Chemistry of Fluorinated Pyrimidines in the Era of Personalized Medicine
Abstract
1. Introduction
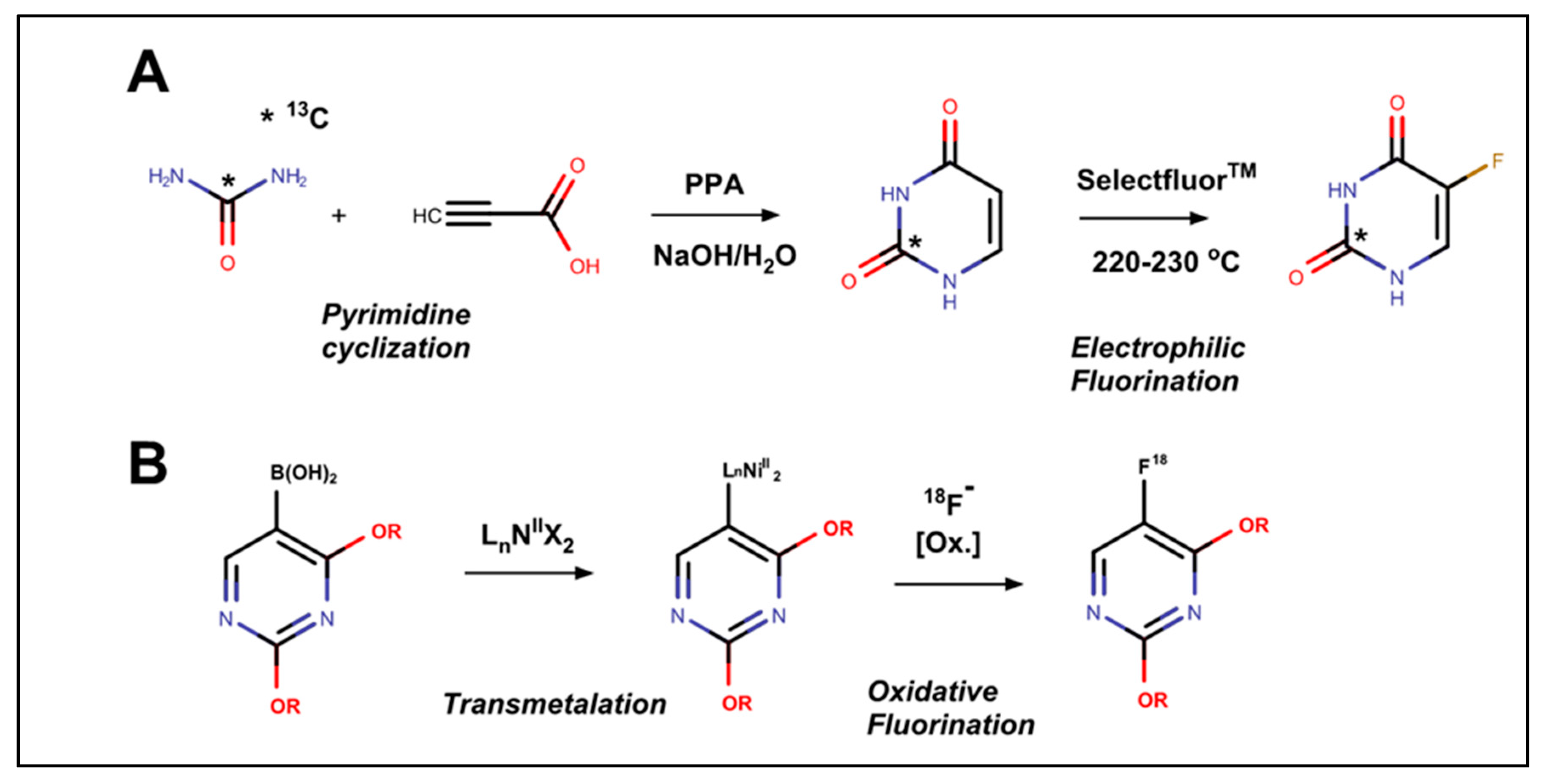
2. Synthesis and Isotopic Labeling of 5-FU
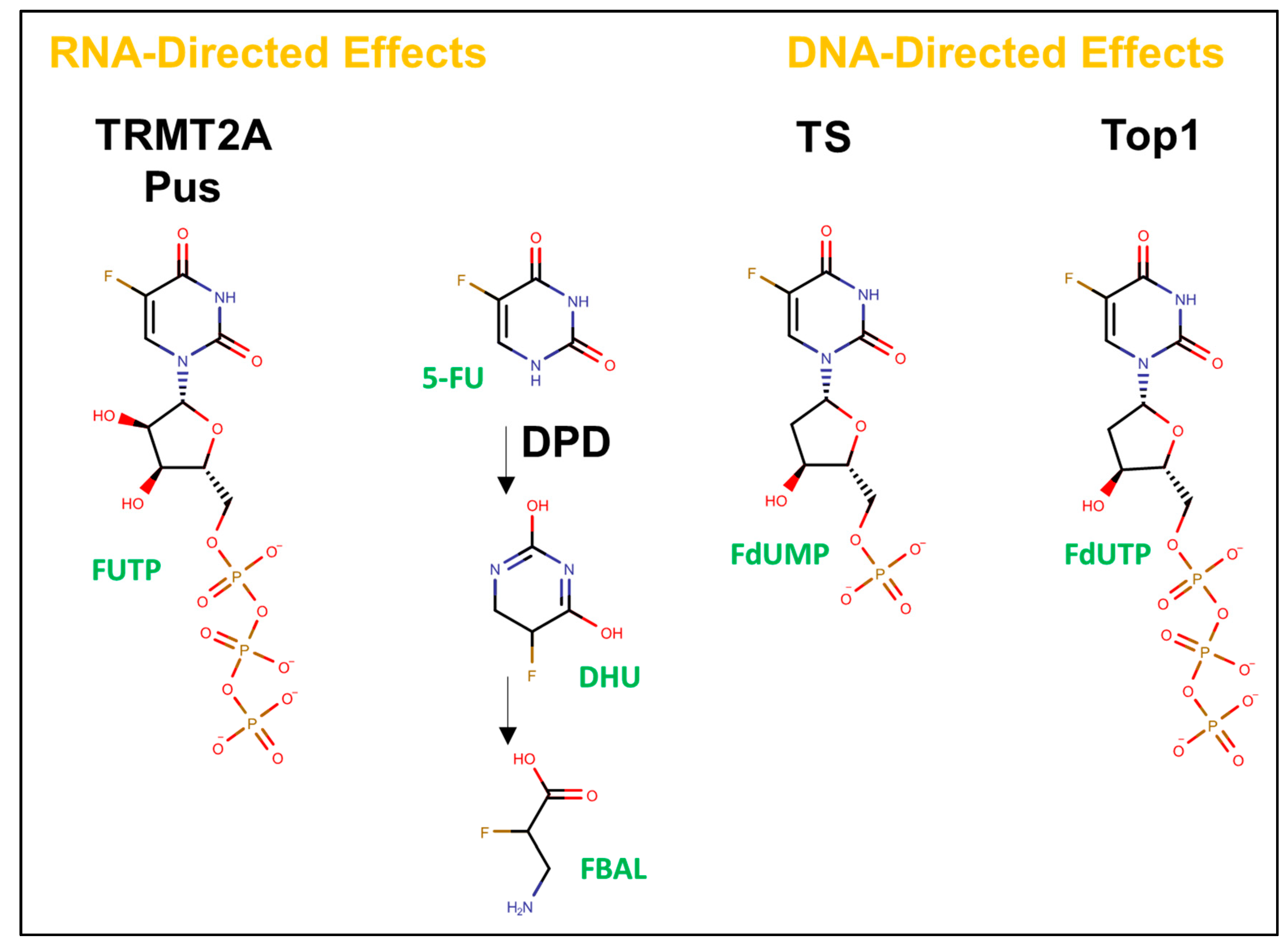
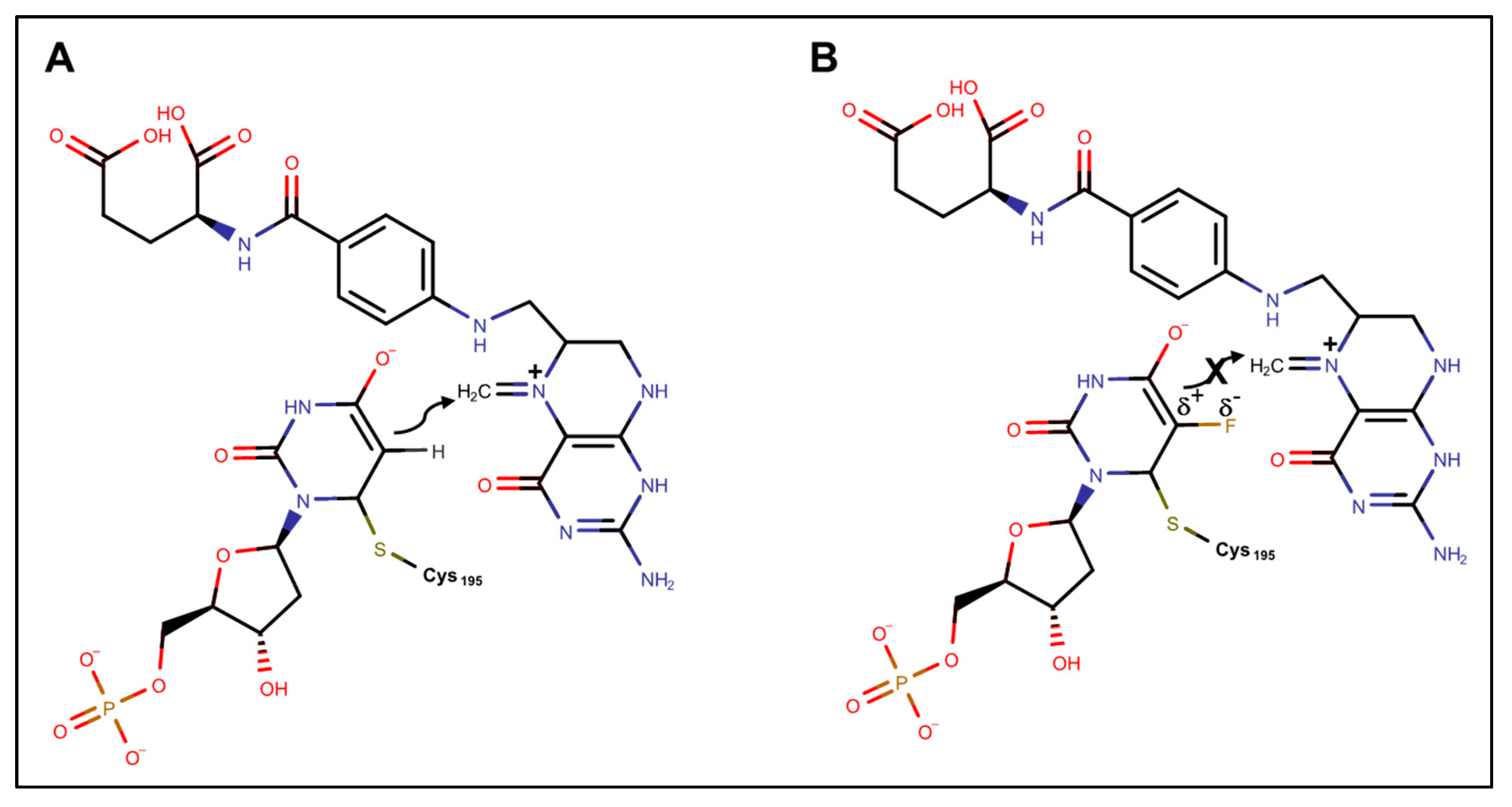
3. TS Inhibition and DNA-Directed Effects of FPs
4. Inhibition of RNA Modification Enzymes and RNA-Mediated Effects of FPs
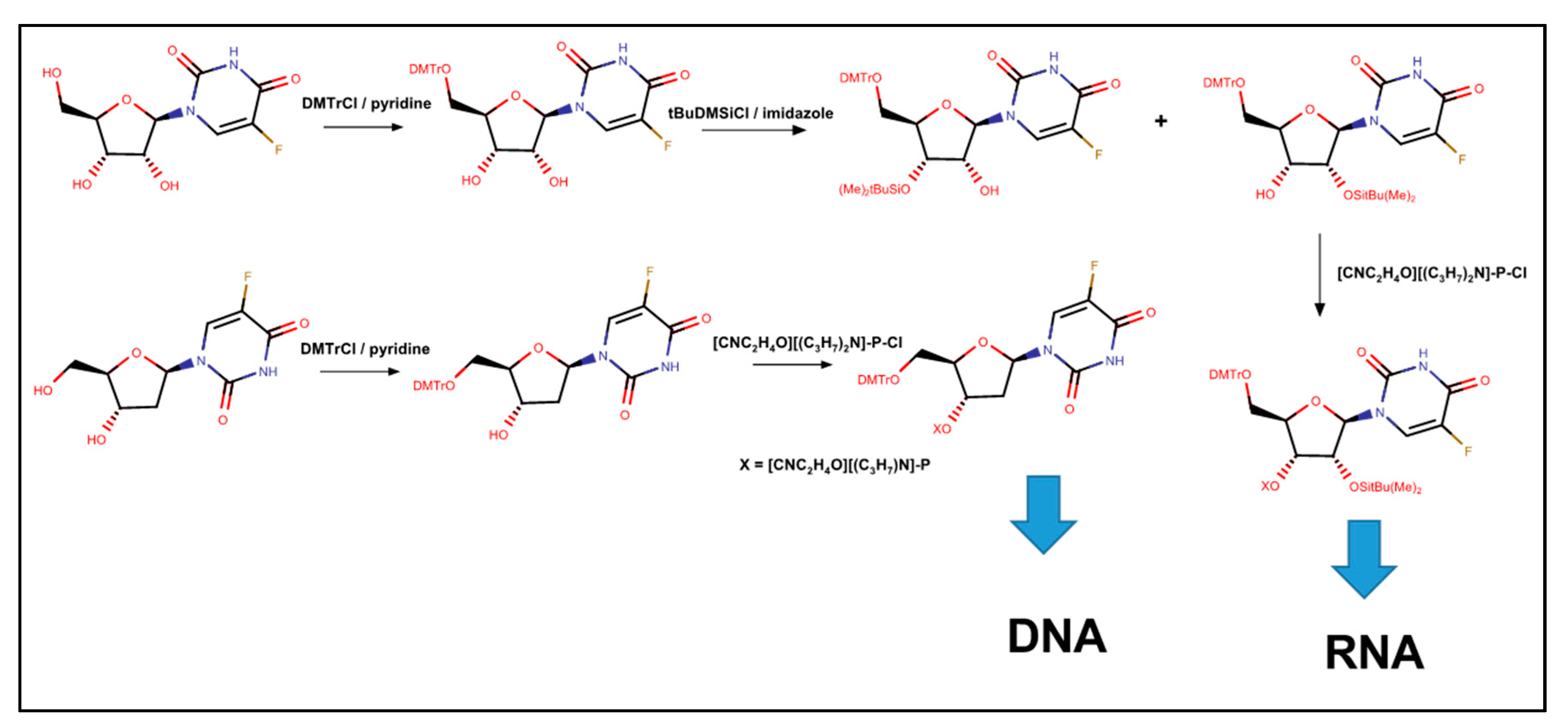
5. Synthesis of 5-FU Substituted RNA and DNA for Biophysical Studies
6. Conclusions
Funding
Conflicts of Interest
References
- Peters, R.A. Mechanism of the toxicity of the active constituent of Dichapetalum cymosum and related compounds. Adv. Enzymol. Relat. Subj. Biochem. 1957, 18, 113–159. [Google Scholar] [CrossRef] [PubMed]
- Heidelberger, C.; Chaudhuri, N.K.; Danneberg, P.; Mooren, D.; Griesbach, L.; Duschinsky, R.; Schnitzer, R.J.; Pleven, E.; Scheiner, J. Fluorinated pyrimidines, a new class of tumour-inhibitory compounds. Nature 1957, 179, 663–666. [Google Scholar] [CrossRef] [PubMed]
- Heidelberger, C.; Griesbach, L.; Cruz, O.; Schnitzer, R.J.; Grunberg, E. Fluorinated pyrimidines. VI. Effects of 5-fluorouridine and 5-fluoro-2′-deoxyuridine on transplanted tumors. Proc. Soc. Exp. Biol. Med. 1958, 97, 470–475. [Google Scholar] [CrossRef] [PubMed]
- Parker, W.B.; Cheng, Y.C. Metabolism and mechanism of action of 5-fluorouracil. Pharmacol. Ther. 1990, 48, 381–395. [Google Scholar] [CrossRef]
- Weckbecker, G. Biochemical pharmacology and analysis of fluoropyrimidines alone and in combination with modulators. Pharmacol. Ther. 1991, 50, 367–424. [Google Scholar] [CrossRef]
- Spears, C.P.; Gustavsson, B.G.; Mitchell, M.S.; Spicer, D.; Berne, M.; Bernstein, L.; Danenberg, P.V. Thymidylate synthetase inhibition in malignant tumors and normal liver of patients given intravenous 5-fluorouracil. Cancer Res. 1984, 44, 4144–4150. [Google Scholar]
- Houghton, J.A.; Harwood, F.G.; Tillman, D.M. Thymineless death in colon carcinoma cells is mediated via fas signaling. Proc. Natl. Acad. Sci. USA 1997, 94, 8144–8149. [Google Scholar] [CrossRef]
- Baasner, B.; Klauke, E. A New Route to the Synthesis of 5-Fluorouracil. J. Fluor. Chem. 1989, 45, 417–430. [Google Scholar] [CrossRef]
