VAE-Sim: A Novel Molecular Similarity Measure Based on a Variational Autoencoder
Abstract
1. Introduction
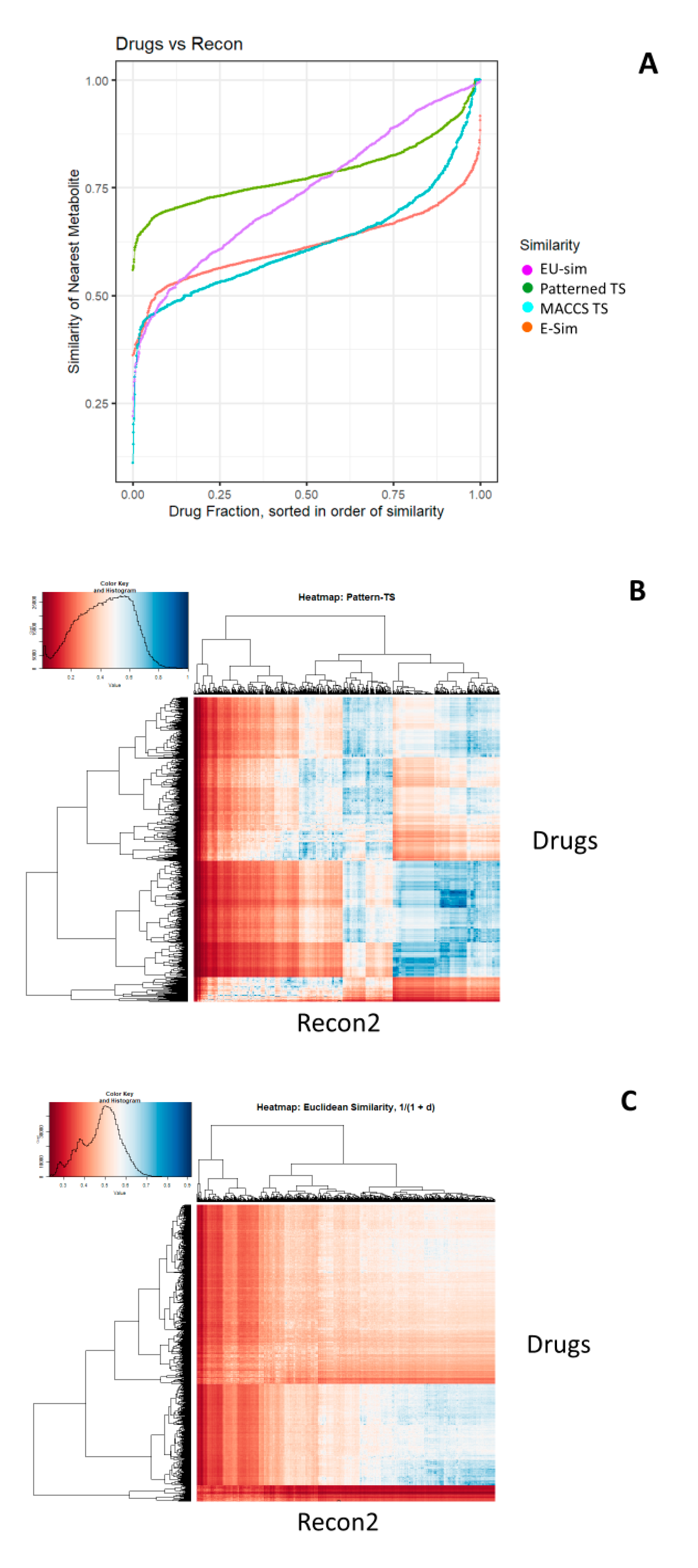
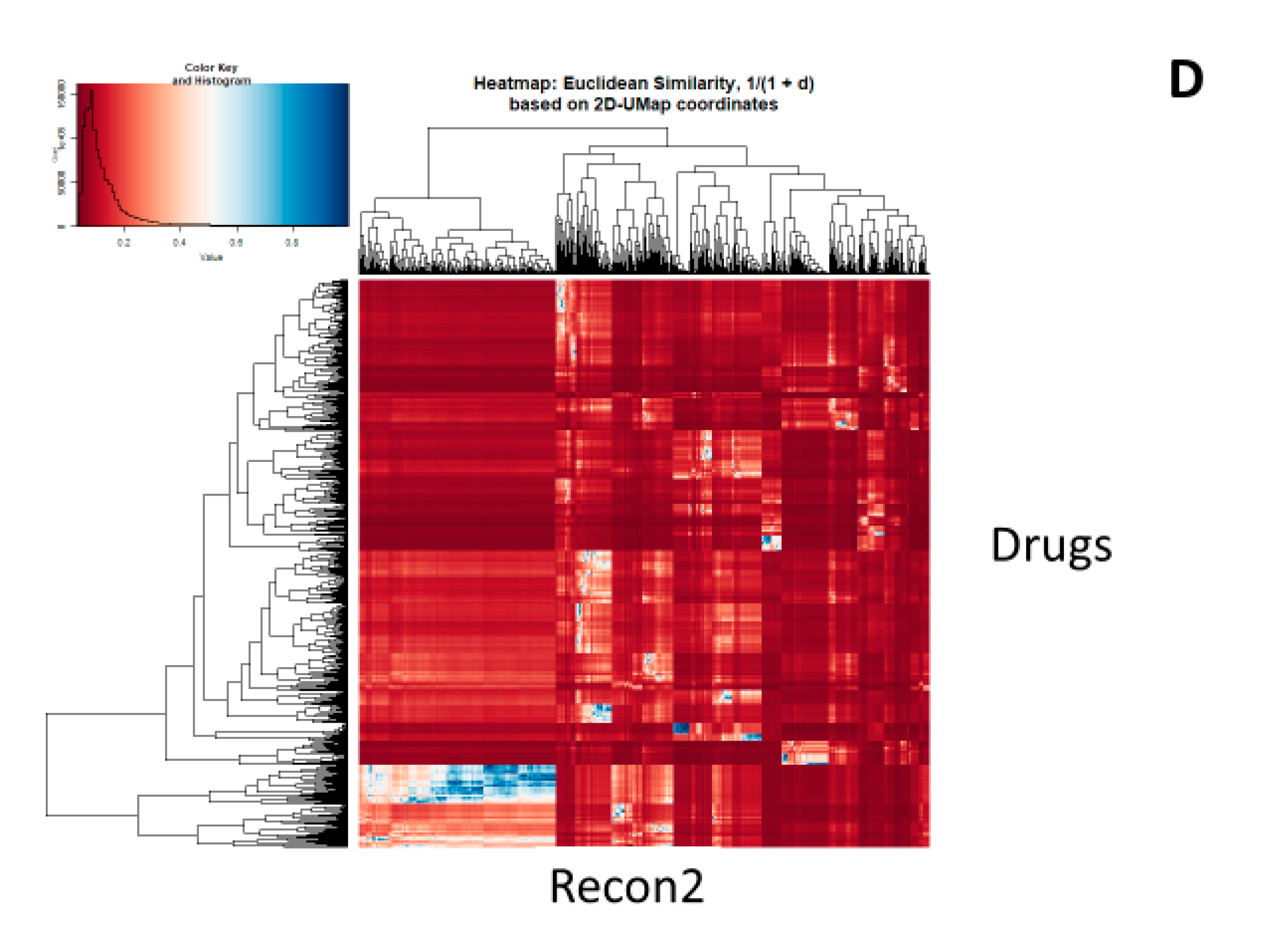
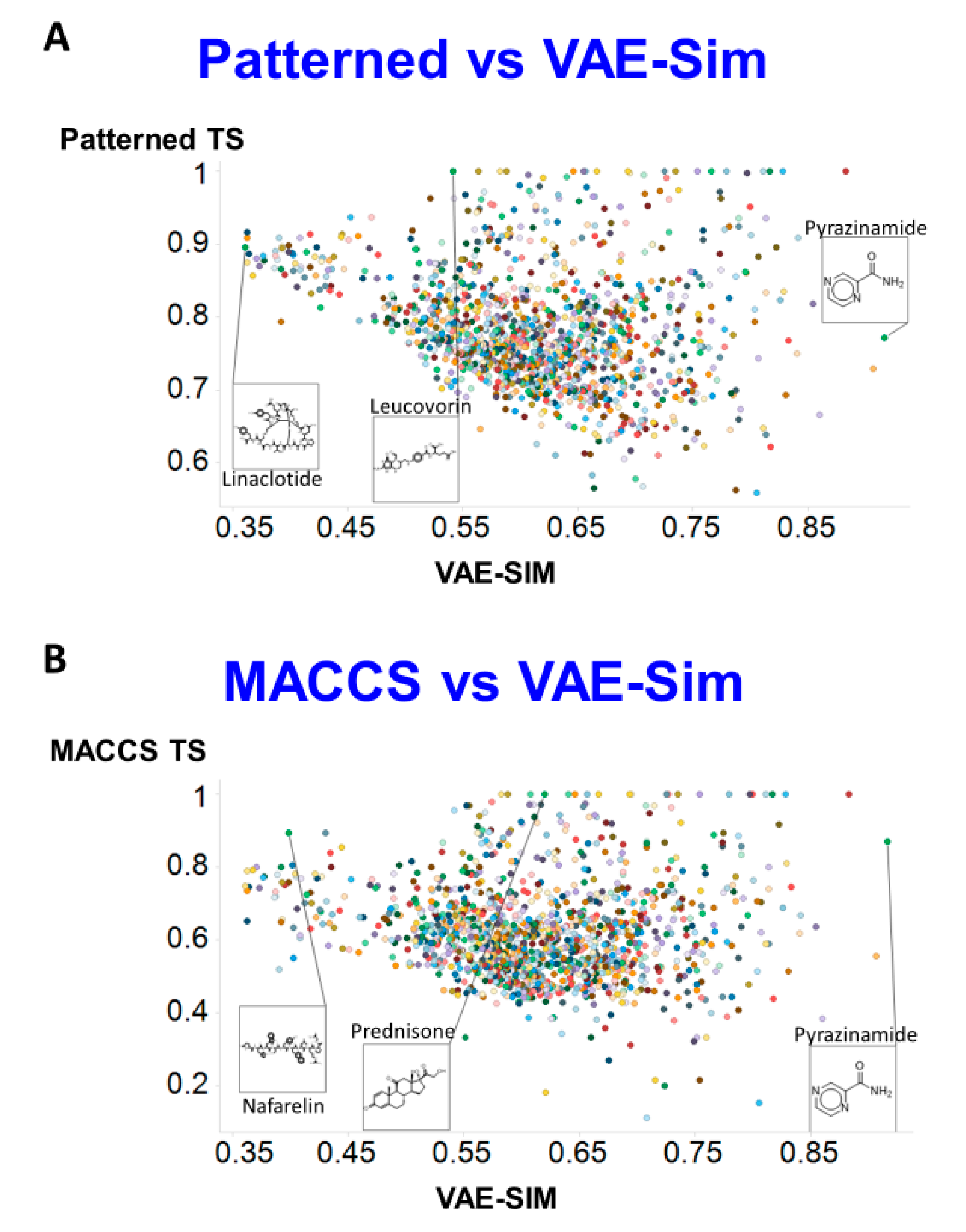
2. Results
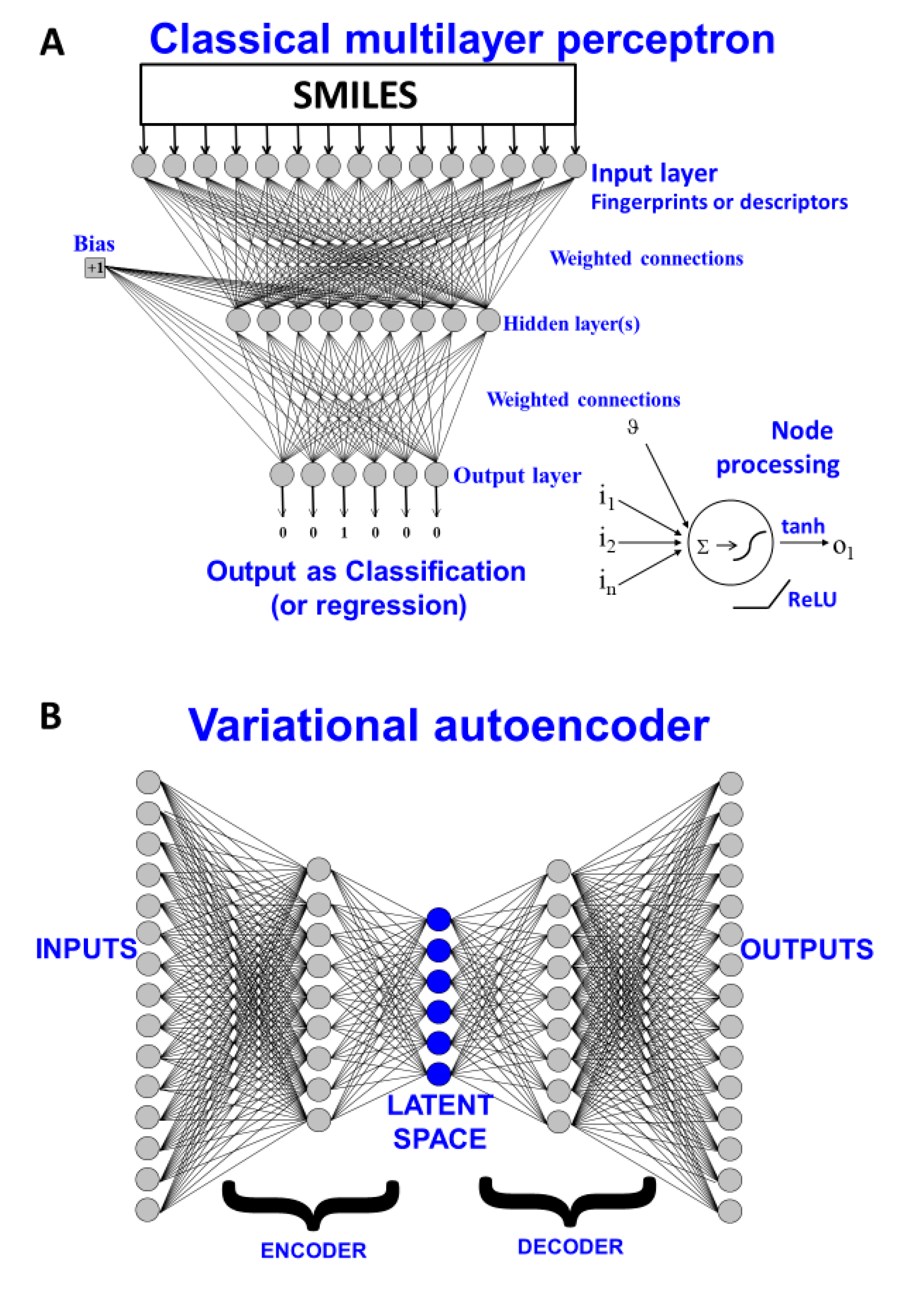
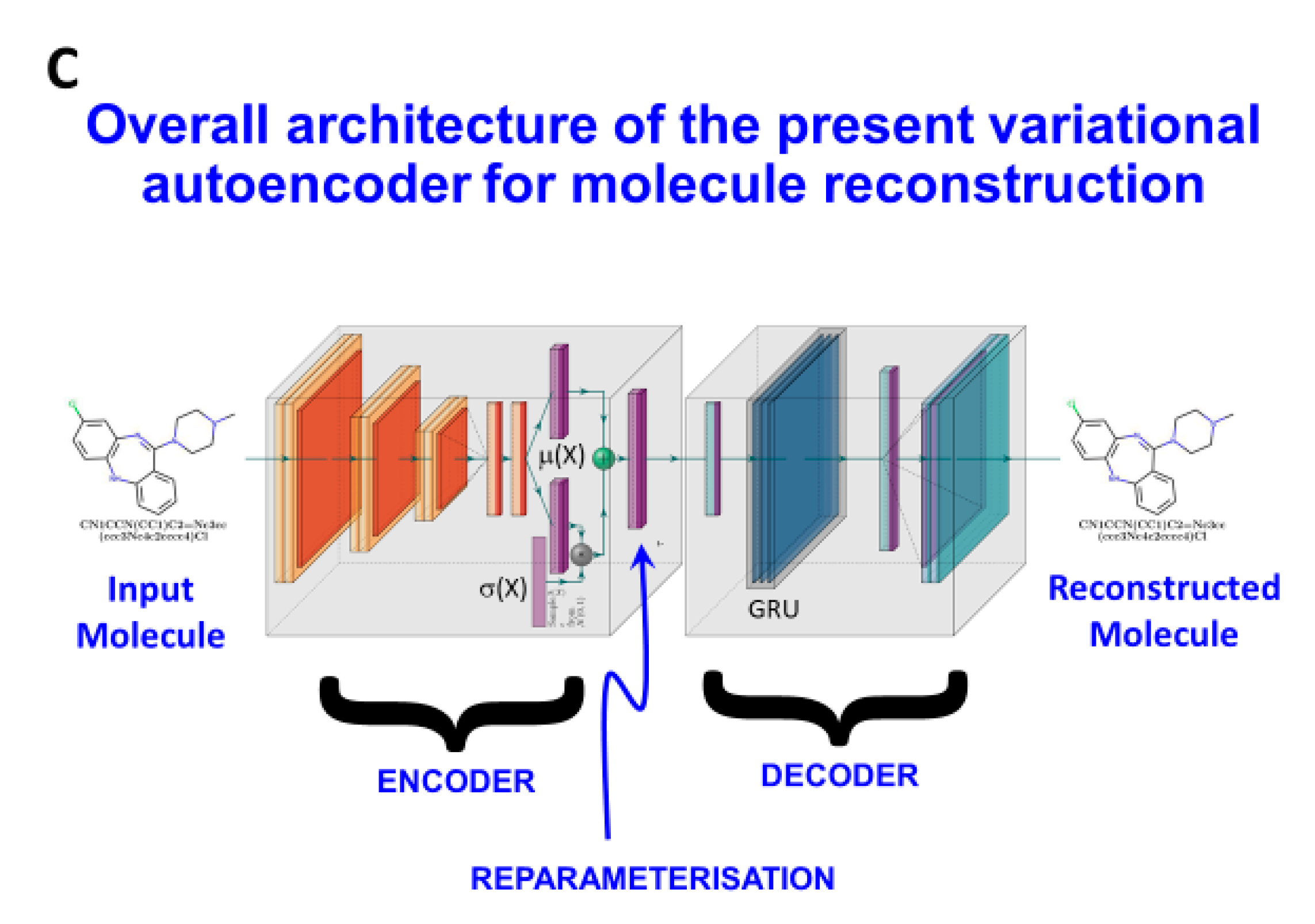
3. Methods
4. Discussion
What Determines the Extent to Which VAEs can Generate Novel Examples?
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Gasteiger, J. Handbook of Chemoinformatics: From Data to Knowledge; Wiley/VCH: Weinheim, Germany, 2003. [Google Scholar]
- Leach, A.R.; Gillet, V.J. An Introduction to Chemoinformatics; Springer: Dordrecht, The Netherlands, 2007. [Google Scholar]
- Maggiora, G.; Vogt, M.; Stumpfe, D.; Bajorath, J. Molecular similarity in medicinal chemistry. J. Med. Chem. 2014, 57, 3186–3204. [Google Scholar] [PubMed]
- Willett, P. Similarity-based data mining in files of two-dimensional chemical structures using fingerprint measures of molecular resemblance. Wires Data Min. Knowl. 2011, 1, 241–251. [Google Scholar] [CrossRef]
- Todeschini, R.; Consonni, V. Molecular Descriptors for Cheminformatics; Wiley-VCH: Weinheim, Germany, 2009. [Google Scholar]
- Ballabio, D.; Manganaro, A.; Consonni, V.; Mauri, A.; Todeschini, R. Introduction to mole db—On-line molecular descriptors database. Math Comput. Chem. 2009, 62, 199–207. [Google Scholar]
- Dehmer, M.; Varmuza, K.; Bonchev, D. Statistical Modelling of Molecular Descriptors in QSAR/QSPR; Wiley-VCH: Weinheim, Germany, 2012. [Google Scholar]
- Bender, A.; Glen, R.C. Molecular similarity: A key technique in molecular informatics. Org. Biomol. Chem. 2004, 2, 3204–3218. [Google Scholar] [CrossRef] [PubMed]
- Nisius, B.; Bajorath, J. Rendering conventional molecular fingerprints for virtual screening independent of molecular complexity and size effects. ChemMedChem 2010, 5, 859–868. [Google Scholar] [CrossRef]
- Owen, J.R.; Nabney, I.T.; Medina-Franco, J.L.; López-Vallejo, F. Visualization of molecular fingerprints. J. Chem. Inf. Model 2011, 51, 1552–1563. [Google Scholar] [CrossRef]
- Riniker, S.; Landrum, G.A. Similarity maps—A visualization strategy for molecular fingerprints and machine-learning methods. J. Cheminform. 2013, 5, 43. [Google Scholar] [CrossRef]
- Vogt, M.; Bajorath, J. Bayesian screening for active compounds in high-dimensional chemical spaces combining property descriptors and molecular fingerprints. Chem. Biol. Drug Des. 2008, 71, 8–14. [Google Scholar] [CrossRef]
- Awale, M.; Reymond, J.L. The polypharmacology browser: A web-based multi-fingerprint target prediction tool using chembl bioactivity data. J. Cheminform. 2017, 9, 11. [Google Scholar] [CrossRef]
- Geppert, H.; Bajorath, J. Advances in 2d fingerprint similarity searching. Expert Opin. Drug Discov. 2010, 5, 529–542. [Google Scholar] [CrossRef]
- Muegge, I.; Mukherjee, P. An overview of molecular fingerprint similarity search in virtual screening. Expert Opin. Drug. Discov. 2016, 11, 137–148. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Sayle, R.A. Comparing structural fingerprints using a literature-based similarity benchmark. J. Cheminform. 2016, 8, 36. [Google Scholar] [CrossRef]
- Willett, P. Similarity searching using 2d structural fingerprints. Meth. Mol. Biol. 2011, 672, 133–158. [Google Scholar]
