Plectrabarbene, a New Abietane Diterpene from Plectranthus barbatus Aerial Parts
Abstract
1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. Molecular Docking of Isolated Compounds
2.3. Possible Biosynthetic Pathway of Compound 2
3. Materials and Methods
3.1. General
3.2. Plant Material
3.3. Extraction and Isolation
3.4. Molecular Docking Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Retief, E. Lamiaceae (Labiatae). In Seed Plants of Southern Africa: Strelitzia; Leistner, O.A., Ed.; National Botanical Institute: Pretoria, South Africa, 2000. [Google Scholar]
- Paton, A.J.; Springate, D.; Suddee, S.; Otieno, D.; Grayer, R.J.; Harley, M.M.; Willis, F.; Simmonds, M.S.; Powell, M.P.; Savolainen, V. Phylogeny and evolution of basils and allies (Ocimeae, Labiatae) based on three plastid DNA regions. Mol. Phylogenet. Evol. 2004, 31, 277–299. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Mogib, M.; Albar, H.A.; Batterjee, S.M. Chemistry of the genus Plectranthus. Molecules 2002, 7, 271–301. [Google Scholar] [CrossRef]
- Rice, L.J.; Brits, G.J.; Potgieter, C.J.; Van Staden, J. Plectranthus: A plant for the future? S. Afr. J. Bot. 2011, 77, 947–959. [Google Scholar] [CrossRef]
- Lukhoba, C.W.; Simmonds, M.S.; Paton, A.J. Plectranthus: A review of ethnobotanical uses. J. Ethnopharmacol. 2006, 103, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Alasbahi, R.H.; Melzig, M.F. Plectranthus barbatus: A review of phytochemistry, ethnobotanical uses and pharmacology–part 1. Planta Med. 2010, 76, 653–661. [Google Scholar] [CrossRef] [PubMed]
- Ammon, H.P.; Kemper, F.H. Ayurveda: 3000 years of Indian traditional medicine. Med. Welt. 1982, 33, 148–153. [Google Scholar] [PubMed]
- Yashaswini, S.; Vasundhara, M. Coleus (Plectranthus barbatus)-A multipurpose medicinal herb. Int. Res. J. Pharm. 2011, 2, 47–58. [Google Scholar]