- An, Q.; Robins, P.; Lindahl, T.; Barnes, D.E. 5-Fluorouracil incorporated into DNA is excised by the Smug1 DNA glycosylase to reduce drug cytotoxicity. Cancer Res. 2007, 67, 940–945. [Google Scholar] [CrossRef]
- Wilson, P.M.; Danenberg, P.V.; Johnston, P.G.; Lenz, H.J.; Ladner, R.D. Standing the test of time: Targeting thymidylate biosynthesis in cancer therapy. Nat. Rev. Clin. Oncol. 2014, 11, 282–298. [Google Scholar] [CrossRef]
- Gmeiner, W.H. Novel chemical strategies for thymidylate synthase inhibition. Curr. Med. Chem. 2005, 12, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Liao, Z.Y.; Sordet, O.; Zhang, H.L.; Kohlhagen, G.; Antony, S.; Gmeiner, W.H.; Pommier, Y. A novel polypyrimidine antitumor agent FdUMP [10] induces thymineless death with topoisomerase I-DNA complexes. Cancer Res. 2005, 65, 4844–4851. [Google Scholar] [CrossRef] [PubMed]
- Gmeiner, W.H. Entrapment of DNA topoisomerase-DNA complexes by nucleotide/nucleoside analogs. Cancer Drug Resist. 2019, 2, 994–1001. [Google Scholar] [CrossRef] [PubMed]
- Spedaliere, C.J.; Mueller, E.G. Not all pseudouridine synthases are potently inhibited by RNA containing 5-fluorouridine. RNA 2004, 10, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Gmeiner, W.H.; Miller, L.D.; Chou, J.W.; Dominijanni, A.; Mutkus, L.; Marini, F.; Ruiz, J.; Dotson, T.; Thomas, K.W.; Parks, G.; et al. Dysregulated Pyrimidine Biosynthesis Contributes to 5-FU Resistance in SCLC Patient-Derived Organoids but Response to a Novel Polymeric Fluoropyrimidine, CF10. Cancers 2020, 12, 788. [Google Scholar] [CrossRef]
- Ginsburg, G.S.; Phillips, K.A. Precision Medicine: From Science to Value. Health Aff. (Millwood) 2018, 37, 694–701. [Google Scholar] [CrossRef]
- Wigle, T.J.; Tsvetkova, E.V.; Welch, S.A.; Kim, R.B. DPYD and Fluorouracil-Based Chemotherapy: Mini Review and Case Report. Pharmaceutics 2019, 11, 199. [Google Scholar] [CrossRef]
- Sadee, W.; Dai, Z. Pharmacogenetics/genomics and personalized medicine. Hum. Mol. Genet. 2005, 14, R207–R214. [Google Scholar] [CrossRef]
- Lunenburg, C.; Henricks, L.M.; Guchelaar, H.J.; Swen, J.J.; Deenen, M.J.; Schellens, J.H.M.; Gelderblom, H. Prospective DPYD genotyping to reduce the risk of fluoropyrimidine-induced severe toxicity: Ready for prime time. Eur. J. Cancer 2016, 54, 40–48. [Google Scholar] [CrossRef]
