Yeast Telomerase RNA Flexibly Scaffolds Protein Subunits: Results and Repercussions
Abstract
1. Introduction
2. Results from Investigating Yeast Telomerase RNA
2.1. Rapid Evolution
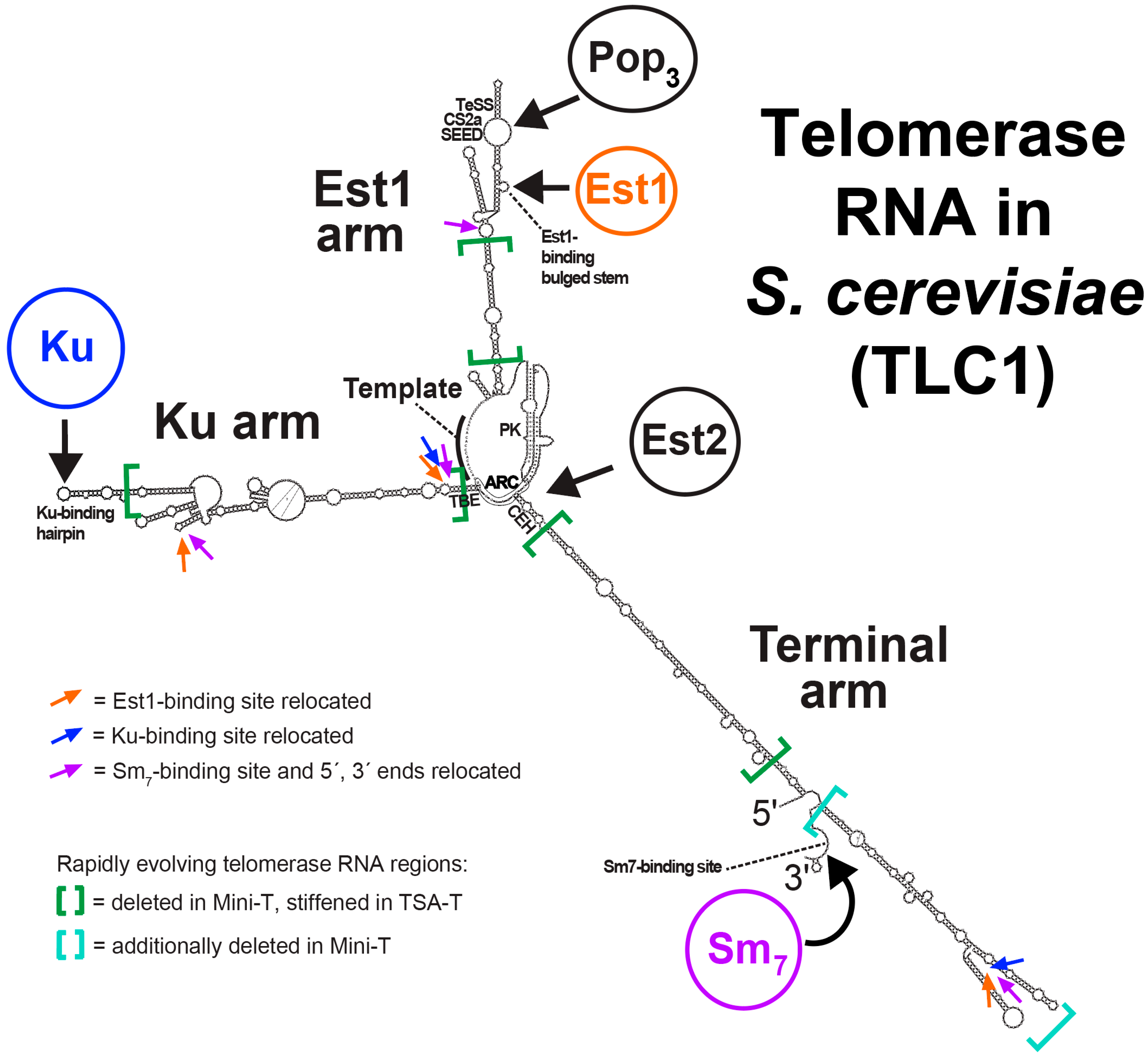
2.2. Identifying S. cerevisiae Telomerase RNA Secondary Structure
2.3. Functional Repositioning of Holoenzyme Subunits and the Ends of TLC1
2.3.1. Est1
2.3.2. Ku
2.3.3. Sm7 and the 5′ and 3′ ends
2.4. Deleting or Stiffening the Presumably Pliable TLC1 RNA Arms
- We deleted the bulk of the three rapidly evolving RNA arms to test whether they were important [25]. These 384–500-nt “Mini-T” RNAs still contained all of the conserved patches of sequences that bind to proteins; they only have the rapidly evolving medial segments of the RNA arms excised (Figure 1).
