Synthesis and Fundamental Evaluation of Radioiodinated Rociletinib (CO-1686) as a Probe to Lung Cancer with L858R/T790M Mutations of Epidermal Growth Factor Receptor (EGFR)
Abstract
1. Introduction
2. Results and Discussion
2.1. Synthesis of Non-Radioactive Compound
2.2. Cell Viability Assays
2.3. EGFR-Tyrosine Kinase (TK) Inhibition Assay
2.4. Radiolabeling of [125I]ICO1686 ([125I]10)
2.5. Determination of Partition Coefficient
2.6. In Vitro Stability Experiments
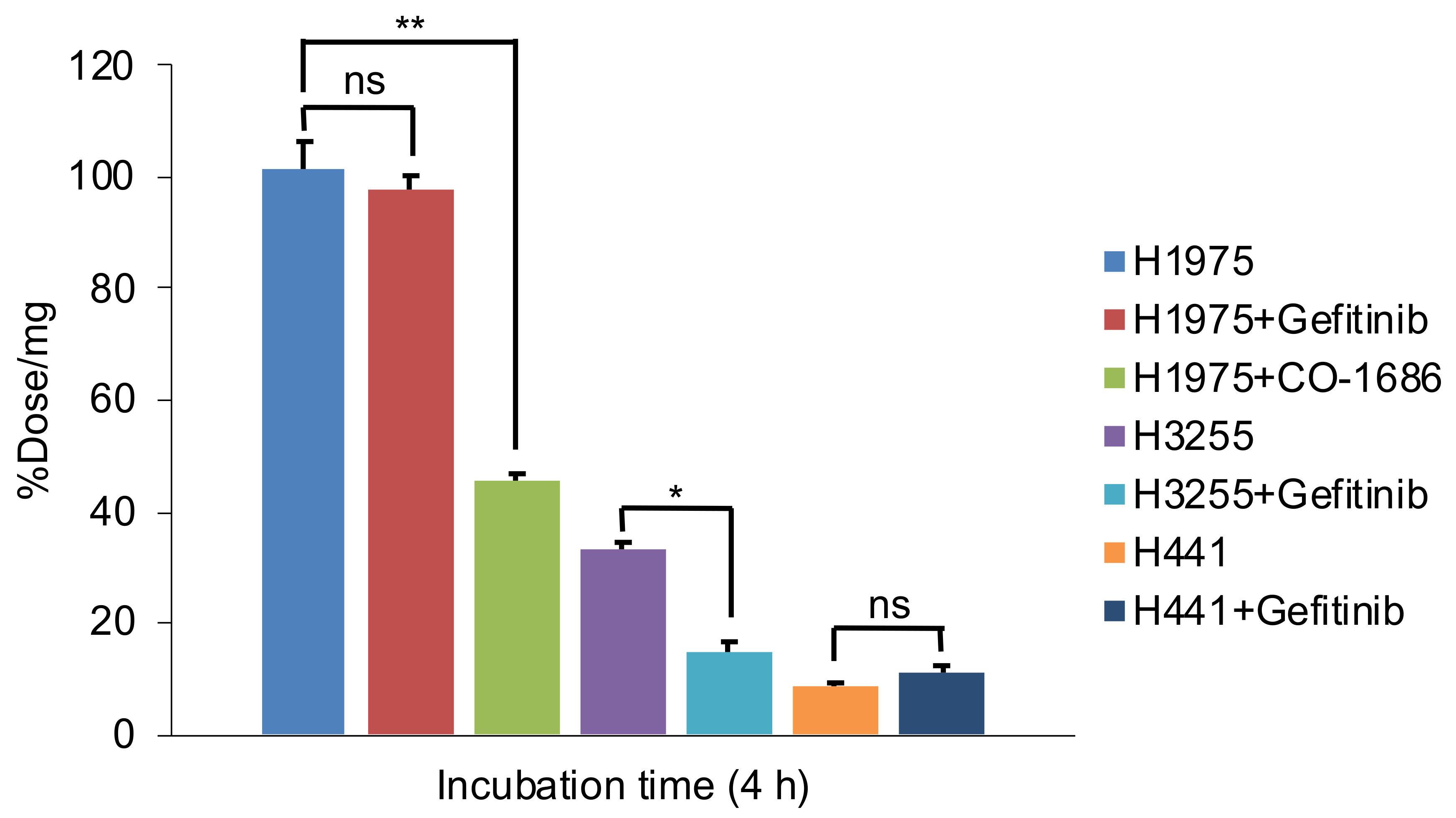
2.7. Cellular Uptake Studies
2.8. Biodistribution Studies
3. Materials and Methods
3.1. General Chemistry
3.2. Synthesis Scheme
3.2.1. Synthesis of 1-Iodo-3,5-dinitrobenzene (1)
3.2.2. Synthesis of 5-Iodobenzene-1,3-diamine (2)
3.2.3. Synthesis of tert-Butyl (3-amino-5-iodophenyl)carbamate (3)
3.2.4. Synthesis of tert-Butyl (3-{[2-chloro-5-(trifluoromethyl)pyrimidin-4-yl]amino}-5-iodophenyl) carbamate (4)
3.2.5. Synthesis of N-(3-{[2-chloro-5-(trifluoromethyl)pyrimidin-4-yl]amino}-5-iodophenyl) acrylamide (5)
3.2.6. Synthesis of 4-Fluoro-2-methoxy-1-nitrobenzene (6)
3.2.7. Synthesis of 1-(Piperazine-1-yl)ethan-1-one (7)
3.2.8. Synthesis of 1-(4-{3-Methoxy-4-nitrophenyl}piperazine-1-yl)ethan-1-one (8)
3.2.9. Synthesis of 1-(4-{4-Amino-3-methoxyphenyl}piperazine-1-yl)ethan-1-one (9)
3.2.10. Synthesis of N-(3-{[2-({4-[4-acetylpiperazin-1-yl]-2-methoxyphenyl}amino)-5- (trifluoromethyl)pyrimidin-4-yl]amino}-5-iodophenyl)acrylamide (10)
3.2.11. Synthesis of N-(3-{[2-({4-[4-acetylpiperazin-1-yl]-2-methoxyphenyl}amino)-5- (trifluoromethyl)pyrimidin-4-yl]amino}-5-{tributylstannyl}phenyl)acrylamide (11)
3.3. Cell Viability Assays (WST-8 Assay)
3.4. Kinase Enzymatic Assays
3.5. Radiosynthesis of [125I]ICO1686 ([125I]10)
3.6. Determination of Partition Coefficient
3.7. In Vitro Stability Experiments
3.8. Cellular Uptake Studies
3.9. Animals
3.10. Biodistribution Studies
3.11. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Rodriquez-Canales, J.; Parra-Cuentas, E.; Witsuba, I.I. Diagnosis and Molecular Classification of Lung Cancer. In Lung Cancer Treatment and Research; Reckamp, K.L., Rosen, S.T., Eds.; Springer International Publishing: Basel, Switzerland, 2016; pp. 25–28. [Google Scholar]