- Durant, J.L.; Leland, B.A.; Henry, D.R.; Nourse, J.G. Reoptimization of mdl keys for use in drug discovery. J. Chem. Inf. Comput. Sci. 2002, 42, 1273–1280. [Google Scholar] [CrossRef]
- Carhart, R.E.; Smith, D.H.; Venkataraghavan, R. Atom pairs as molecular-features in structure activity studies—Definition and applications. J. Chem. Inf. Comp. Sci. 1985, 25, 64–73. [Google Scholar] [CrossRef]
- Nilakantan, R.; Bauman, N.; Dixon, J.S.; Venkataraghavan, R. Topological torsion—A new molecular descriptor for sar applications—Comparison with other descriptors. J. Chem. Inf. Comp. Sci. 1987, 27, 82–85. [Google Scholar] [CrossRef]
- Rogers, D.; Hahn, M. Extended-connectivity fingerprints. J. Chem. Inf. Model. 2010, 50, 742–754. [Google Scholar] [CrossRef]
- Hassan, M.; Brown, R.D.; Varma-O’brien, S.; Rogers, D. Cheminformatics analysis and learning in a data pipelining environment. Mol. Divers. 2006, 10, 283–299. [Google Scholar] [CrossRef]
- Glen, R.C.; Bender, A.; Arnby, C.H.; Carlsson, L.; Boyer, S.; Smith, J. Circular fingerprints: Flexible molecular descriptors with applications from physical chemistry to adme. IDrugs 2006, 9, 199–204. [Google Scholar]
- Riniker, S.; Landrum, G.A. Open-source platform to benchmark fingerprints for ligand-based virtual screening. J. Cheminform. 2013, 5, 26. [Google Scholar] [CrossRef]
- O’Hagan, S.; Kell, D.B. Consensus rank orderings of molecular fingerprints illustrate the ‘most genuine’ similarities between marketed drugs and small endogenous human metabolites, but highlight exogenous natural products as the most important ‘natural’ drug transporter substrates. ADMET & DMPK 2017, 5, 85–125. [Google Scholar]
- Dickens, D.; Rädisch, S.; Chiduza, G.N.; Giannoudis, A.; Cross, M.J.; Malik, H.; Schaeffeler, E.; Sison-Young, R.L.; Wilkinson, E.L.; Goldring, C.E.; et al. Cellular uptake of the atypical antipsychotic clozapine is a carrier-mediated process. Mol. Pharm. 2018, 15, 3557–3572. [Google Scholar] [CrossRef] [PubMed]
- Weininger, D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 1988, 28, 31–36. [Google Scholar] [CrossRef]
- Rumelhart, D.E.; McClelland, J.L. The PDP Research Group. Parallel Distributed Processing. Experiments in the Microstructure of Cognition; M.I.T. Press: Cambridge, MA, USA, 1986. [Google Scholar]
- Goodacre, R.; Kell, D.B.; Bianchi, G. Rapid assessment of the adulteration of virgin olive oils by other seed oils using pyrolysis mass spectrometry and artificial neural networks. J. Sci. Food Agric. 1993, 63, 297–307. [Google Scholar] [CrossRef]
- Goodacre, R.; Timmins, É.M.; Burton, R.; Kaderbhai, N.; Woodward, A.M.; Kell, D.B.; Rooney, P.J. Rapid identification of urinary tract infection bacteria using hyperspectral whole-organism fingerprinting and artificial neural networks. Microbiology UK 1998, 144, 1157–1170. [Google Scholar] [CrossRef]
- Tetko, I.V.; Gasteiger, J.; Todeschini, R.; Mauri, A.; Livingstone, D.; Ertl, P.; Palyulin, V.; Radchenko, E.; Zefirov, N.S.; Makarenko, A.S.; et al. Virtual computational chemistry laboratory—Design and description. J. Comput. Aided Mol. Des. 2005, 19, 453–463. [Google Scholar] [CrossRef]
