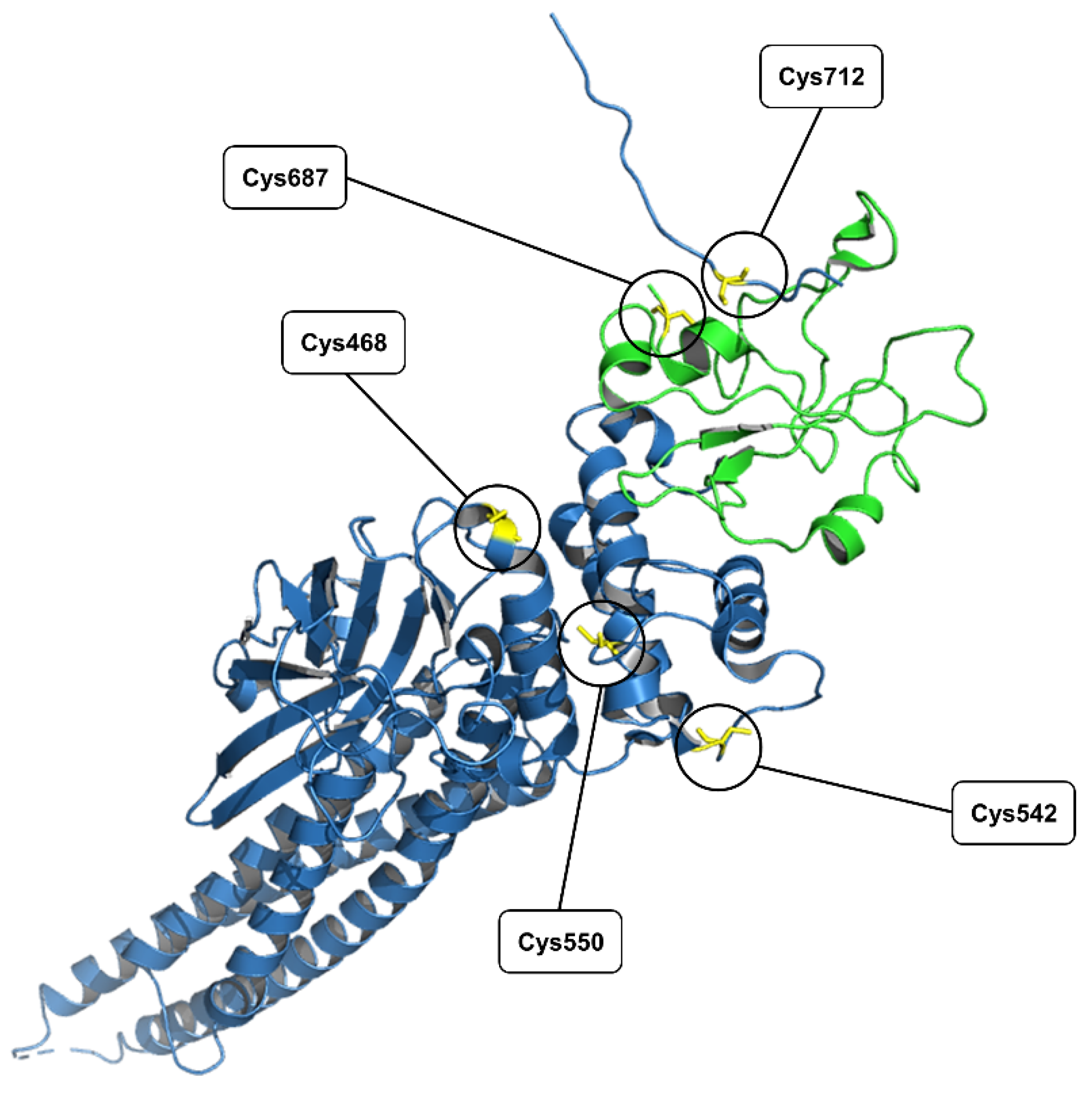
| Compound 1 vs. STAT3 | |||
|---|---|---|---|
| Mutated Cys | IC50 (µM) | Mutated Cys | IC50 (µM) |
| WT | 15.8 ± 0.6 | Cys550A | >30 |
| Cys468A | >30 | Cys687A | >30 |
| Cys542A | >30 | Cys712A | >30 |
| Time (min) | 1 mM HCl, pH 3 (A) | ACN (B) |
|---|---|---|
| 0.00 | 95 | 5 |
| 1.00 | 95 | 5 |
| 16.00 | 30 | 70 |
| 16.50 | 30 | 70 |
| 16.51 | 95 | 5 |
| 22.00 | 95 | 5 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mori, M.; Gilardoni, E.; Regazzoni, L.; Pedretti, A.; Colombo, D.; Parkinson, G.; Asai, A.; Meneghetti, F.; Villa, S.; Gelain, A. Towards the Inhibition of Protein–Protein Interactions (PPIs) in STAT3: Insights into a New Class of Benzothiadiazole Derivatives. Molecules 2020, 25, 3509. https://doi.org/10.3390/molecules25153509
Mori M, Gilardoni E, Regazzoni L, Pedretti A, Colombo D, Parkinson G, Asai A, Meneghetti F, Villa S, Gelain A. Towards the Inhibition of Protein–Protein Interactions (PPIs) in STAT3: Insights into a New Class of Benzothiadiazole Derivatives. Molecules. 2020; 25(15):3509. https://doi.org/10.3390/molecules25153509
Chicago/Turabian StyleMori, Matteo, Ettore Gilardoni, Luca Regazzoni, Alessandro Pedretti, Diego Colombo, Gary Parkinson, Akira Asai, Fiorella Meneghetti, Stefania Villa, and Arianna Gelain. 2020. "Towards the Inhibition of Protein–Protein Interactions (PPIs) in STAT3: Insights into a New Class of Benzothiadiazole Derivatives" Molecules 25, no. 15: 3509. https://doi.org/10.3390/molecules25153509
APA StyleMori, M., Gilardoni, E., Regazzoni, L., Pedretti, A., Colombo, D., Parkinson, G., Asai, A., Meneghetti, F., Villa, S., & Gelain, A. (2020). Towards the Inhibition of Protein–Protein Interactions (PPIs) in STAT3: Insights into a New Class of Benzothiadiazole Derivatives. Molecules, 25(15), 3509. https://doi.org/10.3390/molecules25153509