- Bronte, G.; Rizzo, S.; La Paglia, L.; Adamo, V.; Siragusa, S.; Ficorella, C.; Santini, D.; Bazan, V.; Colucci, G.; Gebbia, N.; et al. Driver mutations and differential sensitivity to targeted therapies: A new approach to the treatment of lung adenocarcinoma. Cancer Treat. Rev. 2010, 36, S21–S29. [Google Scholar] [CrossRef]
- Yarden, Y.; Shilo, B.-Z. SnapShot: EGFR Signaling Pathway. Cell 2007, 131, 1018-e1. [Google Scholar] [CrossRef] [PubMed]
- Gazdar, A.F. Personalized Medicine and Inhibition of EGFR Signaling in Lung Cancer. N. Engl. J. Med. 2009, 361, 1018–1020. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.-E.; Zhu, S.-J.; Liang, L.; Zhao, P.; Choi, H.G.; Yun, C.-H. Structural basis of mutant-selectivity and drug-resistance related to CO-1686. Oncotarget 2017, 8, 53508–53517. [Google Scholar] [CrossRef]
- Singh, M.; Jadhav, H.R. Targeting non-small cell lung cancer with small-molecule EGFR tyrosine kinase inhibitors. Drug Discov. Today 2018, 23, 745–753. [Google Scholar] [CrossRef]
- Lu, X.; Yu, L.; Zhang, Z.; Ren, X.; Smaill, J.B.; Ding, K. Targeting EGFRL858R/T790M and EGFRL858R/T790M/C797S resistance mutations in NSCLC: Current developments in medicinal chemistry. Med. Res. Rev. 2018, 38, 1550–1581. [Google Scholar] [CrossRef]
- Pao, W.; Wang, T.Y.; Riely, G.J.; Miller, V.A.; Pan, Q.; Ladanyi, M.; Zakowski, M.F.; Heelan, R.T.; Kris, M.G.; Varmus, H.E. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005, 2, e17. [Google Scholar] [CrossRef]
- Ballard, P.; Yates, J.W.T.; Yang, Z.; Kim, D.W.; Yang, J.C.H.; Cantarini, M.; Pickup, K.; Jordan, A.; Hickey, M.; Grist, M.; et al. Preclinical comparison of osimertinib with other EGFR-TKIs in EGFR-mutant NSCLC brain metastases models, and early evidence of clinical brain metastases activity. Clin. Cancer Res. 2016. [Google Scholar] [CrossRef]
- Hirsch, F.R.; Varella-Garcia, M.; McCoy, J.; West, H.; Xavier, A.C.; Gumerlock, P.; Bunn, P.A.; Franklin, W.A.; Crowley, J.; Gandara, D.R. Increased epidermal growth factor receptor gene copy number detected by fluorescence in situ hybridization associates with increased sensitivity to gefitinib in patients with bronchioloalveolar carcinoma subtypes: A southwest oncology group study. J. Clin. Oncol. 2005, 23, 6838–6845. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Song, Y.; Kai, W.; Sun, X.; Shen, B. Evaluation of 99mTc-HYNIC-MPG as a novel SPECT radiotracer to detect EGFR-activating mutations in NSCLC. Oncotarget 2017, 8, 40732–40740. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Späth, S.S.; Marjani, S.L.; Zhang, W.; Pan, X. Characterization of cancer genomic heterogeneity by next-generation sequencing advances precision medicine in cancer treatment. Precis. Clin. Med. 2018, 1, 29–48. [Google Scholar] [CrossRef] [PubMed]