- Schultz, C.; Bossolani, M.P.; Torres, L.M.; Lima-Landman, M.T.R.; Lapa, A.J.; Souccar, C. Inhibition of the gastric H+, K+-ATPase by plectrinone A, a diterpenoid isolated from Plectranthus barbatus Andrews. J. Ethnopharmacol. 2007, 111, 1–7. [Google Scholar] [CrossRef]
- De Souza, N.J.; Dohadwalla, A.N.; Reden, U. Forskolin: A labdane diterpenoid with antihypertensive, positive inotropic, platelet aggregation inhibitory, and adenylate cyclase activating properties. Med. Res. Rev. 1983, 3, 201–219. [Google Scholar] [CrossRef]
- Yao, C.S.; Xu, Y.L. The diterpenoid quinones from Coleus forskohlii. Chin. Chem. Let. 2001, 12, 339–342. [Google Scholar]
- Yao, C.S.; Shen, Y.H.; Xu, Y.L. The chemical constituents of Coleus forskohlii. Nat. Prod. Res. Dev. 2002, 14, 1–6. [Google Scholar]
- Amina, M.; Al-Musayeib, N.M.; Alam, P.; Aleanizy, F.S.; Alqahtni, F.Y.; Al-Said, M.S.; Al-Rashidi, N.S.; Shakeel, F. Cytotoxic evaluation and concurrent analysis of two diterpenes in the chloroform extract of Plectranthus barbatus using a validated HPTLC-UV method. Bull. Chem. Soc. Ethiopia. 2018, 32, 407–419. [Google Scholar] [CrossRef]
- Amina, M.; Al-Musayeib, N.M.; Al-Said, M.S.; Al-Zahrani, R.A.; Ibrahim, S.R.; Mohamed, G.A. Barbaterpene and barbatusterol, new constituents from Plectranthus barbatus growing in Saudi Arabia. Lett. Drug Des. Discov. 2018, 15, 851–856. [Google Scholar] [CrossRef]
- Albarran, G.; Boggess, W.; Rassolov, V.; Schuler, R.H. Absorption spectrum, mass spectrometric properties, and electronic structure of 1, 2-benzoquinone. J. Phy. Chem. A. 2010, 114, 7470–7478. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, B. 1H and 13C NMR spectral assignments of some natural abietane diterpenoids. Magn. Reson. Chem. 2003, 41, 741–746. [Google Scholar] [CrossRef]
- Inatani, R.; Fuwa, H.; Seto, H.; Nakatani, N. Structure of a new antioxidative phenolic diterpene isolated from rosemary (Rosmarinus officinalis L.). Agric. Biol. Chem. 1982, 46, 1661–1666. [Google Scholar] [CrossRef]
