Cell Growth Stimulation, Cell Cycle Alternation, and Anti-Apoptosis Effects of Bovine Bone Collagen Hydrolysates Derived Peptides on MC3T3-E1 Cells Ex Vivo
Abstract
1. Introduction
2. Results
2.1. Bovine Bone Collagen Hydrolysate (BBCH) Fractionation via Ultrafiltration
2.2. Identification of Peptides in the BBCH-1 Fraction
2.3. Molecular Docking for Screening Peptides that Interact with EGFR
2.4. In Vitro Effect of Synthesized Peptides on MC3T3-E1 Cell Proliferation
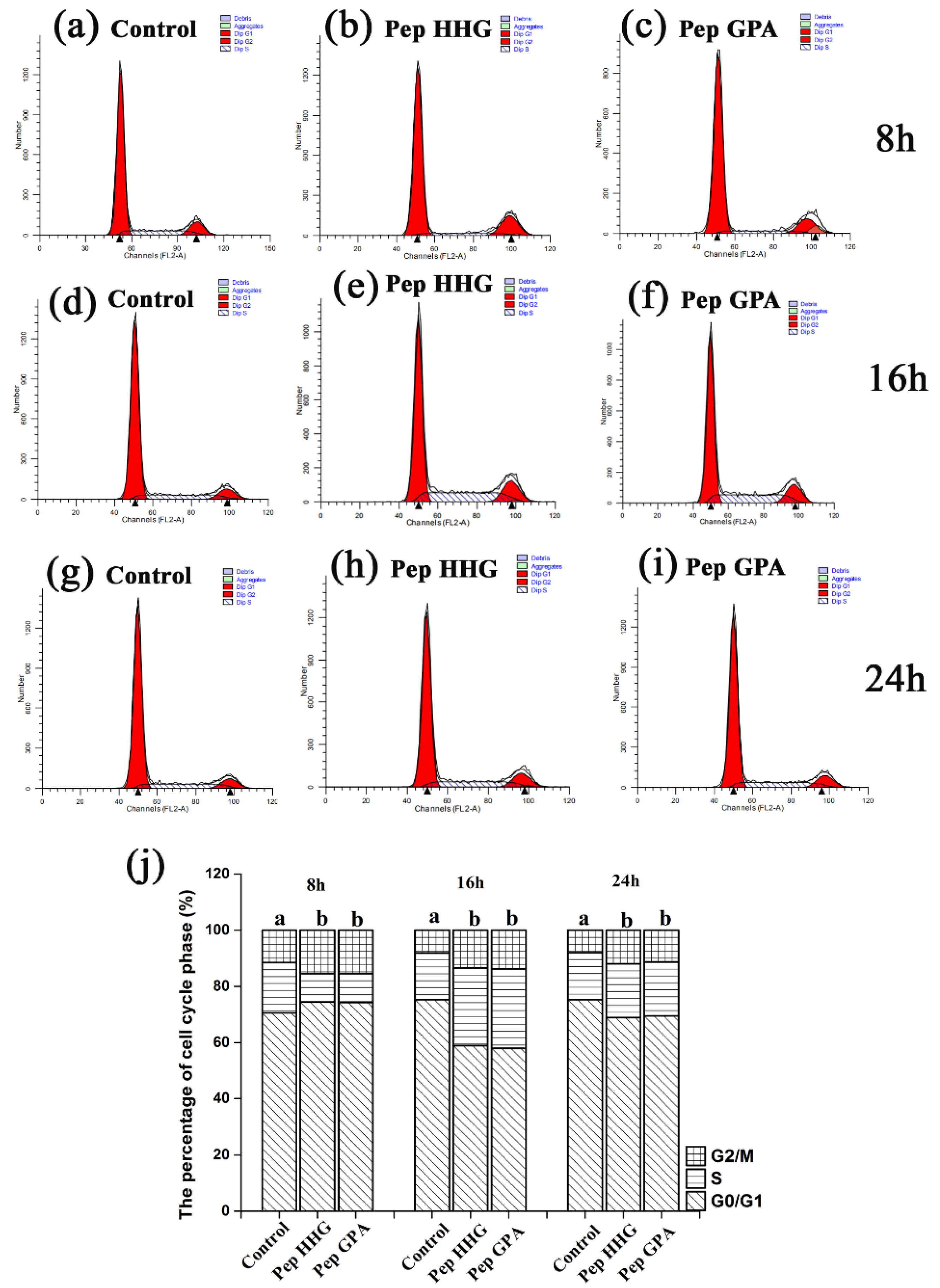
2.5. Effects of Synthesized Peptides on Cell Cycle in MC3T3-E1 Cells
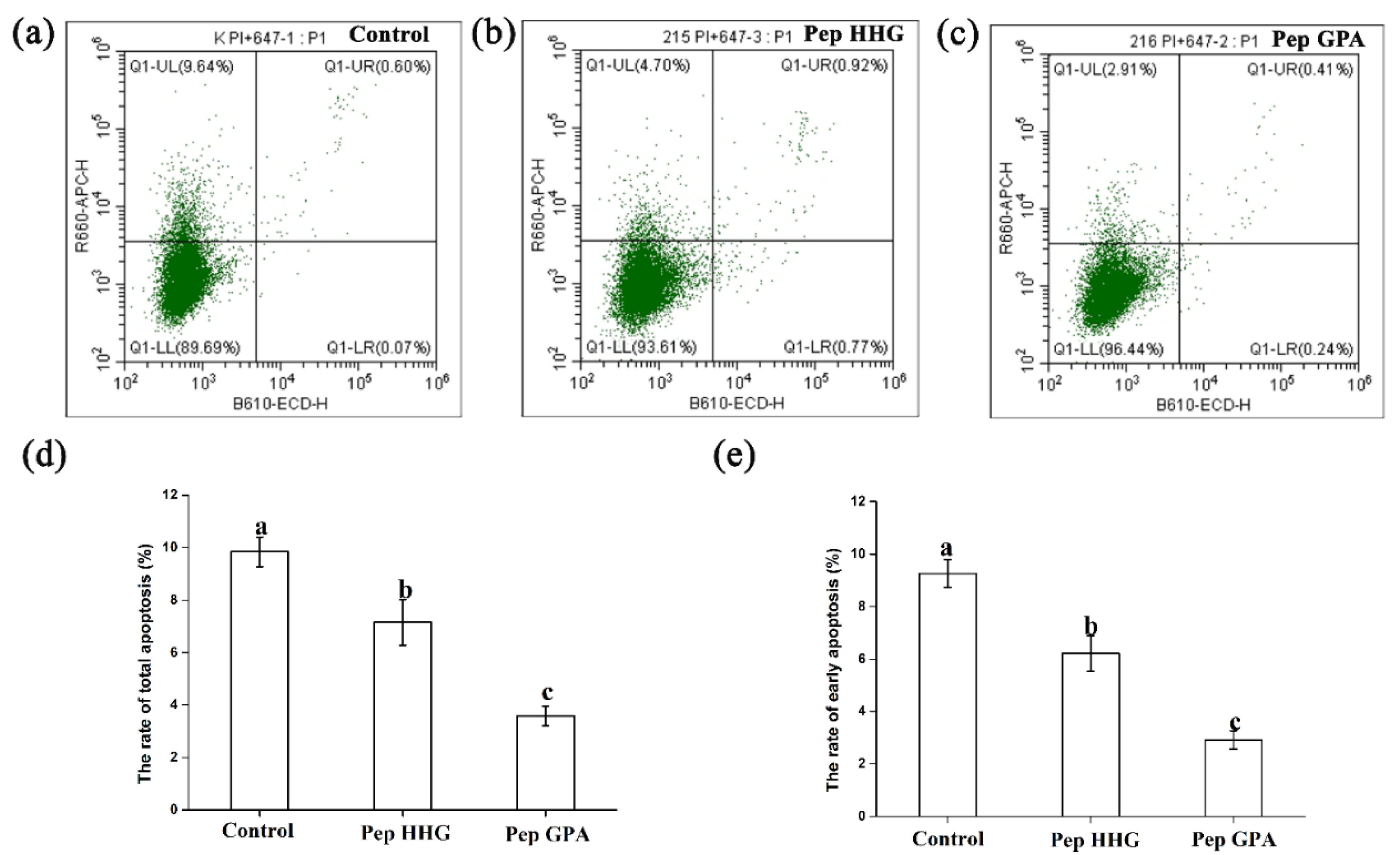
2.6. Anti-Apoptotic Effects of the Synthesized Peptides
2.7. Molecular Docking between Synthesized Peptides and EGFR
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. BBCH Fractionation via Ultrafiltration
4.3. BBCH-1 and BBCH-2 Cell Proliferation Assays
4.4. BBCH-1 Nano-HPLC-MS-MS Analysis
4.5. Molecular Docking
4.6. Peptide Synthesis
4.7. MTT Assay and CCK8 Assay for Synthesized Peptides
4.8. Cell Cycle Analysis
4.9. Apoptosis Analysis
4.10. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Chim, S.M.; Tickner, J.; Chow, S.T.; Kuek, V.; Guo, B.; Zhang, G.; Rosen, V.; Erber, W.; Xu, J. Angiogennic factors in bone local environment. Cytokine Growth Factor Rev. 2013, 24, 297–310. [Google Scholar] [CrossRef] [PubMed]
- Aasetha, J.; Boivinbc, G.; Andersen, O. Osteoporosis and trace elements. J. Trace. Elem. Med. Biol. 2012, 26, 149–152. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.G.; Chiba, F.Y.; de Lima, C.; Mattera, M.S.; Pereira, R.F.; de Cássia Alves Nunes, R.; Tsosura, T.V.S.; Okamoto, R.; Sumida, D.H. Effects of fluoride on insulin signaling and bone metabolism in ovariectomized rats. J. Trace. Elem. Med. Biol. 2017, 39, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Eriksen, E.F.; Díez-Pérez, A.; Boonen, S. Update on long-term treatment with bisphosphoanates for postmenopausal osteoporosis: A systematic review. Bone 2014, 58, 126–135. [Google Scholar] [CrossRef]
- Shi, P.; Liu, M.; Fan, F.J.; Chen, H.; Yu, C.P.; Lu, W.H.; Du, M. Identification and mechanism of peptides with activity promoting osteoblast proliferation from bovine and lactoferrin. Food Biosci. 2018, 22, 19–25. [Google Scholar] [CrossRef]