- We stiffened the arms by converting them all (and each single- and double-arm combination) to perfect double-stranded RNA (dsRNA) rod-like struts by removing all of the bulges and loops that connect all the dsRNA segments [26] (Figure 1). The triple-stiff-arm RNA was called TSA-T. As with Mini-T, TSA-T did not have altered structure in the conserved patches that we knew bound to protein subunits. (N.B., long, perfect stretches of dsRNA do not feed into a siRNA pathway in S. cerevisiae cells, since the organism lacks miRNA-processing components.)
2.5. TLC1 RNA Has Some Important Secondary and Tertiary Structure within Subunit-Binding Modules
3. Repercussions
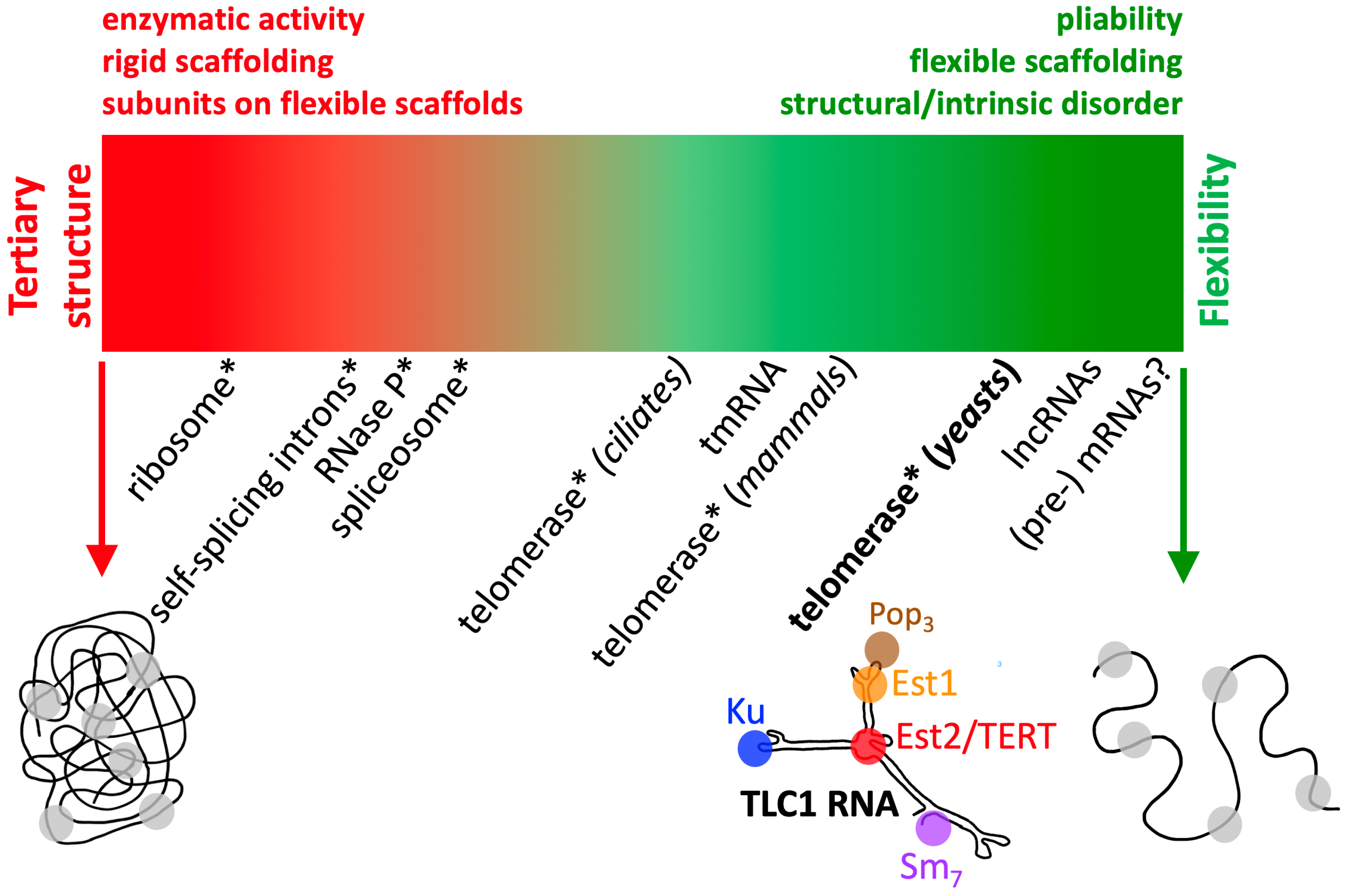
3.1. A Spectrum of RNP Flexibility
3.1.1. Defining the Limits: The Most Rigid RNPs
3.1.2. Defining the Limits: The Most Flexible RNPs
3.2. What TLC1 Flexible Scaffolding Implies for other Telomerase RNPs
4. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Cech, T.R. Crawling Out of the RNA World. Cell 2009, 136, 599–602. [Google Scholar] [CrossRef]
- Greider, C.W.; Blackburn, E. Identification of a specific telomere terminal transferase activity in tetrahymena extracts. Cell 1985, 43, 405–413. [Google Scholar] [CrossRef]
- Singer, M.; Gottschling, D. TLC1: Template RNA component of Saccharomyces cerevisiae telomerase. Science 1994, 266, 404–409. [Google Scholar] [CrossRef]
- Lingner, J.; Hughes, T.R.; Shevchenko, A.; Mann, M.; Lundblad, V.; Cech, T.R. Reverse Transcriptase Motifs in the Catalytic Subunit of Telomerase. Science 1997, 276, 561–567. [Google Scholar] [CrossRef]
- Lundblad, V.; Szostak, J.W. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 1989, 57, 633–643. [Google Scholar] [CrossRef]
- Seto, A.G.; Livengood, A.J.; Tzfati, Y.; Blackburn, E.H.; Cech, T.R. A bulged stem tethers Est1p to telomerase RNA in budding yeast. Genes Dev. 2002, 16, 2800–2812. [Google Scholar] [CrossRef]
- Peterson, S.E.; Stellwagen, A.E.; Diede, S.; Singer, M.S.; Haimberger, Z.W.; Johnson, C.O.; Tzoneva, M.; Gottschling, D.E. The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat. Genet. 2001, 27, 64–67. [Google Scholar] [CrossRef]
- Stellwagen, A.E.; Haimberger, Z.W.; Veatch, J.R.; Gottschling, D.E. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev. 2003, 17, 2384–2395. [Google Scholar] [CrossRef]
- Seto, A.G.; Zaug, A.J.; Sobel, S.G.; Wolin, S.L.; Cech, T.R. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature 1999, 401, 177–180. [Google Scholar] [CrossRef]