- Richard, M.B.; Derek, G.P.; Seamus, O. Intratumor Heterogeneity and Branched Evolution. N. Engl. J. Med. 2012, 366, 2132–2133. [Google Scholar] [CrossRef]
- Ogawa, K.; Takeda, T.; Mishiro, K.; Toyoshima, A.; Shiba, K.; Yoshimura, T.; Shinohara, A.; Kinuya, S.; Odani, A. Radiotheranostics Coupled between an At-211-Labeled RGD Peptide and the Corresponding Radioiodine-Labeled RGD Peptide. ACS Omega 2019, 4, 4584–4591. [Google Scholar] [CrossRef]
- Song, A.; Zhang, J.; Ge, Y.; Wang, C.; Meng, Q.; Tang, Z.; Peng, J.; Liu, K.; Li, Y.; Ma, X. C-2 (E)-4-(Styryl)aniline substituted diphenylpyrimidine derivatives (Sty-DPPYs) as specific kinase inhibitors targeting clinical resistance related EGFRT790M mutant. Bioorganic Med. Chem. 2017, 25, 2724–2729. [Google Scholar] [CrossRef]
- Waaijer, S.J.H.; Kok, I.C.; Eisses, B.; Schröder, C.P.; Jalving, M.; Brouwers, A.H.; Lub-de Hooge, M.N.; de Vries, E.G.E. Molecular Imaging in Cancer Drug Development. J. Nucl. Med. 2018, 59, 726–732. [Google Scholar] [CrossRef]
- Yeh, H.H.; Ogawa, K.; Balatoni, J.; Mukhapadhyay, U.; Pal, A.; Gonzalez-Lepera, C.; Shavrin, A.; Soghomonyan, S.; Flores, L.; Young, D.; et al. Molecular imaging of active mutant L858R EGF receptor (EGFR) kinase-expressing nonsmall cell lung carcinomas using PET/CT. Proc. Natl. Acad. Sci. USA 2011, 108, 1603–1608. [Google Scholar] [CrossRef]
- Su, H.; Seimbille, Y.; Ferl, G.Z.; Bodenstein, C.; Fueger, B.; Kim, K.J.; Hsu, Y.T.; Dubinett, S.M.; Phelps, M.E.; Czernin, J.; et al. Evaluation of [18F]gefitinib as a molecular imaging probe for the assessment of the epidermal growth factor receptor status in malignant tumors. Eur. J. Nucl. Med. Mol. Imaging 2008, 35, 1089–1099. [Google Scholar] [CrossRef]
- Sequist, L.V.; Soria, J.-C.; Goldman, J.W.; Wakelee, H.A.; Gadgeel, S.M.; Varga, A.; Papadimitrakopoulou, V.; Solomon, B.J.; Oxnard, G.R.; Dziadziuszko, R.; et al. Rociletinib in EGFR -Mutated Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2015, 372, 1700–1709. [Google Scholar] [CrossRef]