- Park, S.H.; Song, T.J.; Bae, T.S.; Khang, G.; Choi, B.H.; Park, S.R.; Min, B.H. Comparative analysis of collagens extracted from different animal sources for application of cartilage tissue engineering. Int. J. Precis. Eng. Man. 2012, 13, 2059–2066. [Google Scholar] [CrossRef]
- Tziveleka, L.A.; Loannou, E.; Tsiourvas, D.; Berillis, P.; Foufa, E.; Roussis, V. Collagen from the marine sponges Axinella cannabina and Suberites carnosus: Isolation and morphological, biochemical, and biophysical characterization. Mar. Drugs 2017, 15, 152. [Google Scholar] [CrossRef]
- Moskowitz, R.W. Role of collagen hydrolysate in bone and joint disease. Semin. Arthritis Rheum. 2000, 30, 87–99. [Google Scholar] [CrossRef]
- De Almeida Jackix, E.; Cúneo, F.; Amaya-Farfan, J.; de Assunção, J.V.; Quintaes, K.D. A food supplement of hydrolyzed collagen improves compositional and biodynamic characteristics of vertebrae in ovariectomized rats. J. Med. Food 2010, 13, 1385–1390. [Google Scholar] [CrossRef]
- Liu, J.L.; Zhang, B.; Song, S.J.; Ma, M.; Si, S.Y.; Wang, Y.H.; Xu, B.X.; Feng, K.; Wu, J.G.; Guo, Y.C. Bovine collagen peptides compounds promote the proliferation and differentiation of MC3T3-E1 pre-osteoblasts. PLoS ONE. 2014, 4, e99920. [Google Scholar] [CrossRef]
- Liu, J.L.; Song, S.J.; Si, S.Y.; GAO, J.; Wang, Y.H.; Qin, Y.Y.; Chen, X.N.; Guo, Y.C. Effect of collagen peptide and calcium citrate on bone loss in ovariectomized rats. Chin. J. Osteoporos. Bone Miner. Res. 2015, 8, 334–339. [Google Scholar]
- Wang, Y.L.; Liu, J.L.; Li, B.C.; Wang, Y.H.; Yang, G.A.; Gao, L.M.; Gengen, T.; Wang, F.R.; Guo, Y.C. Effects of collagen peptide combined with Caltrate D on the treatment of postmenopausal osteoporosis. Chin. J. Osteoporos. Bone Miner. Res. 2019, 25, 228–232. [Google Scholar]
- Xie, M.; Dart, D.A.; Guo, T.; Xing, X.F.; Ji, J.F. MicroRNA-1 acts as a tumor suppressor microRNA by inhibiting angiogenesis-related growth factors in human gastric cancer. Gastric Cancer 2017, 21, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.N.; Head, M.S.; Kulkarni, A.; LaLonde, J.M. Validation studies of the site-directed docking program libdock. J. Chem. Inf. Mode 2007, 47, 2159–2171. [Google Scholar] [CrossRef]
- Kang, D.; Pang, X.C.; Lian, W.W.; Xu, L.J.; Wang, J.H.; Jia, H.; Zhang, B.Y.; Du, G.H. Discovery of VEGFR2 inhibitors by integrating naï ve Bayesian classification, molecular docking and drug screening approaches. RSC Adv. 2018, 8, 5286–5297. [Google Scholar] [CrossRef]