- Tzfati, Y.; Fulton, T.B.; Roy, J.; Blackburn, E.H. Template Boundary in a Yeast Telomerase Specified by RNA Structure. Science 2000, 288, 863–867. [Google Scholar] [CrossRef]
- Tzfati, Y.; Knight, Z.; Roy, J.; Blackburn, E.H. A novel pseudoknot element is essential for the action of a yeast telomerase. Genes Dev. 2003, 17, 1779–1788. [Google Scholar] [CrossRef]
- Seto, A.G.; Umansky, K.; Tzfati, Y.; Zaug, A.J.; Blackburn, E.H.; Cech, T.R. A template-proximal RNA paired element contributes to Saccharomyces cerevisiae telomerase activity. RNA 2003, 9, 1323–1332. [Google Scholar] [CrossRef][Green Version]
- Zappulla, D.C.; Cech, T.R. Yeast telomerase RNA: A flexible scaffold for protein subunits. Proc. Natl. Acad. Sci. USA 2004, 101, 10024–10029. [Google Scholar] [CrossRef]
- Dandjinou, A.T.; Levesque, N.; LaRose, S.; Lucier, J.-F.; Elela, S.A.; Wellinger, R.J. A Phylogenetically Based Secondary Structure for the Yeast Telomerase RNA. Curr. Boil. 2004, 14, 1148–1158. [Google Scholar] [CrossRef]
- Livengood, A.J.; Zaug, A.J.; Cech, T.R. Essential Regions of Saccharomyces cerevisiae Telomerase RNA: Separate Elements for Est1p and Est2p Interaction. Mol. Cell. Boil. 2002, 22, 2366–2374. [Google Scholar] [CrossRef]
- Mathews, D.H.; Sabina, J.; Zuker, M.; Turner, D.H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Boil. 1999, 288, 911–940. [Google Scholar] [CrossRef]
- Hofacker, I.L. Vienna RNA secondary structure server. Nucleic Acids Res. 2003, 31, 3429–3431. [Google Scholar] [CrossRef]
- Niederer, R.O.; Zappulla, D.C. Refined secondary-structure models of the core of yeast and human telomerase RNAs directed by SHAPE. RNA 2015, 21, 1053. [Google Scholar] [CrossRef]
- Qiao, F.; Cech, T.R. Triple-helix structure in telomerase RNA contributes to catalysis. Nat. Struct. Mol. Boil. 2008, 15, 634–640. [Google Scholar] [CrossRef]
- Lin, J.; Ly, H.; Hussain, A.; Abraham, M.; Pearl, S.; Tzfati, Y.; Parslow, T.G.; Blackburn, E.H. A universal telomerase RNA core structure includes structured motifs required for binding the telomerase reverse transcriptase protein. Proc. Natl. Acad. Sci. USA 2004, 101, 14713–14718. [Google Scholar] [CrossRef]