- Diasio, R.B. The role of dihydropyrimidine dehydrogenase (DPD) modulation in 5-FU pharmacology. Oncology-Ny 1998, 12, 23–27. [Google Scholar]
- Gmeiner, W.H.; Trump, E.; Wei, C. Enhanced DNA-directed effects of FdUMP [10] compared to 5FU. Nucleosides Nucleotides Nucleic Acids 2004, 23, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Duschinsky, R.; Pleven, E.; Heidelberger, C. The Synthesis of 5-Fluoropyrimidines. J. Am. Chem. Soc. 1957, 79, 4559–4560. [Google Scholar] [CrossRef]
- Barton, D.H.; Hesse, R.H.; To, H.T.; Pechet, M.M. A convenient synthesis of 5-fluorouracil. J. Org. Chem. 1972, 37, 329–330. [Google Scholar] [CrossRef]
- Banks, R.E. Selectfluor (TM) reagent F-TEDA-BF4 in action: Tamed fluorine at your service. J. Fluor. Chem. 1998, 87, 1–17. [Google Scholar] [CrossRef]
- Lal, G.S.; Pastore, W.; Pesaresi, R. A Convenient Synthesis of 5-Fluoropyrimidines Using 1-(Chloromethyl)-4-fluoro-1,4-diazabicyclo[2.2.2]Octane Bis(Tetrafluoroborate)-Selectfluor Reagent. J. Org. Chem. 1995, 60, 7340–7342. [Google Scholar] [CrossRef]
- Hoover, A.J.; Lazari, M.; Ren, H.; Narayanam, M.K.; Murphy, J.M.; van Dam, R.M.; Hooker, J.M.; Ritter, T. A Transmetalation Reaction Enables the Synthesis of [(18)F]5-Fluorouracil from [(18)F]Fluoride for Human PET Imaging. Organometallics 2016, 35, 1008–1014. [Google Scholar] [CrossRef]
- Shani, J.; Young, D.; Schlesinger, T.; Siemsen, J.K.; Chlebowski, R.T.; Bateman, J.R.; Wolf, W. Dosimetry and Preliminary Human Studies of F-18-5-Fluorouracil. Int. J. Nucl. Med. Biol. 1982, 9, 25–35. [Google Scholar] [CrossRef]
- Kesner, A.L.; Hsueh, W.A.; Czernin, J.; Padgett, H.; Phelps, M.E.; Silverman, D.H.S. Radiation dose estimates for [F-18]5-fluorouracil derived from PET-based and tissue-based methods in rats. Mol. Imaging Biol. 2008, 10, 341–348. [Google Scholar] [CrossRef]
- Rangwala, H.S.; Giraldes, J.W.; Gurvich, V.J. Synthesis and purification of [2-C-13]-5-fluorouracil. J. Labelled Compd. Rad. 2011, 54, 340–343. [Google Scholar] [CrossRef]
- Mukherji, D.; Massih, S.A.; Tfayli, A.; Kanso, M.; Faraj, W. Three different polymorphisms of the DPYD gene associated with severe toxicity following administration of 5-FU: A case report. J. Med. Case Rep. 2019, 13, 76. [Google Scholar] [CrossRef]