- Wadood, A.; Riaz, M.; Jamal, S.B.; Shah, M.; Lodhi, M.A. Molecular docking study of p4-benzoxaborole-substituted ligands as inhibitors of hcv ns3/4a protease. Bioinformation 2013, 9, 309–314. [Google Scholar] [CrossRef]
- Fu, Y.; Young, J.F.; Løkke, M.M.; Lametsch, R.; Aluko, R.E.; Therkildsen, M. Revalorisation of bovine collagen as a potential precursor of angiotensin i-converting enzyme (ace) inhibitory peptides based on in silico and in vitro protein digestions. J. Funct. Foods 2016, 24, 196–206. [Google Scholar] [CrossRef]
- Cai, L.; Wu, X.; Zhang, Y.; Li, X.; Ma, S.; Li, J. Purification and characterization of three antioxidant peptides from protein hydrolysate of grass carp (ctenopharyngodon idella) skin. J. Funct. Foods 2015, 16, 234–242. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, R.; Chen, X.; Zeng, Z.; Ma, H.; Chen, S.W. Dipeptidyl peptidase iv-inhibitory peptides derived from silver carp (hypophthalmichthys molitrix) proteins. J. Agric. Food Chem. 2016, 64, 831–839. [Google Scholar] [CrossRef]
- Lin, L.; Lv, S.; Li, B. Angiotensin-I-converting enzyme (ACE)-inhibitory and antihypertensive properties of squid skin gelatin hydrolysates. Food Chem. 2012, 131, 225–230. [Google Scholar] [CrossRef]
- Alemán, A.; Pérez-Santin, E.; Bordenave-Juchereau, S.; Arnaudin, I.; Gómez-Guillén, M.C.; Montero, P. Squid gelatin hydrolysates with antihypertensive, anticancer and antioxidant activity. Food Res. Int. 2011, 44, 1044–1051. [Google Scholar] [CrossRef]
- Yang, J.I.; Ho, H.Y.; Chu, Y.J.; Chow, C.J. Characteristic and antioxidant activity of retorted gelatin hydrolysates from cobia (Rachycentron canadum) skin. Food Chem. 2008, 110, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Li, B.; Hou, H.; Zhang, H.; Zhao, X. Isolation and identification calcium-chelating peptides from pacific cod skin gelatin and their binding properties with calcium. Food Funct. 2017, 8, 4441–4448. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Kim, M.G.; Leem, K.H. Osteogenic activity of collagen peptide via ERK/MAPK pathway mediated boosting of collagen synthesis and its therapeutic efficacy in osteoporotic bone by back-scattered electron imaging and microarchitecture analysis. Molecules 2013, 18, 15474–15489. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.L.; Si, S.Y.; Qin, Y.Y.; Zhang, B.; Song, S.J.; Guo, Y.C. The effect of different molecular weight collagen peptides on mc3t3-e1 cells differentiation. Bio. Med. Mater. Eng. 2015, 26, S2041–S2047. [Google Scholar] [CrossRef] [PubMed]