- Liu, F.; Theimer, C.A. Telomerase Activity Is Sensitive to Subtle Perturbations of the TLC1 Pseudoknot 3′ Stem and Tertiary Structure. J. Mol. Boil. 2012, 423, 719–735. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Kim, Y.; Cruickshank, C.; Theimer, C.A. Thermodynamic characterization of the Saccharomyces cerevisiae telomerase RNA pseudoknot domain in vitro. RNA 2012, 18, 973–991. [Google Scholar] [CrossRef] [PubMed]
- Zappulla, D.C.; Goodrich, K.J.; Arthur, J.R.; Gurski, L.A.; Denham, E.M.; Stellwagen, A.E.; Cech, T.R. Ku can contribute to telomere lengthening in yeast at multiple positions in the telomerase RNP. RNA 2010, 17, 298–311. [Google Scholar] [CrossRef] [PubMed]
- Hass, E.P.; Zappulla, D.C. Repositioning the Sm-Binding Site in Saccharomyces cerevisiae Telomerase RNA Reveals RNP Organizational Flexibility and Sm-Directed 3′-End Formation. Non-Coding RNA 2020, 6, 9. [Google Scholar] [CrossRef]
- Zappulla, D.C.; Goodrich, K.; Cech, T.R. A miniature yeast telomerase RNA functions in vivo and reconstitutes activity in vitro. Nat. Struct. Mol. Boil. 2005, 12, 1072–1077. [Google Scholar] [CrossRef]
- Lebo, K.J.; Zappulla, D.C. Stiffened yeast telomerase RNA supports RNP function in vitro and in vivo. RNA 2012, 18, 1666–1678. [Google Scholar] [CrossRef]
- Lin, K.-W.; McDonald, K.R.; Guise, A.J.; Chan, A.; Cristea, I.M.; Zakian, V.A. Proteomics of yeast telomerase identified Cdc48-Npl4-Ufd1 and Ufd4 as regulators of Est1 and telomere length. Nat. Commun. 2015, 6, 8290. [Google Scholar] [CrossRef]
- Lemieux, B.; Laterreur, N.; Perederina, A.; Noël, J.-F.; Dubois, M.-L.; Krasilnikov, A.S.; Wellinger, R.J. Active Yeast Telomerase Shares Subunits with Ribonucleoproteins RNase P and RNase MRP. Cell 2016, 165, 1171–1181. [Google Scholar] [CrossRef]
- Mefford, M.A.; Rafiq, Q.; Zappulla, D.C. RNA connectivity requirements between conserved elements in the core of the yeast telomerase RNP. EMBO J. 2013, 32, 2980–2993. [Google Scholar] [CrossRef]
- Mefford, M.A.; Hass, E.P.; Zappulla, D.C. A 4-base pair core-enclosing helix in telomerase RNA is essential and binds to the TERT catalytic protein subunit. bioRxiv 2020. [Google Scholar] [CrossRef]
- Laterreur, N.; Eschbach, S.H.; Lafontaine, D.A.; Wellinger, R.J. A new telomerase RNA element that is critical for telomere elongation. Nucleic Acids Res. 2013, 41, 7713–7724. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Garcia, P.D.; Leach, R.W.; Wadsworth, G.M.; Choudhary, K.; Li, H.; Aviran, S.; Kim, H.D.; Zakian, V.A. Stability and nuclear localization of yeast telomerase depend on protein components of RNase P/MRP. Nat. Commun. 2020, 11, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Evans, S.K.; Lundblad, V. The Est1 subunit of Saccharomyces cerevisiae telomerase makes multiple contributions to telomere length maintenance. Genetics 2002, 162, 1101–1115. [Google Scholar]