- O’Boyle, N.; Dalke, A. Deepsmiles: An Adaptation of Smiles for use in Machine-learning of Chemical Structures. ChemRxiv. 2018, p. 7097960.v7097961. Available online: https://chemrxiv.org/articles/preprint/DeepSMILES_An_Adaptation_of_SMILES_for_Use_in_Machine-Learning_of_Chemical_Structures/7097960 (accessed on 29 July 2020).
- Segler, M.H.S.; Kogej, T.; Tyrchan, C.; Waller, M.P. Generating focussed molecule libraries for drug discovery with recurrent neural networks. ACS Central Sci. 2017, 4, 120–131. [Google Scholar] [CrossRef]
- Jin, W.; Barzilay, R.; Jaakkola, T. Junction Tree Variational Autoencoder for Molecular Graph Generation. arXiv 2018, arXiv:1802.04364v04362. [Google Scholar]
- Kajino, H. Molecular Hypergraph Grammar with its Application to Molecular Optimization. arXiv 2018, arXiv:02745v02741. [Google Scholar]
- Panteleev, J.; Gao, H.; Jia, L. Recent applications of machine learning in medicinal chemistry. Bioorg. Med. Chem. Lett. 2018, 28, 2807–2815. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, S.; Fulle, S.; Turk, S. Mol2vec: Unsupervised machine learning approach with chemical intuition. J. Chem. Inf. Model. 2018, 58, 27–35. [Google Scholar] [CrossRef]
- Shibayama, S.; Marcou, G.; Horvath, D.; Baskin, I.I.; Funatsu, K.; Varnek, A. Application of the mol2vec technology to large-size data visualization and analysis. Mol. Inform. 2020, 39, e1900170. [Google Scholar] [CrossRef]
- Duvenaud, D.; Maclaurin, D.; Aguilera-Iparraguirre, J.; Gómez-Bombarelli, R.; Hirzel, T.; Aspuru-Guzik, A.; Adams, R.P. Convolutional networks on graphs for learning molecular fingerprints. Adv. NIPS 2015, 2, 2224–2232. [Google Scholar]
- Kearnes, S.; McCloskey, K.; Berndl, M.; Pande, V.; Riley, P. Molecular graph convolutions: Moving beyond fingerprints. J. Comput. Aided Mol. Des. 2016, 30, 595–608. [Google Scholar] [CrossRef]
- Gupta, A.; Müller, A.T.; Huisman, B.J.H.; Fuchs, J.A.; Schneider, P.; Schneider, G. Generative recurrent networks for de novo drug design. Mol. Inform. 2018, 37, 1700111. [Google Scholar] [CrossRef]
- Schneider, G. Generative models for artificially-intelligent molecular design. Mol. Inf. 2018, 37, 188031. [Google Scholar] [CrossRef]
- Grisoni, F.; Schneider, G. De novo molecular design with generative long short-term memory. Chimia 2019, 73, 1006–1011. [Google Scholar] [CrossRef]
- Arús-Pous, J.; Blaschke, T.; Ulander, S.; Reymond, J.L.; Chen, H.; Engkvist, O. Exploring the gdb-13 chemical space using deep generative models. J. Cheminform. 2019, 11, 20. [Google Scholar] [CrossRef]
- Jørgensen, P.B.; Schmidt, M.N.; Winther, O. Deep generative models for molecular science. Mol. Inf. 2018, 37, 1700133. [Google Scholar] [CrossRef]
- Li, Y.; Hu, J.; Wang, Y.; Zhou, J.; Zhang, L.; Liu, Z. Deepscaffold: A comprehensive tool for scaffold-based de novo drug discovery using deep learning. J. Chem. Inf. Model 2020, 60, 77–91. [Google Scholar] [CrossRef]
- Lim, J.; Hwang, S.Y.; Moon, S.; Kim, S.; Kim, W.Y. Scaffold-based molecular design with a graph generative model. Chem. Sci. 2020, 11, 1153–1164. [Google Scholar] [CrossRef]
- Moret, M.; Friedrich, L.; Grisoni, F.; Merk, D.; Schneider, G. Generative molecular design in low data regimes. Nat. Mach. Intell. 2020, 2, 171–180. [Google Scholar] [CrossRef]