- Tran, P.N.; Klempner, S.J. Profile of rociletinib and its potential in the treatment of non-small-cell lung cancer. Lung Cancer Targets Ther. 2016, 7, 91–97. [Google Scholar] [CrossRef]
- Hirano, T.; Yasuda, H.; Tani, T.; Hamamoto, J.; Oashi, A.; Ishioka, K.; Arai, D.; Nukaga, S.; Miyawaki, M.; Kawada, I.; et al. In vitro modeling to determine mutation specificity of EGFR tyrosine kinase inhibitors against clinically relevant EGFR mutants in non-small-cell lung cancer. Oncotarget 2015, 6, 38789–38803. [Google Scholar] [CrossRef] [PubMed]
- Walter, A.O.; Sjin, R.T.T.; Haringsma, H.J.; Sun, J.; Ohashi, K.; Lee, K.; Dubrovskiy, A.; Labenski, M.; Wang, Z.; Zhu, Z.; et al. Discovery of a mutant-selective covalent inhibitor of EGFR that overcomes T790M mediated resistance in NSCLC. Cancer Discov. 2013, 3, 1404–1415. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Song, Z.; Jin, Y.; Tang, Z.; Kang, J.; Ma, X. Novel Selective and Potent EGFR Inhibitor that Overcome T790M-Mediated Resistance in Non-Small Cell Lung Cancer. Molecules 2016, 21, 1462. [Google Scholar] [CrossRef]
- Liu, X.; Testa, B.; Fahr, A. Lipophilicity and its relationship with passive drug permeation. Pharm. Res. 2011, 28, 962–977. [Google Scholar] [CrossRef]
- Pal, A.; Balatoni, J.A.; Mukhopadhyay, U.; Ogawa, K.; Gonzalez-Lepera, C.; Shavrin, A.; Volgin, A.; Tong, W.; Alauddin, M.M.; Gelovani, J.G. Radiosynthesis and initial in vitro evaluation of [18F]F- PEG 6-IPQA-A novel PET radiotracer for imaging EGFR expression-activity in lung carcinomas. Mol. Imaging Biol. 2011, 13, 853–861. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, L.; Wei, A.; Jia, X.; Liu, X. CM082, a novel angiogenesis inhibitor, enhances the antitumor activity of gefitinib on epidermal growth factor receptor mutant non-small cell lung cancer in vitro and in vivo. Thorac. Cancer 2020, 11, 1566–1577. [Google Scholar] [CrossRef]
- Makino, A.; Miyazaki, A.; Tomoike, A.; Kimura, H.; Arimitsu, K.; Hirata, M.; Ohmomo, Y.; Nishii, R.; Okazawa, H.; Kiyono, Y.; et al. PET probe detecting non-small cell lung cancer susceptible to epidermal growth factor receptor tyrosine kinase inhibitor therapy. Bioorganic Med. Chem. 2018, 26, 1609–1613. [Google Scholar] [CrossRef]