- Hou, T.; Liu, Y.; Guo, D.; Li, B.; He, H. Collagen peptides from crucian skin improve bioavailability and structural characterization by HPLC-ESI-MS/MS. J. Agric Food Chem. 2017, 65, 8847–8854. [Google Scholar] [CrossRef] [PubMed]
- León-López, A.; Morales-Peñaloza, A.; Martínez-Juárez, V.M.; Vargas-Torres, A.; Zeugolis, D.I.; Aguirre-Álvarez, G. Hydrolyzed Collagen—Sources and Applications. Molecules 2019, 24, 4031. [Google Scholar] [CrossRef]
- Nomura, Y.; Oohashi, K.; Watanabe, M.; Kasugai, S. Increase in bone mineral density through oral administration of shark gelatin to ovariectomized rats. Nutrition 2005, 21, 1120–1126. [Google Scholar] [CrossRef]
- Ye, M.L.; Jia, W.; Zhang, C.H.; Shen, Q.S.; Zhu, L.Y.; Wang, L.S. Preparation, identification and molecular docking study of novel osteoblast proliferation-promoting peptides from Yak (Bos grunniens) bones. RSC Adv. 2019, 9, 14627–14637. [Google Scholar] [CrossRef]
- Meisel, H.; FitzGerald, R.J. Biofuntional peptides from milk proteins: Mineral binding and cytomodulatory effects. Curr. Pharm. Des. 2003, 9, 1289–1295. [Google Scholar]
- Manolagas, S.C. Birth and death of bone cells: Basic regulatory mechanisms and implications for the pathogenesis and treatment of osteoporosis. Endocr. Rev. 2000, 21, 115–137. [Google Scholar] [PubMed]
- Drissi, H.; Hushka, D.; Aslam, F.; Nguyen, Q.; Buffone, E.; Koff, A.; van Wijnen, A.; Lian, J.B.; Stein, J.L.; Stein, G.S. The cell cycle regulator p27kip1 contributes to growth and differentiation of osteoblasts. Cancer Res. 1999, 59, 3705–3711. [Google Scholar] [PubMed]
- Castro, G.A.; Maria, D.A.; Bouhallab, S.; Sgarbieri, V.C. In vitro impact of a whey protein isolate (WPI) and collagen hydrolysates on B16F10 melanoma cells proliferation. J. Dermatol Sci. 2009, 56, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Zhao, X.H. In vitro responses of hFOB1.19 cells towards chum salmon (Oncorhynchus keta) skin gelatin hydrolysates in cell proliferation, cycle progression and apoptosis. J. Funct. Foods 2013, 5, 279–288. [Google Scholar] [CrossRef]