- van Deursen, R.; Ertl, P.; Tetko, I.V.; Godin, G. Gen: Highly efficient smiles explorer using autodidactic generative examination networks. J. Cheminform. 2020, 12, 22. [Google Scholar] [CrossRef]
- Walters, W.P.; Murcko, M. Assessing the impact of generative ai on medicinal chemistry. Nat Biotechnol 2020, 38, 143–145. [Google Scholar] [CrossRef]
- Yan, C.; Wang, S.; Yang, J.; Xu, T.; Huang, J. Re-balancing Variational Autoencoder Loss for Molecule Sequence Generation. arXiv 2019, arXiv:1910.00698v00691. [Google Scholar]
- Winter, R.; Montanari, F.; Noé, F.; Clevert, D.A. Learning continuous and data-driven molecular descriptors by translating equivalent chemical representations. Chem. Sci. 2019, 10, 1692–1701. [Google Scholar] [CrossRef]
- Samanta, B.; De, A.; Ganguly, N.; Gomez-Rodriguez, M. Designing Random Graph Models using Variational Autoencoders with Applications to Chemical Design. arXiv 2018, arXiv:1802.05283. [Google Scholar]
- Krenn, M.; Häse, F.; Nigam, A.; Friederich, P.; Aspuru-Guzik, A. Self-Referencing Embedded Strings (selfies): A 100% Robust Molecular String Representation. arXiv 2019, arXiv:1905.13741. [Google Scholar]
- Sattarov, B.; Baskin, I.I.; Horvath, D.; Marcou, G.; Bjerrum, E.J.; Varnek, A. De novo molecular design by combining deep autoencoder recurrent neural networks with generative topographic mapping. J. Chem. Inf. Model. 2019, 59, 1182–1196. [Google Scholar] [CrossRef]
- Bengio, Y.; Courville, A.; Vincent, P. Representation learning: A review and new perspectives. IEEE Trans. Patt. Anal. Mach. Intell. 2013, 35, 1798–1828. [Google Scholar] [CrossRef]
- Bousquet, O.; Gelly, S.; Tolstikhin, I.; Simon-Gabriel, C.-J.; Schoelkopf, B. From Optimal Transport to Generative Modeling: The Vegan Cookbook. arXiv 2017, arXiv:1705.07642. [Google Scholar]
- Husain, H.; Nock, R.; Williamson, R.C. Adversarial Networks and Autoencoders: The Primal-dual Relationship and Generalization Bounds. arXiv 2019, arXiv:1902.00985. [Google Scholar]
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozairy, S.; Courville, A.; Bengio, Y. Generative adversarial nets. arXiv 2014, arXiv:1406.2661v1401. [Google Scholar]
- Polykovskiy, D.; Zhebrak, A.; Vetrov, D.; Ivanenkov, Y.; Aladinskiy, V.; Mamoshina, P.; Bozdaganyan, M.; Aliper, A.; Zhavoronkov, A.; Kadurin, A. Entangled conditional adversarial autoencoder for de novo drug discovery. Mol. Pharm. 2018, 15, 4398–4405. [Google Scholar] [CrossRef]
- Arjovsky, M.; Chintala, S.; Bottou, L. Wasserstein gan. arXiv 2017, arXiv:1701.07875v07873. [Google Scholar]
- Goodfellow, I. Generative adversarial networks. arXiv 2017, arXiv:1701.00160v00161. [Google Scholar]
- Foster, D. Generative Deep Learning; O’Reilly: Sebastopol, CA, USA, 2019. [Google Scholar]
- Langr, J.; Bok, V. Gans in Action; Manning: Shelter Island, NY, USA, 2019. [Google Scholar]
- Prykhodko, O.; Johansson, S.V.; Kotsias, P.C.; Arús-Pous, J.; Bjerrum, E.J.; Engkvist, O.; Chen, H.M. A de novo molecular generation method using latent vector based generative adversarial network. J. Cheminform. 2019, 11, 74. [Google Scholar] [CrossRef]
- Zhao, J.J.; Kim, Y.; Zhang, K.; Rush, A.M.; LeCun, Y. Adversarially Regularized Autoencoders for Generating Discrete Structures. arXiv 2017, arXiv:1706.04223v04221. [Google Scholar]
- Kingma, D.; Welling, M. Auto-encoding variational bayes. arXiv 2014, arXiv:1312.6114v1310. [Google Scholar]