- Enzell, C.R.; Wahlberg, I. Mass spectrometric studies of diterpenes. Acta. Chem. Scand. 1969, 23, 871–890. [Google Scholar] [CrossRef][Green Version]
- Bajpai, V.K.; Kang, S.C. Isolation and characterization of biologically active secondary metabolites from Metasequoia glyptostroboides MIKI EX Hu. J. Food Saf. 2011, 31, 276–283. [Google Scholar] [CrossRef]
- Yueh-Hsiung, K.Y.-H.; Chen, C.-H.; Huang, S.-L. New Diterpenes from the Heartwood of Chamaecyparis obtusa var. formosana. J. Nat. Prod. 1998, 61, 829–831. [Google Scholar]
- Khanna, I. Drug discovery in pharmaceutical industry: Productivity challenges and trends. Drug Discov. Today. 2012, 17, 1088–1102. [Google Scholar] [CrossRef]
- Geromichalos, G.D.; Alifieris, C.E.; Geromichalou, E.G.; Trafalis, D.T. Overview on the current status of virtual high-throughput screening and combinatorial chemistry approaches in multi-target anticancer drug discovery; Part I. J. Buon. 2016, 21, 764–779. [Google Scholar] [PubMed]
- Cheng, T.; Li, Q.; Zhou, Z.; Wang, Y.; Bryant, S.H. Structure-based virtual screening for drug discovery: A problem-centric review. The AAPS J. 2012, 14, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Lauro, G.; Romano, A.; Riccio, R.; Bifulco, G. Inverse virtual screening of antitumor targets: Pilot study on a small database of natural bioactive compounds. J. Nat. Prod. 2011, 74, 1401–1407. [Google Scholar] [CrossRef] [PubMed]
- Glaab, E. Building a virtual ligand screening pipeline using free software: A survey. Brief. Bioinform. 2016, 17, 352–366. [Google Scholar] [CrossRef] [PubMed]
- Brückner, K.; Božić, D.; Manzano, D.; Papaefthimiou, D.; Pateraki, I.; Scheler, U.; Ferrer, A.; de Vos, R.C.H.; Kanellis, A.K.; Tissier, A. Characterization of two genes for the biosynthesis of abietane-type diterpenes in rosemary (Rosmarinus officinalis) glandular trichomes. Phytochemistry 2014, 101, 52–64. [Google Scholar]