- Afzal, S.; Gusella, M.; Vainer, B.; Vogel, U.B.; Andersen, J.T.; Broedbaek, K.; Petersen, M.; Jimenez-Solem, E.; Bertolaso, L.; Barile, C.; et al. Combinations of polymorphisms in genes involved in the 5-Fluorouracil metabolism pathway are associated with gastrointestinal toxicity in chemotherapy-treated colorectal cancer patients. Clin. Cancer Res. 2011, 17, 3822–3829. [Google Scholar] [CrossRef] [PubMed]
- Schmoll, H.J. Dihydropyrimidine dehydrogenase inhibition as a strategy for the oral administration of 5-fluorouracil: Utility in the treatment of advanced colorectal cancer. Anticancer Drugs 2003, 14, 695–702. [Google Scholar] [CrossRef] [PubMed]
- Aguado, C.; Garcia-Paredes, B.; Sotelo, M.J.; Sastre, J.; Diaz-Rubio, E. Should capecitabine replace 5-fluorouracil in the first-line treatment of metastatic colorectal cancer? World J. Gastroenterol. 2014, 20, 6092–6101. [Google Scholar] [CrossRef] [PubMed]
- Kobayakawa, M.; Kojima, Y. Tegafur/gimeracil/oteracil (S-1) approved for the treatment of advanced gastric cancer in adults when given in combination with cisplatin: A review comparing it with other fluoropyrimidine-based therapies. Onco. Targets Ther. 2011, 4, 193–201. [Google Scholar] [CrossRef]
- Wempen, I.; Fox, J.J. Pyrimidines.2. Synthesis of 6-Fluorouracil. J. Med. Chem. 1964, 7, 207–209. [Google Scholar] [CrossRef]
- Eckstein, J.W.; Foster, P.G.; Finer-Moore, J.; Wataya, Y.; Santi, D.V. Mechanism-based inhibition of thymidylate synthase by 5-(trifluoromethyl)-2′-deoxyuridine 5′-monophosphate. Biochemistry 1994, 33, 15086–15094. [Google Scholar] [CrossRef]
- Heidelberger, C.; Parsons, D.G.; Remy, D.C. Syntheses of 5-Trifluoromethyluracil and 5-Trifluoromethyl-2′-Deoxyuridine. J. Med. Chem. 1964, 7, 1–5. [Google Scholar] [CrossRef]
- Uraguchi, D.; Yamamoto, K.; Ohtsuka, Y.; Tokuhisa, K.; Yamakawa, T. Catalytic trifluoromethylation of uracil to 5-trifluoromethyluracil by use of CF3I and its industrial applications. Appl. Catal. Gen. 2008, 342, 137–143. [Google Scholar] [CrossRef]
- Song, W.T.; Lv, Z.T.; Wang, L.; Yao, S.Z. Preparation method of trifluridine. Patent CN104761602A, 13 June 2017. [Google Scholar]
- Chan, B.M.; Hochster, H.S.; Lenz, H.J. The safety and efficacy of trifluridine-tipiracil for metastatic colorectal cancer: A pharmacy perspective. Am. J. Health Syst. Pharm. 2019, 76, 339–348. [Google Scholar] [CrossRef]