- Rezende, D.J.; Mohamed, S.; Wierstra, D. Stochastic Backpropagation and Approximate Inference in Deep Generative Models. arXiv 2014, arXiv:1401.4082v1403. [Google Scholar]
- Doersch, C. Tutorial on Variational Autoencoders. arXiv 2016, arXiv:1606.05908v05902. [Google Scholar]
- Benhenda, M. Chemgan Challenge for Drug Discovery: Can ai Reproduce Natural Chemical Diversity? arXiv 2017, arXiv:1708.08227v08223. [Google Scholar]
- Griffiths, R.-R.; Hernández-Lobato, J.M. Constrained Bayesian Optimization for Automatic Chemical Design. arXiv 2017, arXiv:1709.05501v05505. [Google Scholar]
- Aumentado-Armstrong, T. Latent Molecular Optimization for Targeted Therapeutic Design. arXiv 2018, arXiv:1809.02032. [Google Scholar]
- Blaschke, T.; Olivecrona, M.; Engkvist, O.; Bajorath, J.; Chen, H.M. Application of generative autoencoder in de novo molecular design. Mol. Inform. 2018, 37, 1700123. [Google Scholar] [CrossRef]
- Gómez-Bombarelli, R.; Wei, J.N.; Duvenaud, D.; Hernández-Lobato, J.M.; Sánchez-Lengeling, B.; Sheberla, D.; Aguilera-Iparraguirre, J.; Hirzel, T.D.; Adams, R.P.; Aspuru-Guzik, A. Automatic chemical design using a data-driven continuous representation of molecules. ACS Cent. Sci. 2018, 4, 268–276. [Google Scholar] [CrossRef]
- Tschannen, M.; Bachem, O.; Lucic, M. Recent Advances in Autoencoder-based Representation Learning. arXiv 2018, arXiv:1812.05069v05061. [Google Scholar]
- Kingma, D.P.; Welling, M. An Introduction to Variational Autoencoders. arXiv 2019, arXiv:1906.02691v02691. [Google Scholar]
- Rezende, D.J.; Viola, F. Taming vaes. arXiv 2018, arXiv:1810.00597v00591. [Google Scholar]
- Hutson, M. Core progress in ai has stalled in some fields. Science 2020, 368, 927. [Google Scholar] [CrossRef]
- Burgess, C.P.; Higgins, I.; Pal, A.; Matthey, L.; Watters, N.; Desjardins, G.; Lerchner, A. Understanding disentangling in β-vae. arXiv 2018, arXiv:1804.03599. [Google Scholar]
- Taghanaki, S.A.; Havaei, M.; Lamb, A.; Sanghi, A.; Danielyan, A.; Custis, T. Jigsaw-vae: Towards Balancing Features in Variational Autoencoders. arXiv 2020, arXiv:2005.05496. [Google Scholar]
- Caterini, A.; Cornish, R.; Sejdinovic, D.; Doucet, A. Variational Inference with Continuously-Indexed Normalizing Flows. arXiv 2020, arXiv:2007.05426. [Google Scholar]
- Nielsen, D.; Jaini, P.; Hoogeboom, E.; Winther, O.; Welling, M. Survae flows: Surjections to bridge the Gap between Vaes and Flows. arXiv 2020, arXiv:2007.02731. [Google Scholar]
- Li, Y.; Yu, S.; Principe, J.C.; Li, X.; Wu, D. Pri-vae: Principle-of-relevant-information Variational Autoencoders. arXiv 2020, arXiv:2007.06503. [Google Scholar]
- Wolpert, D.H.; Macready, W.G. No free lunch theorems for optimization. IEEE Trans. Evol. Comput. 1997, 1, 67–82. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention is All You Need. arXiv 2017, arXiv:1706.03762. [Google Scholar]
- Devlin, J.; Chang, M.-W.; Lee, K.; Toutanova, K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar]
- Dai, B.; Wipf, D. Diagnosing and Enhancing vae Models. arXiv 2019, arXiv:1903.05789v05782. [Google Scholar]
- Asperti, A.; Trentin, M. Balancing Reconstruction Error and Kullback-leibler Divergence in Variational Autoencoders. arXiv 2020, arXiv:2002.07514v07511. [Google Scholar]