- Taggart, A.K.P.; Dulloo, A.G. Est1p As a Cell Cycle-Regulated Activator of Telomere-Bound Telomerase. Science 2002, 297, 1023–1026. [Google Scholar] [CrossRef] [PubMed]
- Ting, N.S.; Yu, Y.; Pohorelic, B.; Lees-Miller, S.P.; Beattie, T.L. Human Ku70/80 interacts directly with hTR, the RNA component of human telomerase. Nucleic Acids Res. 2005, 33, 2090–2098. [Google Scholar] [CrossRef] [PubMed]
- Fisher, T.S.; Taggart, A.K.P.; Zakian, V.A. Cell cycle-dependent regulation of yeast telomerase by Ku. Nat. Struct. Mol. Boil. 2004, 11, 1198–1205. [Google Scholar] [CrossRef]
- Hass, E.P.; Zappulla, D.C. The Ku subunit of telomerase binds Sir4 to recruit telomerase to lengthen telomeres in S. cerevisiae. eLife 2015, 4. [Google Scholar] [CrossRef]
- Chen, H.; Xue, J.; Churikov, D.; Hass, E.P.; Shi, S.; Lemon, L.D.; Luciano, P.; Bertuch, A.A.; Zappulla, D.C.; Geli, V.; et al. Structural Insights into Yeast Telomerase Recruitment to Telomeres. Cell 2018, 172, 331–343.e13. [Google Scholar] [CrossRef]
- Spokoini-Stern, R.; Stamov, D.; Jessel, H.; Aharoni, L.; Haschke, H.; Giron, J.; Unger, R.; Segal, E.; Abu-Horowitz, A.; Bachelet, I. Visualizing the structure and motion of the long noncoding RNA HOTAIR. RNA 2020, 26, 629–636. [Google Scholar] [CrossRef]
- Uhlenbeck, O.C. Keeping RNA happy. RNA 1995, 1, 4–6. [Google Scholar]
- Zappulla, D.C.; Roberts, J.N.; Goodrich, K.J.; Cech, T.R.; Wuttke, D.S. Inhibition of yeast telomerase action by the telomeric ssDNA-binding protein, Cdc13p. Nucleic Acids Res. 2008, 37, 354–367. [Google Scholar] [CrossRef] [PubMed]
- Lei, M.; Zaug, A.J.; Podell, E.R.; Cech, T.R. Switching Human Telomerase On and Off with hPOT1 Protein in Vitro. J. Boil. Chem. 2005, 280, 20449–20456. [Google Scholar] [CrossRef] [PubMed]
- Bajon, E.; Laterreur, N.; Wellinger, R.J. A Single Templating RNA in Yeast Telomerase. Cell Rep. 2015, 12, 441–448. [Google Scholar] [CrossRef]
- Förstemann, K.; Lingner, J. Telomerase limits the extent of base pairing between template RNA and telomeric DNA. EMBO Rep. 2005, 6, 361–366. [Google Scholar] [CrossRef]