- Zhang, M.R.; Kumata, K.; Hatori, A.; Takai, N.; Toyohara, J.; Yamasaki, T.; Yanamoto, K.; Yui, J.; Kawamura, K.; Koike, S.; et al. [11C]Gefitinib ([11C]Iressa): Radiosynthesis, In Vitro uptake, and In Vivo imaging of intact murine fibrosarcoma. Mol. Imaging Biol. 2010, 12, 181–191. [Google Scholar] [CrossRef]
- Slobbe, P.; Windhorst, A.D.; van Walsum, M.S.; Schuit, R.C.; Smit, E.F.; Niessen, H.G.; Solca, F.; Stehle, G.; van Dongen, G.A.M.S.; Poot, A.J. Development of [18F]afatinib as new TKI-PET tracer for EGFR positive tumors. Nucl. Med. Biol. 2014, 41, 749–757. [Google Scholar] [CrossRef]
- Song, Y.; Xiao, Z.; Wang, K.; Wang, X.; Zhang, C.; Fang, F.; Sun, X.; Shen, B. Development and Evaluation of 18F-IRS for Molecular Imaging Mutant EGF Receptors in NSCLC. Sci. Rep. 2017, 7, 3121. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Yuan, L.; Yin, L.; Jiang, Y.; Gai, Y.; Liu, Q.; Wang, Y.; Zhang, Y.; Lan, X. Synthesis and Preclinical Evaluation of 18F-PEG3-FPN for the Detection of Metastatic Pigmented Melanoma. Mol. Pharm. 2017, 14, 3896–3905. [Google Scholar] [CrossRef] [PubMed]
- Lux, J.; Chan, M.; Vander Elst, L.; Schopf, E.; Mahmoud, E.; Laurent, S.; Almutairi, A. Metal chelating crosslinkers form nanogels with high chelation stability. J. Mater. Chem. B 2013, 1, 6359–6364. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Liu, X.; Bai, J.; Long, W. Protein Tyrosine Kinase Modulators and Methods of Use. WO2015003658, 15 January 2015. [Google Scholar]
- Poot, A.; Slobbe, P.; Windhorst, A.; Haumann, R.; Schuit, R.; Van Dongen, G. [11C]nintedanib as TKI-PET tracer for angiogenesis imaging in vivo. J. Nucl. Med. 2016, 57, 1153. [Google Scholar]
- Zhou, W.; Ercan, D.; Chen, L.; Yun, C.H.; Li, D.; Capelletti, M.; Cortot, A.B.; Chirieac, L.; Iacob, R.E.; Padera, R.; et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature 2009, 462, 1070–1074. [Google Scholar] [CrossRef] [PubMed]
- Dulla, B.; Vijayavardhini, S.; Rambau, D.; Anuradha, V.; Rao, M.; Pal, M. Catalyst / Surfactant Free Chemoselective Acylation of Amines in Water. Curr. Green Chem. 2013, 1, 73–79. [Google Scholar] [CrossRef]
- Koura, M.; Yamaguchi, Y.; Kurobuchi, S.; Sumida, H.; Watanabe, Y.; Enomoto, T.; Matsuda, T.; Okuda, A.; Koshizawa, T.; Matsumoto, Y.; et al. Discovery of a 2-hydroxyacetophenone derivative as an outstanding linker to enhance potency and β-selectivity of liver X receptor agonist. Bioorganic Med. Chem. 2016, 24, 3436–3446. [Google Scholar] [CrossRef]