- Goodacre, R.; Pygall, J.; Kell, D.B. Plant seed classification using pyrolysis mass spectrometry with unsupervised learning: The application of auto-associative and kohonen artificial neural networks. Chemometr. Intell. Lab. Syst. 1996, 34, 69–83. [Google Scholar] [CrossRef]
- Yao, X. Evolving artificial neural networks. Proc. IEEE 1999, 87, 1423–1447. [Google Scholar]
- Floreano, D.; Dürr, P.; Mattiussi, C. Neuroevolution: From architectures to learning. Evol. Intell. 2008, 1, 47–62. [Google Scholar] [CrossRef]
- Vassiliades, V.; Christodoulou, C. Toward nonlinear local reinforcement learning rules through neuroevolution. Neural Comput. 2013, 25, 3020–3043. [Google Scholar] [CrossRef]
- Stanley, K.O.; Clune, J.; Lehman, J.; Miikkulainen, R. Designing neural networks through neuroevolution. Nat. Mach. Intell. 2019, 1, 24–35. [Google Scholar] [CrossRef]
- Iba, H.; Noman, N. Deep Neural Evolution: Deep Learning with Evolutionary Computation; Springer: Berlin, Germany, 2020. [Google Scholar]
- Le Cun, Y.; Denker, J.S.; Solla, S.A. Optimal brain damage. Adv. Neural Inf. Proc. Syst. 1990, 2, 598–605. [Google Scholar]
- Dietterich, T.G. Ensemble methods in machine learning. LNCS 2000, 1857, 1–15. [Google Scholar]
- Hinton, G.E.; Srivastava, N.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R.R. Improving neural networks by preventing co-adaptation of feature detectors. arXiv 2012, arXiv:1207.0580. [Google Scholar]
- Keskar, N.S.; Mudigere, D.; Nocedal, J.; Smelyanskiy, M.; Tang, P.T.P. On large-batch training for deep learning: Generalization gap and sharp minima. arXiv 2017, arXiv:1609.04836v04832. [Google Scholar]
- O’Hagan, S.; Swainston, N.; Handl, J.; Kell, D.B. A ‘rule of 0.5′ for the metabolite-likeness of approved pharmaceutical drugs. Metabolomics 2015, 11, 323–339. [Google Scholar] [CrossRef]
- O’Hagan, S.; Kell, D.B. Understanding the foundations of the structural similarities between marketed drugs and endogenous human metabolites. Front. Pharmacol. 2015, 6, 105. [Google Scholar] [CrossRef]
- O’Hagan, S.; Kell, D.B. Metmaxstruct: A tversky-similarity-based strategy for analysing the (sub)structural similarities of drugs and endogenous metabolites. Front. Pharmacol. 2016, 7, 266. [Google Scholar] [CrossRef] [PubMed]
- O’Hagan, S.; Kell, D.B. Analysis of drug-endogenous human metabolite similarities in terms of their maximum common substructures. J. Cheminform. 2017, 9, 18. [Google Scholar] [CrossRef] [PubMed]
- O’Hagan, S.; Kell, D.B. Analysing and navigating natural products space for generating small, diverse, but representative chemical libraries. Biotechnol. J. 2018, 13, 1700503. [Google Scholar] [CrossRef] [PubMed]
- O’Hagan, S.; Kell, D.B. Structural Similarities between Some Common Fluorophores used in Biology and Marketed drugs, Endogenous Metabolites, and Natural Products. bioRxiv 2019, 834325. Available online: https://www.biorxiv.org/content/10.1101/834325v1.abstract (accessed on 29 July 2020).
- Samanta, S.; O’Hagan, S.; Swainston, N.; Roberts, T.J.; Kell, D.B. Vae-sim: A novel Molecular Similarity Measure Based on a Variational Autoencoder. bioRxiv 2020, 172908. Available online: https://www.biorxiv.org/content/10.1101/2020.06.26.172908v1.abstract (accessed on 29 July 2020).