- Ignea, C.; Athanasakoglou, A.; Ioannou, E.; Georgantea, P.; Trikka, F.A.; Loupassaki, S.; Roussis, V.; Makris, A.M.; Kampranis, S.C. Carnosic acid biosynthesis elucidated by a synthetic biology platform. PNAS 2016, 113, 3681–3686. [Google Scholar] [CrossRef]
- Habtemariam, S. The Therapeutic Potential of Rosemary (Rosmarinus officinalis) Diterpenes for Alzheimer’s Disease. Evid. Based Complement. Alternat. Med. 2016, 2016, 2680409. [Google Scholar] [CrossRef]
- Dong, Y.; Morris-Natschke, S.L.; Lee, K.H. Biosynthesis, total syntheses, and antitumor activity of tanshinones and their analogs as potential therapeutic agents. Nat. Prod. Rep. 2011, 28, 529–542. [Google Scholar] [CrossRef]
- Wu, Y.B.; Ni, Z.Y.; Shi, Q.W.; Dong, M.; Kiyota, H.Y.; Gu, C.; Cong, B. Constituents from Salvia species and their biological activities. Chem. Rev. 2012, 112, 5967–6026. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. Auto Dock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar]
- Abagyan, R.; Totrov, M.; Kuznetsov, D. ICM—A new method for protein modeling and design: Applications to docking and structure prediction from the distorted native conformation. J. Comput. Chem. 1994, 15, 488–506. [Google Scholar] [CrossRef]
- Baxter, J. Local optima avoidance in depot location. J. Oper. Res. Soc. 1981, 32, 815–819. [Google Scholar] [CrossRef]
- Blum, C.; Roli, A.; Sampels, M. Hybrid Metaheuristics: An Emerging Approach to Optimization; Springer: Berlin/Heidelberg, Germany, 2008. [Google Scholar]
Sample Availability: Samples of the compounds 2 are available from the authors. |
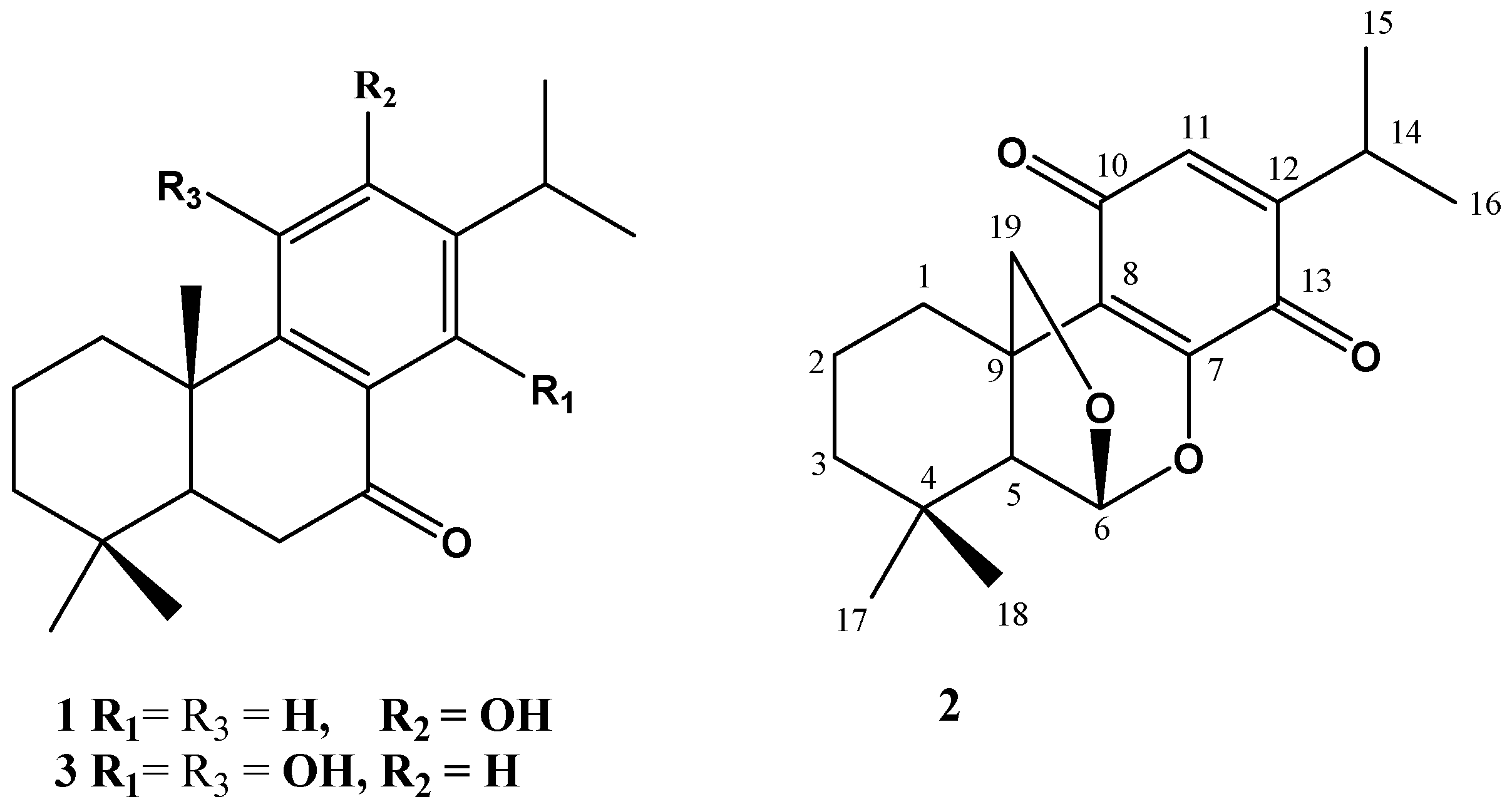
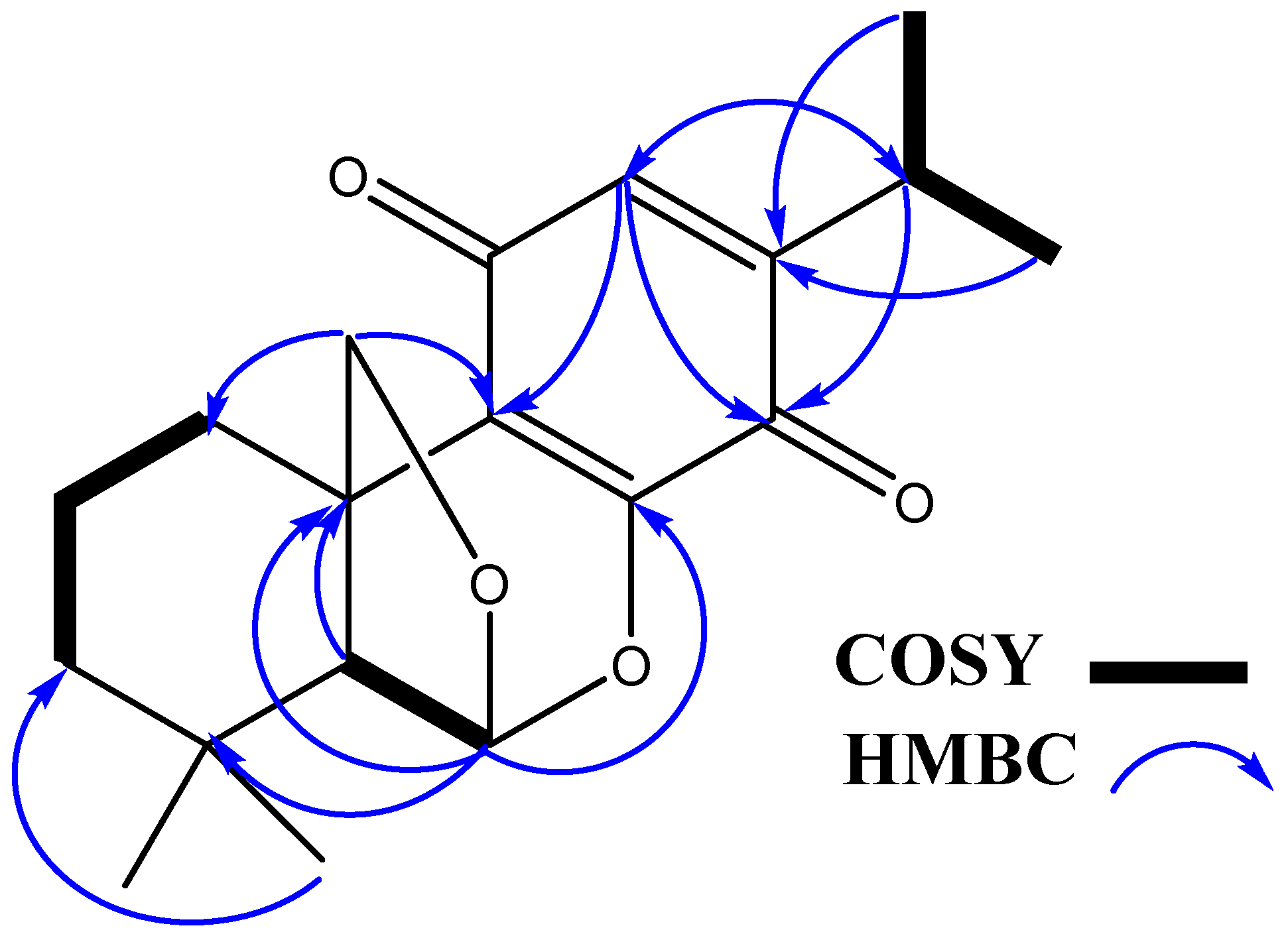



| No. | δH [mult., J (Hz)] | δC (mult.) | HMBC |
|---|---|---|---|
| 1 | 2.74 brd (14.0) 1.62 m | 24.9 CH2 | 2, 3, 5, 9, 19 |
| 2 | 1.66 m | 18.0 CH2 | 9, 19 |