- Xing, L.; Boyce, B.F. Regulation of apoptosis in osteoclasts and osteoblastic cells. Biochem. Biophys. Res. Commun. 2005, 328, 709–720. [Google Scholar] [CrossRef]
- Burgess, A.W. EGFR family: Structure physiology signalling and therapeutic targets. Growth Factors 2008, 26, 263–274. [Google Scholar] [CrossRef]
- Martinelli, E.; Ciardiello, D.; Martini, T.; Troiani, T.; Cardone, C.; Vitiello, P.P.; Normanno, N.; Rachiglio, A.M.; Maiello, E.; Latiano, T.; et al. Implementing anti-epidermal growth factor receptor (EGFR) therapy in metastatic colorectal cancer: Challenges and future perspectives. Ann. Oncol. 2020, 31, 30–40. [Google Scholar] [CrossRef]
- Schlessinger, J. Receptor tyrosine kinases: Legacy of the first two decades. Cold Spring Harb. Perspect. Biol. 2014, 6, a008912. [Google Scholar] [CrossRef]
- Pripp, A.H.; Ardoe, Y. Modelling relationship between angiotensin-(I)-converting enzyme inhibition and the bitter taste of peptides. Food Chem. 2007, 102, 880–888. [Google Scholar] [CrossRef]
- Ogiso, H.; Ishitani, R.; Nureki, O.; Fukai, S.; Yamanaka, M.; Kim, J.H.; Saito, K.; Sakamoto, A.; Inoue, M.; Shirouzu, M.; et al. Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell 2002, 110, 775–787. [Google Scholar] [CrossRef]
Sample Availability: Bovine bone collagen hydrolysates were available from the authors. |



| Rank | Modification (s) | Peptide Sequence | Cdocker_Energy | Calculated Mass | Length (Amino Acids) | Parent Protein |
|---|---|---|---|---|---|---|
| 1 | Deamidation | HHGDQGAPGAVGPAGPRGPAGPSGPAGKDGR | 270.659 | 2785.3391 | 31 | Collagen alpha-2(I) chain |
| 2 | Deamidation | HHGDQGAPGAVGPAGPRGPAGPSGPAGK | 226.104 | 2457.1897 | 28 | Collagen alpha-2(I) chain |
| 3 | Deamidation | GPAGANGDRGEAGPAGPAGPAGPR | 219.025 | 2056.9673 | 24 | Collagen alpha-2(I) chain |
| 4 | Carbamylation | GAAGLPGPKGDRGDAGPK | 177.177 | 1662.8438 | 18 | Collagen alpha-1(I) chain |
| 5 | Deamidation | ANGDRGEAGPAGPAGPAGPR | 162.153 | 1774.8346 | 20 | Collagen alpha-2(I) chain |
| 6 | None | APGAVGPAGPRGPAGPSGPAGK | 142.022 | 1824.9594 | 22 | Collagen alpha-2(I) chain |
| 7 | Sulfation | GAPGAVGPAGPRGPAGPSGPAGK | 133.665 | 1961.9376 | 23 | Collagen alpha-2(I) chain |
| 8 | Acetylation | GARGEPGPAGLPGPPGER | 126.109 | 1712.8594 | 18 | Collagen alpha-1(I) chain |