- Lebo, K.J.; Niederer, R.O.; Zappulla, D.C. A second essential function of the Est1-binding arm of yeast telomerase RNA. RNA 2015, 21, 862–876. [Google Scholar] [CrossRef]
- Dalby, A.B.; Goodrich, K.J.; Pfingsten, J.S.; Cech, T.R. RNA recognition by the DNA end-binding Ku heterodimer. RNA 2013, 19, 841–851. [Google Scholar] [CrossRef]
- Mason, D.X.; Goneska, E.; Greider, C.W. Stem-Loop IV of Tetrahymena Telomerase RNA Stimulates Processivity in trans. Mol. Cell. Boil. 2003, 23, 5606–5613. [Google Scholar] [CrossRef]
- Mefford, M.A.; Zappulla, D.C. Physical connectivity mapping by circular permutation of human telomerase RNA reveals new regions critical for activity and processivity. Mol. Cell. Boil. 2015, 36, MCB.00794-15. [Google Scholar] [CrossRef]
- Shay, J.; Bacchetti, S. A survey of telomerase activity in human cancer. Eur. J. Cancer 1997, 33, 787–791. [Google Scholar] [CrossRef]
- Tomas-Loba, A.; Flores, I.; Fernández-Marcos, P.; Cayuela, M.L.; Maraver, A.; Tejera, A.M.; Borrás, C.; Matheu, A.; Klatt, P.; Flores, J.M.; et al. Telomerase Reverse Transcriptase Delays Aging in Cancer-Resistant Mice. Cell 2008, 135, 609–622. [Google Scholar] [CrossRef]
- Blasco, M.A.; Lee, H.-W.; Hande, M.P.; Samper, E.; Lansdorp, P.M.; DePinho, R.A.; Greider, C.W. Telomere Shortening and Tumor Formation by Mouse Cells Lacking Telomerase RNA. Cell 1997, 91, 25–34. [Google Scholar] [CrossRef]
- Nguyen, T.H.D.; Tam, J.; Wu, R.A.; Greber, B.J.; Toso, D.; Nogales, E.; Collins, K. Cryo-EM structure of substrate-bound human telomerase holoenzyme. Nature 2018, 557, 190–195. [Google Scholar] [CrossRef]
- Chen, J.J.-L.; Blasco, M.A.; Greider, C.W. Secondary Structure of Vertebrate Telomerase RNA. Cell 2000, 100, 503–514. [Google Scholar] [CrossRef]
- Zappulla, D.C.; Cech, T. RNA as a Flexible Scaffold for Proteins: Yeast Telomerase and Beyond. Cold Spring Harb. Symp. Quant. Boil. 2006, 71, 217–224. [Google Scholar] [CrossRef]
- Ban, N. The Complete Atomic Structure of the Large Ribosomal Subunit at 2.4 A Resolution. Science 2000, 289, 905–920. [Google Scholar] [CrossRef]
- Brodersen, D.E.; Clemons, W.M., Jr.; Carter, A.P.; Wimberly, B.T.; Ramakrishnan, V. Crystal structure of the 30 s ribosomal subunit from Thermus thermophilus: Structure of the proteins and their interactions with 16 s RNA. J. Mol. Boil. 2002, 316, 725–768. [Google Scholar] [CrossRef]
- Available online: https://www.nobelprize.org/prizes/chemistry/2009/ramakrishnan/lecture/ (accessed on 14 June 2020).