- Dai, H.; Tian, Y.; Dai, B.; Skiena, S.; Song, L. Syntax-Directed Variational Autoencoder for Structured data. arXiv 2018, arXiv:1802.08786v08721. [Google Scholar]
- Kusner, M.J.; Paige, B.; Hernández-Lobato, J.M. Grammar Variational Autoencoder. arXiv 2017, arXiv:1703.01925v01921. [Google Scholar]
- Kingma, D.P.; Ba, J.L. Adam: A Method for Stochastic Optimization. arXiv 2015, arXiv:1412.6980v1418. [Google Scholar]
- Glorot, X.; Bengio, Y. Understanding the difficulty of training deep feedforward neural networks. Proc. AISTATs 2010, 9, 249–256. [Google Scholar]
- O’Hagan, S.; Kell, D.B. The knime workflow environment and its applications in genetic programming and machine learning. Genetic Progr. Evol. Mach. 2015, 16, 387–391. [Google Scholar] [CrossRef][Green Version]
- McInnes, L.; Healy, J.; Melville, J. Umap: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv 2018, arXiv:1802.03426v03422. [Google Scholar]
- McInnes, L.; Healy, J.; Saul, N.; Großberger, L. Umap: Uniform manifold approximation and projection. J. Open Source Software 2018. [Google Scholar] [CrossRef]
- Citraro, R.; Leo, A.; Aiello, R.; Pugliese, M.; Russo, E.; De Sarro, G. Comparative analysis of the treatment of chronic antipsychotic drugs on epileptic susceptibility in genetically epilepsy-prone rats. Neurotherapeutics 2015, 12, 250–262. [Google Scholar] [CrossRef]
- Thorn, C.F.; Muller, D.J.; Altman, R.B.; Klein, T.E. Pharmgkb summary: Clozapine pathway, pharmacokinetics. Pharmacogenet. Genomics 2018, 28, 214–222. [Google Scholar] [CrossRef]
- Hopkins, A.L.; Mason, J.S.; Overington, J.P. Can we rationally design promiscuous drugs? Curr. Opin. Struct. Biol. 2006, 16, 127–136. [Google Scholar] [CrossRef]
- Mestres, J.; Gregori-Puigjané, E.; Valverde, S.; Solé, R.V. The topology of drug-target interaction networks: Implicit dependence on drug properties and target families. Mol. Biosyst. 2009, 5, 1051–1057. [Google Scholar] [CrossRef]
- Mestres, J.; Gregori-Puigjané, E. Conciliating binding efficiency and polypharmacology. Trends Pharmacol. Sci. 2009, 30, 470–474. [Google Scholar] [CrossRef] [PubMed]
- Oprea, T.I.; Bauman, J.E.; Bologa, C.G.; Buranda, T.; Chigaev, A.; Edwards, B.S.; Jarvik, J.W.; Gresham, H.D.; Haynes, M.K.; Hjelle, B.; et al. Drug repurposing from an academic perspective. Drug Discov. Today Ther. Strateg. 2011, 8, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Dimova, D.; Hu, Y.; Bajorath, J. Matched molecular pair analysis of small molecule microarray data identifies promiscuity cliffs and reveals molecular origins of extreme compound promiscuity. J. Med. Chem. 2012, 55, 10220–10228. [Google Scholar]
- Peters, J.U.; Hert, J.; Bissantz, C.; Hillebrecht, A.; Gerebtzoff, G.; Bendels, S.; Tillier, F.; Migeon, J.; Fischer, H.; Guba, W.; et al. Can we discover pharmacological promiscuity early in the drug discovery process? Drug Discov. Today 2012, 17, 325–335. [Google Scholar] [CrossRef]
- Hu, Y.; Gupta-Ostermann, D.; Bajorath, J. Exploring compound promiscuity patterns and multi-target activity spaces. Comput. Struct. Biotechnol. J. 2014, 9, e201401003. [Google Scholar] [CrossRef] [PubMed]
- Bajorath, J. Molecular similarity concepts for informatics applications. Methods Mol. Biol. 2017, 1526, 231–245. [Google Scholar] [PubMed]
- Eckert, H.; Bajorath, J. Molecular similarity analysis in virtual screening: Foundations, limitations and novel approaches. Drug Discov. Today 2007, 12, 225–233. [Google Scholar] [CrossRef]
- Medina-Franco, J.L.; Maggiora, G.M. Molecular similarity analysis. In Chemoinformatics for Drug Discovery; Bajorath, J., Ed.; Wiley: Hoboken, NJ, USA, 2014; pp. 343–399. [Google Scholar]