- Effendi, N.; Ogawa, K.; Mishiro, K.; Takarada, T.; Yamada, D.; Kitamura, Y.; Shiba, K.; Maeda, T.; Odani, A. Synthesis and evaluation of radioiodinated 1-{2-[5-(2-methoxyethoxy)-1H-benzo[d]imidazol-1-yl]quinolin-8-yl}piperidin-4-amine derivatives for platelet-derived growth factor receptor β (PDGFRβ) imaging. Bioorganic Med. Chem. 2017, 25, 5576–5585. [Google Scholar] [CrossRef]
- Ogawa, K.; Shiba, K.; Akhter, N.; Yoshimoto, M.; Washiyama, K.; Kinuya, S.; Kawai, K.; Mori, H. Evaluation of radioiodinated vesamicol analogs for sigma receptor imaging in tumor and radionuclide receptor therapy. Cancer Sci. 2009, 100, 2188–2192. [Google Scholar] [CrossRef]
Sample Availability: Sample of the compound ICO1686 available from the authors. |


| Cell Lines | Mutation Status | IC50 (μM) | ||
|---|---|---|---|---|
| 10 | CO-1686 | Gefitinib | ||
| H1975 | L858R/T790M | 0.20 ± 0.05 | 0.14 ± 0.05 | >10 |
| H3255 | L858R | 0.50 ± 0.21 | 0.15 ± 0.02 | 0.02 ± 0.02 |
| H441 | Wild-type | 1.84 ± 0.44 | 0.26 ± 0.04 | >10 |
| Compound | IC50 Value (μM) | SI (WT: L858R/T790M) | |
|---|---|---|---|
| Wild-Type (WT) | L858R/T790M | ||
| CO-1686 | 1.68 ± 0.15 | 0.04 ± 0.01 | 42 |
| 10 | >10 | 0.31 ± 0.15 | >32 |
| Tissues | Time after Injection | |||
|---|---|---|---|---|
| 10 min | 1 h | 4 h | 24 h | |
| [125I]10 | – | – | – | – |
| Blood | 1.06 (0.19) | 0.49 (0.05) | 0.33 (0.07) | 0.05 (0.01) |
| Liver | 26.69 (0.81) | 18.57 (2.72) | 7.85 (0.92) | 0.80 (0.10) |
| Kidney | 7.48 (1.55) | 3.76 (0.40) | 2.37 (0.17) | 0.25 (0.01) |
| Small intestine | 9.46 (2.19) | 23.96 (2.18) | 9.81 (1.03) | 0.10 (0.01) |
| Large intestine | 0.57 (0.04) | 0.53 (0.08) | 53.44 (6.73) | 0.34 (0.08) |
| Spleen | 2.14 (0.50) | 0.90 (0.17) | 0.58 (0.11) | 0.08 (0.04) |
| Pancreas | 1.93 (0.37) | 0.99 (0.26) | 0.41 (0.03) | 0.10 (0.11) |
| Lung | 4.47 (1.39) | 1.33 (0.08) | 0.86 (0.10) | 0.26 (0.03) |
| Heart | 3.11 (0.64) | 0.79 (0.13) | 0.35 (0.09) | 0.09 (0.05) |
| Stomach ‡ | 3.52 (2.70) | 5.10 (1.18) | 1.38 (0.56) | 0.08 (0.02) |
| Bone | 1.16 (0.21) | 0.50 (0.22) | 0.29 (0.11) | 0.07 (0.05) |
| Muscle | 1.40 (0.16) | 0.44 (0.05) | 0.13 (0.03) | 0.04 (0.04) |
| Brain | 0.11 (0.02) | 0.05 (0.02) | 0.03 (0.01) | 0.01 (0.01) |
| Urine | – | – | – | 4.07 (0.83) |
| Feces | – | – | – | 69.93 (15.71) |
| Tissues | Time after Injection | ||