- Longley, D.B.; Harkin, D.P.; Johnston, P.G. 5-fluorouracil: Mechanisms of action and clinical strategies. Nat. Rev. Cancer 2003, 3, 330–338. [Google Scholar] [CrossRef]
- Liu, L.; Santi, D.V. 5-Fluoro-2′-deoxycytidine 5′-monophosphate is a mechanism-based inhibitor of thymidylate synthase. Biochem. Biophys. Acta 1994, 1209, 89–94. [Google Scholar] [CrossRef]
- Brody, J.R.; Gallmeier, E.; Yoshimura, K.; Hucl, T.; Kulesza, P.; Canto, M.I.; Hruban, R.H.; Schulick, R.D.; Kern, S.E. A proposed clinical test for monitoring fluoropyrimidine therapy: Detection and stability of thymidylate synthase ternary complexes. Cancer Biol. Ther. 2006, 5, 923–927. [Google Scholar] [CrossRef] [PubMed]
- Wolmark, N.; Rockette, H.; Mamounas, E.; Jones, J.; Wieand, S.; Wickerham, D.L.; Bear, H.D.; Atkins, J.N.; Dimitrov, N.V.; Glass, A.G.; et al. Clinical trial to assess the relative efficacy of fluorouracil and leucovorin, fluorouracil and levamisole, and fluorouracil, leucovorin, and levamisole in patients with Dukes’ B and C carcinoma of the colon: Results from National Surgical Adjuvant Breast and Bowel Project C-04. J. Clin. Oncol. 1999, 17, 3553–3559. [Google Scholar] [PubMed]
- Nadal, J.C.; Van Groeningen, C.J.; Pinedo, H.M.; Peters, G.J. In vivo potentiation of 5-fluorouracil by leucovorin in murine colon carcinoma. Biomed. Pharmacother. 1988, 42, 387–393. [Google Scholar] [PubMed]
- Grem, J.L.; Quinn, M.G.; Keith, B.; Monahan, B.P.; Hamilton, J.M.; Xu, Y.; Harold, N.; Nguyen, D.; Takimoto, C.H.; Rowedder, A.; et al. A phase I and pharmacologic study of weekly gemcitabine in combination with infusional 5-fluorodeoxyuridine and oral calcium leucovorin. Cancer Chemother. Pharmacol. 2003, 52, 487–496. [Google Scholar] [CrossRef]
- Lee, J.J.; Beumer, J.H.; Chu, E. Therapeutic drug monitoring of 5-fluorouracil. Cancer Chemother. Pharmacol. 2016, 78, 447–464. [Google Scholar] [CrossRef]
- Salonga, D.; Danenberg, K.D.; Johnson, M.; Metzger, R.; Groshen, S.; Tsao-Wei, D.D.; Lenz, H.J.; Leichman, C.G.; Leichman, L.; Diasio, R.B.; et al. Colorectal tumors responding to 5-fluorouracil have low gene expression levels of dihydropyrimidine dehydrogenase, thymidylate synthase, and thymidine phosphorylase. Clin. Cancer Res. 2000, 6, 1322–1327. [Google Scholar]