| 3 | 1.50 brd (14.0) 1.33 m | 38.8 CH2 | 1, 2, 4, 5, 18 |
| 4 | - | 29.8 C | - |
| 5 | 2.10 brs | 53.3 CH | 6, 8, 9, 17, 18, 19 |
| 6 | 5.74 brs | 102.6 CH | 4, 5, 7, 8, 9, 19 |
| 7 | - | 128.4 C | - |
| 8 | - | 152.1 C | - |
| 9 | - | 42.5 C | - |
| 10 | - | 186.0 C | - |
| 11 | 6.32 s | 131.0 CH | 7, 8, 9, 12, 13, 14, 15, 16 |
| 12 | - | 150.8 C | - |
| 13 | - | 181.5 C | - |
| 14 | 2.97 m | 26.1 CH | 11, 12, 13, 15, 16 |
| 15 | 1.12 d (7.0) | 20.2 CH3 | 12, 14 |
| 16 | 1.13 d (7.0) | 20.2 CH3 | 12, 14 |
| 17 | 1.11 s | 32.4 CH3 | 4, 5, 18 |
| 18 | 1.02 s | 21.1 CH3 | 3, 4, 5, 17 |
| 19 | 4.27 d (14.0) 4.21 d (14.0) | 80.9 CH2 | 1, 5, 6, 7, 8, 9 |
| Name of Bond and Amino Acid Involved in Interaction | Type of Interaction | Distance (Å) | Binding Energy (kcal mol−1) |
|---|---|---|---|
| Compound (1) | |||
| ASP70: OD2-drug TRP82-drug C18 PHE329-drug C19 PHE329-drug C20 TYR332-drug TYR332-drug HIS438-drug C19 H2O 734:H2-drug O1 H2O 765:H1-drug O1 H2O1006:H1-drug O1 | Pi-Anion interaction Pi-Alkyl interaction Pi-Alkyl interaction Pi-Alkyl interaction Pi-Pi interaction Pi-Alkyl interaction Pi-Alkyl interaction Hydrogen bond Hydrogen bond Hydrogen bond | 4.25 4.42 4.73 4.82 4.80 5.69 5.23 2.41 2.15 2.67 | −6.3 |
| Compound (2) | |||
| ASP70: OD2- drug H16 TRP82- drug C18 TRP82- drug C18 TRP82- drug C19 TRP82- drug C19 GLY116: HA1- drug O4 TYR332- drug C19 TYR332-drug H2O 734: H2-drug O2 H2O 734: H2-drug O1 H2O 1002: H2- drug O4 | Carbon hydrogen bond Pi-Alkyl interaction Pi-Alkyl interaction Pi-Alkyl interaction Pi-Alkyl interaction Carbon hydrogen bond Pi-Alkyl interaction Pi-Alkyl interaction Hydrogen bond Hydrogen bond Hydrogen bond | 1.93 3.71 3.72 4.90 4.80 2.19 4.10 3.95 2.26 2.8 1.94 | −4.7 |
| Compound (3) | |||