|---|---|---|---|
| 1 h | 4 h | Blocking (1 h) | |
| [125I]10 | – | – | – |
| Blood | 1.74 (0.18) | 0.77 (0.07) | 1.79 (0.21) |
| Liver | 28.12 (5.43) | 12.40 (1.06) | 21.77 (2.83) |
| Kidney | 5.26 (1.50) | 1.62 (0.26) | 4.19 (1.28) |
| Small intestine | 85.58 (7.68) | 9.82 (3.17) | 60.68 (6.67) |
| Large intestine | 18.65 (19.88) | 144.21 (10.97) | 8.96 (7.48) |
| Spleen | 3.20 (0.62) | 0.71 (0.19) | 2.81 (0.77) |
| Pancreas | 1.99 (0.52) | 0.49 (0.12) | 3.87 (0.87) |
| Lung | 4.67 (0.85) | 1.02 (0.28) | 3.40 (0.93) |
| Heart | 1.46 (0.31) | 0.60 (0.67) | 1.53 (0.58) |
| Stomach ‡ | 1.04 (0.36) | 0.48 (0.19) | 1.44 (0.57) |
| Bone | 0.50 (0.40) | 0.24 (0.20) | 1.07 (0.36) |
| Muscle | 0.86 (0.21) | 0.27 (0.08) | 1.09 (0.42) |
| Brain | 0.11 (0.08) | 0.04 (0.02) | 0.10 (0.02) |
| H1975 | 1.77 (0.43) | 0.43 (0.08) | 1.65 (0.64) |
| H3255 | 1.63 (0.23) | 0.70 (0.13) | 1.47 (0.71) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fawwaz, M.; Mishiro, K.; Nishii, R.; Sawazaki, I.; Shiba, K.; Kinuya, S.; Ogawa, K. Synthesis and Fundamental Evaluation of Radioiodinated Rociletinib (CO-1686) as a Probe to Lung Cancer with L858R/T790M Mutations of Epidermal Growth Factor Receptor (EGFR). Molecules 2020, 25, 2914. https://doi.org/10.3390/molecules25122914
Fawwaz M, Mishiro K, Nishii R, Sawazaki I, Shiba K, Kinuya S, Ogawa K. Synthesis and Fundamental Evaluation of Radioiodinated Rociletinib (CO-1686) as a Probe to Lung Cancer with L858R/T790M Mutations of Epidermal Growth Factor Receptor (EGFR). Molecules. 2020; 25(12):2914. https://doi.org/10.3390/molecules25122914
Chicago/Turabian StyleFawwaz, Muammar, Kenji Mishiro, Ryuichi Nishii, Izumi Sawazaki, Kazuhiro Shiba, Seigo Kinuya, and Kazuma Ogawa. 2020. "Synthesis and Fundamental Evaluation of Radioiodinated Rociletinib (CO-1686) as a Probe to Lung Cancer with L858R/T790M Mutations of Epidermal Growth Factor Receptor (EGFR)" Molecules 25, no. 12: 2914. https://doi.org/10.3390/molecules25122914
APA StyleFawwaz, M., Mishiro, K., Nishii, R., Sawazaki, I., Shiba, K., Kinuya, S., & Ogawa, K. (2020). Synthesis and Fundamental Evaluation of Radioiodinated Rociletinib (CO-1686) as a Probe to Lung Cancer with L858R/T790M Mutations of Epidermal Growth Factor Receptor (EGFR). Molecules, 25(12), 2914. https://doi.org/10.3390/molecules25122914