- Mori, R.; Yoshida, K.; Futamura, M.; Suetsugu, T.; Shizu, K.; Tanahashi, T.; Tanaka, Y.; Matsuhashi, N.; Yamaguchi, K. The inhibition of thymidine phosphorylase can reverse acquired 5FU-resistance in gastric cancer cells. Gastric. Cancer 2019, 22, 497–505. [Google Scholar] [CrossRef]
- Freel Meyers, C.L.; Hong, L.; Joswig, C.; Borch, R.F. Synthesis and biological activity of novel 5-fluoro-2′-deoxyuridine phosphoramidate prodrugs. J. Med. Chem. 2000, 43, 4313–4318. [Google Scholar] [CrossRef]
- Liu, J.; Skradis, A.; Kolar, C.; Kolath, J.; Anderson, J.; Lawson, T.; Talmadge, J.; Gmeiner, W.H. Increased cytotoxicity and decreased in vivo toxicity of FdUMP[10] relative to 5-FU. Nucleosides Nucleotides 1999, 18, 1789–1802. [Google Scholar] [CrossRef]
- Liu, J.; Kolar, C.; Lawson, T.A.; Gmeiner, W.H. Targeted drug delivery to chemoresistant cells: Folic acid derivatization of FdUMP[10] enhances cytotoxicity toward 5-FU-resistant human colorectal tumor cells. J. Org. Chem. 2001, 66, 5655–5663. [Google Scholar] [CrossRef]
- Pardee, T.S.; Gomes, E.; Jennings-Gee, J.; Caudell, D.; Gmeiner, W.H. Unique dual targeting of thymidylate synthase and topoisomerase1 by FdUMP[10] results in high efficacy against AML and low toxicity. Blood 2012, 119, 3561–3570. [Google Scholar] [CrossRef] [PubMed]
- Pardee, T.S.; Stadelman, K.; Jennings-Gee, J.; Caudell, D.L.; Gmeiner, W.H. The poison oligonucleotide F10 is highly effective against acute lymphoblastic leukemia while sparing normal hematopoietic cells. Oncotarget 2014, 5, 4170–4179. [Google Scholar] [CrossRef] [PubMed]
- Gmeiner, W.H.; Lema-Tome, C.; Gibo, D.; Jennings-Gee, J.; Milligan, C.; Debinski, W. Selective anti-tumor activity of the novel fluoropyrimidine polymer F10 towards G48a orthotopic GBM tumors. J. Neurooncol. 2014, 116, 447–454. [Google Scholar] [CrossRef][Green Version]
- Gmeiner, W.H.; Willingham, M.C.; Bourland, J.D.; Hatcher, H.C.; Smith, T.L.; D’Agostino, R.B., Jr.; Blackstock, W. F10 Inhibits Growth of PC3 Xenografts and Enhances the Effects of Radiation Therapy. J. Clin. Oncol. Res. 2014, 2, 1028. [Google Scholar] [PubMed]
- Gmeiner, W.H.; Dominijanni, A.; Caudell, D.; D’Agostino, R.; Pasche, B.; Smith, T.L.; Mani, C.; Palle, K.; Haber, A.; Brody, J. Efficacy of the Polymeric Fluoropyrimidine CF10 in Colorectal Cancer thru Increased Replication Stress. in preparation.