- Singh, G.; Pratt, G.; Yeo, G.W.; Moore, M.J. The Clothes Make the mRNA: Past and Present Trends in mRNP Fashion. Annu. Rev. Biochem. 2015, 84, 325–354. [Google Scholar] [CrossRef]
- Guttman, M.; Donaghey, J.; Carey, B.W.; Garber, M.; Grenier, J.K.; Munson, G.; Young, G.; Lucas, A.B.; Ach, R.; Bruhn, L.; et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011, 477, 295–300. [Google Scholar] [CrossRef]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martín, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genes Dev. 2012, 22, 1775–1789. [Google Scholar] [CrossRef]
- Novikova, I.V.; Hennelly, S.P.; Tung, C.-S.; Sanbonmatsu, K.Y. Rise of the RNA Machines: Exploring the Structure of Long Non-Coding RNAs. J. Mol. Boil. 2013, 425, 3731–3746. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Rice, K.; Wang, Y.; Chen, W.; Zhong, Y.; Nakayama, Y.; Zhou, Y.; Klibanski, A. Maternally expressed gene 3 (MEG3) noncoding ribonucleic acid: Isoform structure, expression, and functions. Endocrinol. 2009, 151, 939–947. [Google Scholar] [CrossRef] [PubMed]
- Uroda, T.; Anastasakou, E.; Rossi, A.; Teulon, J.-M.; Pellequer, J.-L.; Annibale, P.; Pessey, O.; Inga, A.; Chillón, I.; Marcia, M. Conserved Pseudoknots in lncRNA MEG3 Are Essential for Stimulation of the p53 Pathway. Mol. Cell 2019, 75, 982–995.e9. [Google Scholar] [CrossRef] [PubMed]
- Sauerwald, A.; Sandin, S.; Cristofari, G.; Scheres, S.H.; Lingner, J.; Rhodes, D. Structure of active dimeric human telomerase. Nat. Struct. Mol. Boil. 2013, 20, 454–460. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Brown, A.F.; Wu, J.; Xue, J.; Bley, C.; Rand, D.P.; Wu, L.; Zhang, R.; Chen, J.J.-L.; Lei, M. Structural basis for protein-RNA recognition in telomerase. Nat. Struct. Mol. Boil. 2014, 21, 507–512. [Google Scholar] [CrossRef]
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zappulla, D.C. Yeast Telomerase RNA Flexibly Scaffolds Protein Subunits: Results and Repercussions. Molecules 2020, 25, 2750. https://doi.org/10.3390/molecules25122750
Zappulla DC. Yeast Telomerase RNA Flexibly Scaffolds Protein Subunits: Results and Repercussions. Molecules. 2020; 25(12):2750. https://doi.org/10.3390/molecules25122750
Chicago/Turabian StyleZappulla, David C. 2020. "Yeast Telomerase RNA Flexibly Scaffolds Protein Subunits: Results and Repercussions" Molecules 25, no. 12: 2750. https://doi.org/10.3390/molecules25122750
APA StyleZappulla, D. C. (2020). Yeast Telomerase RNA Flexibly Scaffolds Protein Subunits: Results and Repercussions. Molecules, 25(12), 2750. https://doi.org/10.3390/molecules25122750