| ASP70: OD2- drug TRP82-drug C18 TRP82-drug C18 TRP82-drug TRP82-drug TRP82-drug C20 TRP82-drug C20 TYR332-drug C16 TYR332-drug C1 7TYR332-drug GLY116: HA1-drug O3 THR120: OG1-drugO3 H2O 855:H2-drug O1 H2O 765:H1-drug O3 | Pi-Anion interaction Pi-Alkyl interaction Pi-Alkyl interaction Pi-Alkyl interaction Pi-Alkyl interaction Pi-Alkyl interaction Pi-Alkyl interaction Pi-Alkyl interaction Pi-Alkyl interaction Pi-Pi interaction Carbon hydrogen bond Un favorable bond Hydrogen bond Hydrogen bond | 4.93 3.44 3.58 4.69 4.41 3.29 4.64 3.38 3.94 5.72 2.56 2.95 2.30 2.60 | −2.6 |
| Donepezil (Reference) | |||
| H2O 732: O-drug OAY H2O 1002: O-drug OAY H2O 1006: O-drug HAJ1 H2O 1006: O-drug HAV2 ASP70: OD1-drug: NAK TRP82-drug TRP82-drug TRP82-drug TRP82-drug | Hydrogen bond Hydrogen bond Hydrogen bond Hydrogen bond Ionic bond Pi-Pi Interaction Pi-Pi Interaction Pi-Pi Interaction Pi-Pi Interaction | 2.53 2.6 72.94 2.83 4.43 3.65 4.18 4.36 4.88 | −7.32 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al Musayeib, N.M.; Amina, M.; Al-Hamoud, G.A.; Mohamed, G.A.; Ibrahim, S.R.M.; Shabana, S. Plectrabarbene, a New Abietane Diterpene from Plectranthus barbatus Aerial Parts. Molecules 2020, 25, 2365. https://doi.org/10.3390/molecules25102365
Al Musayeib NM, Amina M, Al-Hamoud GA, Mohamed GA, Ibrahim SRM, Shabana S. Plectrabarbene, a New Abietane Diterpene from Plectranthus barbatus Aerial Parts. Molecules. 2020; 25(10):2365. https://doi.org/10.3390/molecules25102365
Chicago/Turabian StyleAl Musayeib, Nawal M., Musarat Amina, Gadah Abdulaziz Al-Hamoud, Gamal A. Mohamed, Sabrin R.M. Ibrahim, and Samah Shabana. 2020. "Plectrabarbene, a New Abietane Diterpene from Plectranthus barbatus Aerial Parts" Molecules 25, no. 10: 2365. https://doi.org/10.3390/molecules25102365
APA StyleAl Musayeib, N. M., Amina, M., Al-Hamoud, G. A., Mohamed, G. A., Ibrahim, S. R. M., & Shabana, S. (2020). Plectrabarbene, a New Abietane Diterpene from Plectranthus barbatus Aerial Parts. Molecules, 25(10), 2365. https://doi.org/10.3390/molecules25102365