- Pommier, Y. Drugging topoisomerases: Lessons and challenges. ACS Chem. Biol. 2013, 8, 82–95. [Google Scholar] [CrossRef]
- Das, B.B.; Huang, S.Y.; Murai, J.; Rehman, I.; Ame, J.C.; Sengupta, S.; Das, S.K.; Majumdar, P.; Zhang, H.; Biard, D.; et al. PARP1-TDP1 coupling for the repair of topoisomerase I-induced DNA damage. Nucleic Acids Res. 2014, 42, 4435–4449. [Google Scholar] [CrossRef]
- Gmeiner, W.H.; Gearhart, P.J.; Pommier, Y.; Nakamura, J. F10 cytotoxicity via topoisomerase I cleavage complex repair consistent with a unique mechanism for thymineless death. Future Oncol. 2016. [Google Scholar] [CrossRef]
- Mani, C.; Pai, S.; Papke, C.M.; Palle, K.; Gmeiner, W.H. Thymineless Death by the Fluoropyrimidine Polymer F10 Involves Replication Fork Collapse and Is Enhanced by Chk1 Inhibition. Neoplasia 2018, 20, 1236–1245. [Google Scholar] [CrossRef]
- Gmeiner, W.H.; Debinski, W.; Milligan, C.; Caudell, D.; Pardee, T.S. The applications of the novel polymeric fluoropyrimidine F10 in cancer treatment: Current evidence. Future Oncol. 2016. [Google Scholar] [CrossRef]
- Dominijanni, A.; Gmeiner, W.H. Improved potency of F10 relative to 5-fluorouracil in colorectal cancer cells with p53 mutations. Cancer Drug Resist. 2018, 1, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Bunz, F.; Hwang, P.M.; Torrance, C.; Waldman, T.; Zhang, Y.; Dillehay, L.; Williams, J.; Lengauer, C.; Kinzler, K.W.; Vogelstein, B. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J. Clin. Investig. 1999, 104, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Christensen, S.; Van der Roest, B.; Besselink, N.; Janssen, R.; Boymans, S.; Martens, J.W.M.; Yaspo, M.L.; Priestley, P.; Kuijk, E.; Cuppen, E.; et al. 5-Fluorouracil treatment induces characteristic T>G mutations in human cancer. Nat. Commun. 2019, 10, 4571. [Google Scholar] [CrossRef] [PubMed]
- Gmeiner, W.H. Fluoropyrimidine Modulation of the Anti-Tumor Immune Response-Prospects for Improved Colorectal Cancer Treatment. Cancers (Basel) 2020, 12, 1641. [Google Scholar] [CrossRef]
- Yarchoan, M.; Hopkins, A.; Jaffee, E.M. Tumor Mutational Burden and Response Rate to PD-1 Inhibition. N Engl. J. Med. 2017, 377, 2500–2501. [Google Scholar] [CrossRef]
- Sahasrabudhe, P.V.; Gmeiner, W.H. Solution structures of 5-fluorouracil-substituted RNA duplexes containing G-U wobble base pairs. Biochemistry 1997, 36, 5981–5991. [Google Scholar] [CrossRef]
- Suzuki, T.; Kamiya, H. Mutations induced by 8-hydroxyguanine (8-oxo-7,8-dihydroguanine), a representative oxidized base, in mammalian cells. Genes Environ. 2017, 39, 2. [Google Scholar] [CrossRef]
- Wyatt, M.D.; Wilson, D.M., 3rd. Participation of DNA repair in the response to 5-fluorouracil. Cell Mol. Life Sci. 2009, 66, 788–799. [Google Scholar] [CrossRef]
- Grogan, B.C.; Parker, J.B.; Guminski, A.F.; Stivers, J.T. Effect of the thymidylate synthase inhibitors on dUTP and TTP pool levels and the activities of DNA repair glycosylases on uracil and 5-fluorouracil in DNA. Biochemistry 2011, 50, 618–627. [Google Scholar] [CrossRef]
- Parker, J.B.; Stivers, J.T. Dynamics of uracil and 5-fluorouracil in DNA. Biochemistry 2011, 50, 612–617. [Google Scholar] [CrossRef]
- Melvin, R.L.; Thompson, W.G.; Godwin, R.C.; Gmeiner, W.H.; Salsbury, F.R., Jr. MutSalpha’s Multi-Domain Allosteric Response to Three DNA Damage Types Revealed by Machine Learning. Front. Phys. 2017, 5. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.W.; Saif, M.W.; El-Rayes, B.F.; Fakih, M.G.; Cartwright, T.H.; Posey, J.A.; King, T.R.; von Borstel, R.W.; Bamat, M.K. Emergency use of uridine triacetate for the prevention and treatment of life-threatening 5-fluorouracil and capecitabine toxicity. Cancer 2017, 123, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, D.M.; Watson, A.J.; Potten, C.S.; Jackman, A.L.; Hickman, J.A. Inhibition by uridine but not thymidine of p53-dependent intestinal apoptosis initiated by 5-fluorouracil: Evidence for the involvement of RNA perturbation. Proc. Natl. Acad. Sci. USA 1997, 94, 1795–1799. [Google Scholar] [CrossRef] [PubMed]
- Kealey, J.T.; Gu, X.; Santi, D.V. Enzymatic mechanism of tRNA (m5U54)methyltransferase. Biochimie 1994, 76, 1133–1142. [Google Scholar] [CrossRef]
- Gu, X.; Matsuda, A.; Ivanetich, K.M.; Santi, D.V. Interaction of tRNA (uracil-5-)-methyltransferase with NO2Ura-tRNA. Nucleic Acids Res. 1996, 24, 1059–1064. [Google Scholar] [CrossRef][Green Version]
- Chang, Y.H.; Nishimura, S.; Oishi, H.; Kelly, V.P.; Kuno, A.; Takahashi, S. TRMT2A is a novel cell cycle regulator that suppresses cell proliferation. Biochem. Biophys. Res. Commun. 2019, 508, 410–415. [Google Scholar] [CrossRef]
- Choudhury, S.A.; Asefa, B.; Webb, A.; Ramotar, D.; Chow, T.Y. Functional and genetic analysis of the Saccharomyces cerevisiae RNC1/TRM2: Evidences for its involvement in DNA double-strand break repair. Mol. Cell Biochem. 2007, 300, 215–226. [Google Scholar] [CrossRef]