- Zhang, B.; Vogt, M.; Maggiora, G.M.; Bajorath, J. Comparison of bioactive chemical space networks generated using substructure- and fingerprint-based measures of molecular similarity. J. Comput. Aided Mol. Des. 2015, 29, 595–608. [Google Scholar] [CrossRef]
- Hornik, K.; Stinchcombe, M.; White, H. Multilayer feedforward networks are universal approximators. Neural Netw. 1989, 2, 359–366. [Google Scholar] [CrossRef]
- Hornik, K. Approximation capabilities of multilayer feedforward networks. Neural Netw. 1991, 4, 251–257. [Google Scholar] [CrossRef]
- Everitt, B.S. Cluster Analysis; Edward Arnold: London, UK, 1993. [Google Scholar]
- Jain, A.K.; Dubes, R.C. Algorithms for Clustering Data; Prentice Hall: Englewood Cliffs, NJ, USA, 1988. [Google Scholar]
- Kaufman, L.; Rousseeuw, P.J. Finding Groups in Data. An Introduction to Cluster Analysis; Wiley: New York, NY, USA, 1990. [Google Scholar]
- Handl, J.; Knowles, J.; Kell, D.B. Computational cluster validation in post-genomic data analysis. Bioinformatics 2005, 21, 3201–3212. [Google Scholar] [CrossRef] [PubMed]
- MacCuish, J.D.; MacCuish, N.E. Clustering in Bioinformatics And Drug Discovery; CRC Press: Boca Raton, FL, USA, 2011. [Google Scholar]
- Hong, S.H.; Ryu, S.; Lim, J.; Kim, W.Y. Molecular generative model based on an adversarially regularized autoencoder. J. Chem. Inf. Model. 2020, 60, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Bozkurt, A.; Esmaeili, B.; Brooks, D.H.; Dy, J.G.; van de Meent, J.-W. Evaluating Combinatorial Generalization in Variational Autoencoders. arXiv 2019, arXiv:1911.04594v04591. [Google Scholar]
- Bozkurt, A.; Esmaeili, B.; Brooks, D.H.; Dy, J.G.; van de Meent, J.-W. Can Vaes Generate novel Examples? arXiv 2018, arXiv:1812.09624v09621. [Google Scholar]
Sample Availability: Samples of the compounds are not available from the authors. |
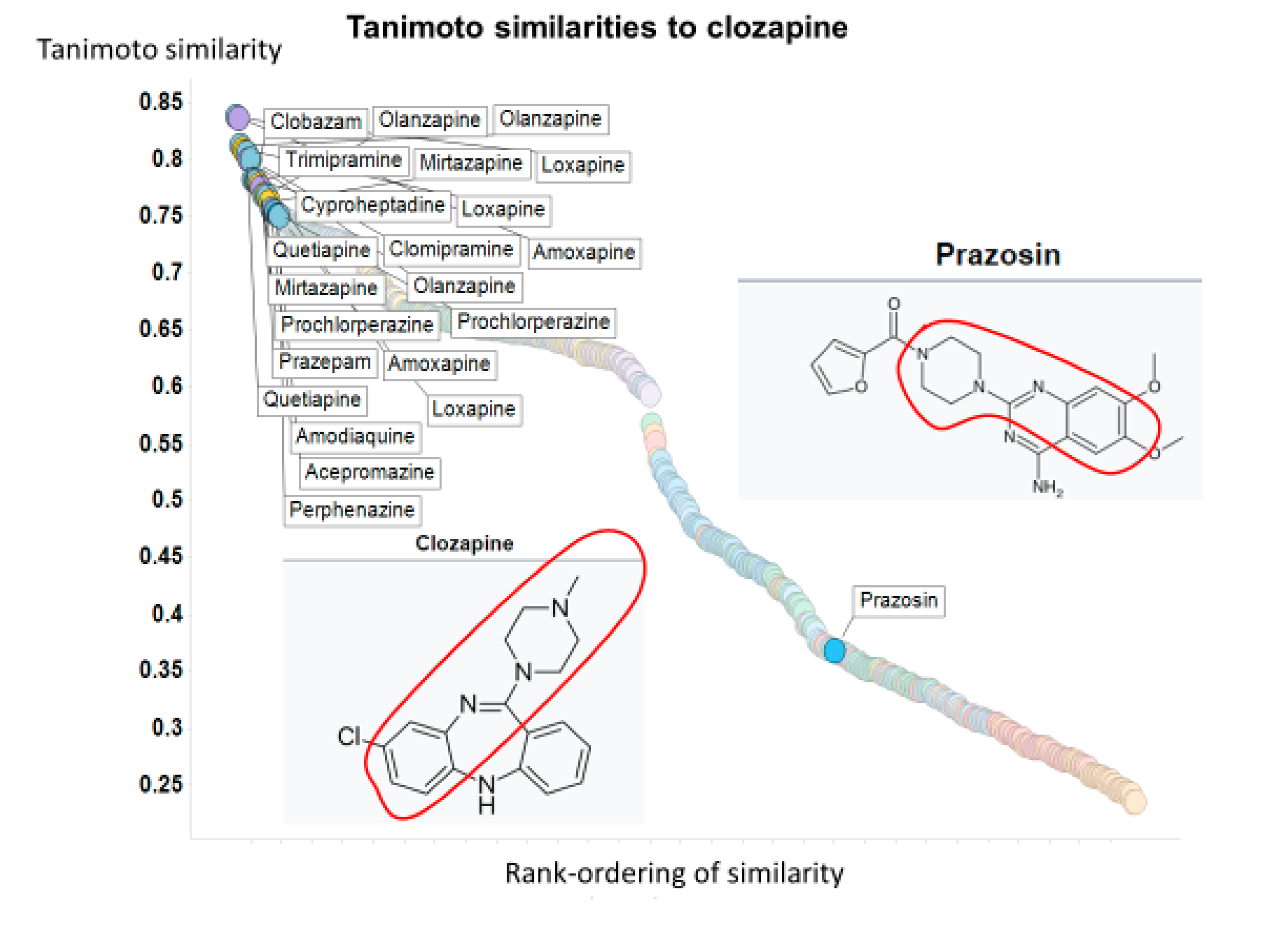
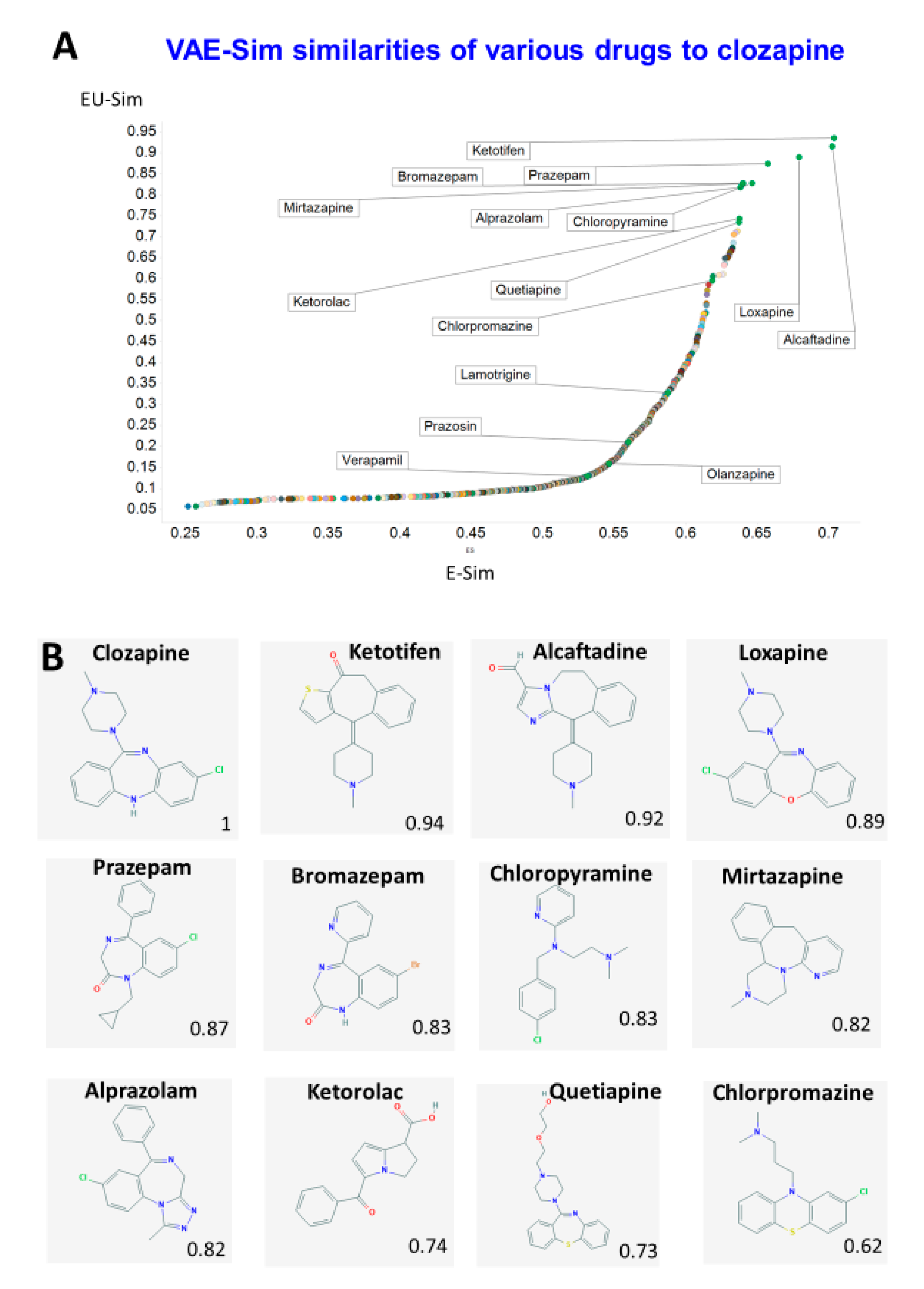
| Drug | % Inhiclozapine Uptake | TS Atom Pair | TS Avalon | TS Feat Morgan | TS Layered | TS MACCS | TS Morgan | TS Pattern | TS RDKit | TS Torsion |
|---|---|---|---|---|---|---|---|---|---|---|
| Olanzapine | 41 | 0.68 | 0.47 | 0.55 | 0.77 | 0.8 | 0.53 | 0.81 | 0.74 | 0.66 |
| Chlorpromazine | 75 | 0.53 | - | 0.35 | - | 0.66 | 0.3 | 0.74 | - | 0.33 |
| Quetiapine | 65 | 0.51 | 0.57 | 0.42 | 0.78 | - | 0.35 | 0.8 | - | 0.48 |
| Prazosin | 94 | - | - | - | - | - | - | - | - | 0.37 |
| Lamotrigine | 26 | - | - | - | - | - | - | - | - | - |
| Indatraline | 35 | - | - | - | - | - | - | - | - | - |
| Veraapamil | 83 | - | - | - | - | - | - | - | - | - |
| Rhein | 39 | - | - | - | - | - | - | - | - | - |
| Data Partition | Total Samples | Valid Reconstructed Samples | Accuracy |
|---|---|---|---|
| Train | 3,101,207 | 2,964,749 | 95.60 |
| Validation | 1,240,483 | 1,170,827 | 94.38 |
| Test | 1,860,725 | 1,757,079 | 94.42 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Samanta, S.; O’Hagan, S.; Swainston, N.; Roberts, T.J.; Kell, D.B. VAE-Sim: A Novel Molecular Similarity Measure Based on a Variational Autoencoder. Molecules 2020, 25, 3446. https://doi.org/10.3390/molecules25153446
Samanta S, O’Hagan S, Swainston N, Roberts TJ, Kell DB. VAE-Sim: A Novel Molecular Similarity Measure Based on a Variational Autoencoder. Molecules. 2020; 25(15):3446. https://doi.org/10.3390/molecules25153446
Chicago/Turabian StyleSamanta, Soumitra, Steve O’Hagan, Neil Swainston, Timothy J. Roberts, and Douglas B. Kell. 2020. "VAE-Sim: A Novel Molecular Similarity Measure Based on a Variational Autoencoder" Molecules 25, no. 15: 3446. https://doi.org/10.3390/molecules25153446
APA StyleSamanta, S., O’Hagan, S., Swainston, N., Roberts, T. J., & Kell, D. B. (2020). VAE-Sim: A Novel Molecular Similarity Measure Based on a Variational Autoencoder. Molecules, 25(15), 3446. https://doi.org/10.3390/molecules25153446