- Hussain, S. On a New Proposed Mechanism of 5-Fluorouracil-Mediated Cytotoxicity. Trends Cancer 2020, 6, 365–368. [Google Scholar] [CrossRef]
- Yu, Y.T.; Meier, U.T. RNA-guided isomerization of uridine to pseudouridine--pseudouridylation. RNA Biol. 2014, 11, 1483–1494. [Google Scholar] [CrossRef]
- Spenkuch, F.; Motorin, Y.; Helm, M. Pseudouridine: Still mysterious, but never a fake (uridine)! RNA Biol. 2014, 11, 1540–1554. [Google Scholar] [CrossRef]
- Gu, X.; Liu, Y.; Santi, D.V. The mechanism of pseudouridine synthase I as deduced from its interaction with 5-fluorouracil-tRNA. Proc. Natl. Acad. Sci. USA 1999, 96, 14270–14275. [Google Scholar] [CrossRef] [PubMed]
- Gmeiner, W.H.; Sahasrabudhe, P.; Pon, R.T. Synthesis of 5′-O-(4,4′-Dimethoxytrityl)-2′-O-(Tert-Butyldimethylsilyl)-5-Fluorouridine 3′-(Cyanoethyl N,N-Diisopropylphosphoramidite) and Its Use in the Synthesis of Rna. J. Org. Chem. 1994, 59, 5779–5783. [Google Scholar] [CrossRef]
- Mioduszewska, K.; Dolzonek, J.; Wyrzykowski, D.; Kubik, L.; Wiczling, P.; Sikorska, C.; Tonski, M.; Kaczynski, Z.; Stepnowski, P.; Bialk-Bielinska, A. Overview of experimental and computational methods for the determination of the pKa values of 5-fluorouracil, cyclophosphamide, ifosfamide, imatinib and methotrexate. Trac-Trend Anal. Chem. 2017, 97, 283–296. [Google Scholar] [CrossRef]
- Wielinska, J.; Nowacki, A.; Liberek, B. 5-Fluorouracil-Complete Insight into Its Neutral and Ionised Forms. Molecules 2019, 24. [Google Scholar] [CrossRef]
- Othmani, H.; Ben Said, R.; Terzi, N.; Jaidane, N.E.; Al Mogren, M.M.; Elmarghany, A.; Hochlaf, M. Structural, energetic and spectroscopic characterisation of 5-fluorouracil anticarcinogenic drug isomers, tautomers and ions. Mol. Phys. 2019, 117, 1589–1603. [Google Scholar] [CrossRef]
- Markova, N.; Enchev, V.; Timtcheva, I. Oxo-hydroxy tautomerism of 5-fluorouracil: Water-assisted proton transfer. J. Phys. Chem. A 2005, 109, 1981–1988. [Google Scholar] [CrossRef]
- Markova, N.; Enchev, V.; Ivanova, G. Tautomeric Equilibria of 5-Fluorouracil Anionic Species in Water. J. Phys. Chem. A 2010, 114, 13154–13162. [Google Scholar] [CrossRef]
- Yakovchuk, P.; Protozanova, E.; Frank-Kamenetskii, M.D. Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res. 2006, 34, 564–574. [Google Scholar] [CrossRef]
- Sahasrabudhe, P.V.; Pon, R.T.; Gmeiner, W.H. Effects of site-specific substitution of 5-fluorouridine on the stabilities of duplex DNA and RNA. Nucleic Acids Res. 1995, 23, 3916–3921. [Google Scholar] [CrossRef]
- Sahasrabudhe, P.V.; Pon, R.T.; Gmeiner, W.H. Solution structures of 5-fluorouracil-substituted DNA and RNA decamer duplexes. Biochemistry 1996, 35, 13597–13608. [Google Scholar] [CrossRef]
- Graber, D.; Moroder, H.; Micura, R. 19F NMR spectroscopy for the analysis of RNA secondary structure populations. J. Am. Chem. Soc. 2008, 130, 17230–17231. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Devany, M.; Greenbaum, N.L. Measurement of chemical exchange between RNA conformers by 19F NMR. Biochem. Biophys. Res. Commun. 2014, 453, 692–695. [Google Scholar] [CrossRef] [PubMed]
- Hennig, M.; Scott, L.G.; Sperling, E.; Bermel, W.; Williamson, J.R. Synthesis of 5-fluoropyrimidine nucleotides as sensitive NMR probes of RNA structure. J. Am. Chem. Soc. 2007, 129, 14911–14921. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Salsbury, F.R., Jr.; Horita, D.A.; Gmeiner, W.H. Zn2+ selectively stabilizes FdU-substituted DNA through a unique major groove binding motif. Nucleic Acids Res. 2011, 39, 4490–4498. [Google Scholar] [CrossRef]
- Ghosh, S.; Salsbury, F.R., Jr.; Horita, D.A.; Gmeiner, W.H. Cooperative stabilization of Zn(2+):DNA complexes through netropsin binding in the minor groove of FdU-substituted DNA. J. Biomol. Struct. Dyn. 2013, 31, 1301–1310. [Google Scholar] [CrossRef]
- Egli, M. The steric hypothesis for DNA replication and fluorine hydrogen bonding revisited in light of structural data. Acc. Chem. Res. 2012, 45, 1237–1246. [Google Scholar] [CrossRef]
- Punt, C.J.; Koopman, M.; Vermeulen, L. From tumour heterogeneity to advances in precision treatment of colorectal cancer. Nat. Rev. Clin. Oncol. 2016. [Google Scholar] [CrossRef]
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gmeiner, W.H. Chemistry of Fluorinated Pyrimidines in the Era of Personalized Medicine. Molecules 2020, 25, 3438. https://doi.org/10.3390/molecules25153438
Gmeiner WH. Chemistry of Fluorinated Pyrimidines in the Era of Personalized Medicine. Molecules. 2020; 25(15):3438. https://doi.org/10.3390/molecules25153438
Chicago/Turabian StyleGmeiner, William H. 2020. "Chemistry of Fluorinated Pyrimidines in the Era of Personalized Medicine" Molecules 25, no. 15: 3438. https://doi.org/10.3390/molecules25153438
APA StyleGmeiner, W. H. (2020). Chemistry of Fluorinated Pyrimidines in the Era of Personalized Medicine. Molecules, 25(15), 3438. https://doi.org/10.3390/molecules25153438