| 9 | Sulfation | GPRGPAGPSGPAGKDGR | 125.316 | 1612.7375 | 17 | Collagen alpha-2(I) chain |
| 10 | None | KGDIGPAGLPGPR | 112.306 | 1233.6829 | 13 | Collagen alpha-1(X) chain |
| 11 | Sulphone | STGISVPGPMGPSGPR | 107.425 | 1527.7351 | 16 | Collagen alpha-1(I) chain |
| 12 | Hydroxylation | ARGPSGPQGPSGPPGPK | 106.07 | 1558.7852 | 17 | Collagen alpha-1(I) chain |
| 13 | Oxidation | STGISVPGPMGPSGPR | 105.231 | 1511.7402 | 16 | Collagen alpha-1(I) chain |
| 14 | Sulphone | TGISVPGPMGPSGPR | 104.886 | 1440.7031 | 15 | Collagen alpha-1(I) chain |
| 15 | Dihydroxy | STGISVPGPMGPSGPR | 101.712 | 1527.7351 | 16 | Collagen alpha-1(I) chain |
| 16 | Oxidation | TGISVPGPMGPSGPR | 98.1569 | 1424.7081 | 15 | Collagen alpha-1(I) chain |
| 17 | Dihydroxy | GFPGLPGPSGEPGK | 94.1413 | 1327.6407 | 14 | Collagen alpha-1(I) chain |
| 18 | Hydroxylation | ARGPSGPQGPSGPPGPK | 92.3435 | 1558.7852 | 17 | Collagen alpha-1(I) chain |
| 19 | None | GDIGPAGLPGPR | 90.1347 | 1105.5879 | 12 | Collagen alpha-1(X) chain |
| 20 | Sulphone | GISVPGPMGPSGPR | 87.741 | 1339.6554 | 14 | Collagen alpha-1(I) chain |
| 21 | Hydroxylation | RGPSGPQGPSGPPGPK | 87.5512 | 1487.748 | 16 | Collagen alpha-1(I) chain |
| 22 | Oxidation | GISVPGPMGPSGPR | 87.3568 | 1323.6605 | 14 | Collagen alpha-1(I) chain |
| 23 | Hydroxylation | GLTGPIGPPGPAG | 84.569 | 1105.5768 | 13 | Collagen alpha-1(I) chain |
| 24 | Hydroxylation | GLTGPIGPPGPAGA | 81.6947 | 1176.6139 | 14 | Collagen alpha-1(I) chain |
| 25 | Hydroxylation | GPSGPQGPSGPPGPK | 78.866 | 1331.647 | 15 | Collagen alpha-1(I) chain |
| 26 | None | AGPAGPAGPAGPR | 77.6665 | 1074.557 | 13 | Collagen alpha-2(I) chain |
| 27 | Dihydroxy | GISVPGPMGPSGPR | 71.7777 | 1339.6554 | 14 | Collagen alpha-1(I) chain |
| 28 | Oxidation | VPGPMGPSGPR | 63.247 | 1066.5229 | 11 | Collagen alpha-1(I) chain |
| 29 | Oxidation | PGPMGPSGPR | 46.9511 | 967.4545 | 10 | Collagen alpha-1(I) chain |
| Libdock | MOEdock | CDOCKER | ||||||
|---|---|---|---|---|---|---|---|---|
| Pose | Reference | RMSD (Å) | Pose | Reference | RMSD (Å) | Pose | Reference | RMSD (Å) |
| 1IVO 1 | 1IVO 4 | 15.0634 | 1IVO 1 | 1IVO 6 | 15.2038 | 1IVO 1 | 1IVO 5 | 12.9719 |
| 1IVO 2 | 1IVO 4 | 18.8533 | 1IVO 2 | 1IVO 6 | 15.3858 | 1IVO 2 | 1IVO 5 | 14.228 |
| 1IVO 3 | 1IVO 4 | 17.3093 | 1IVO 3 | 1IVO 6 | 25.5119 | 1IVO 3 | 1IVO 5 | 21.2551 |
| 1IVO 4 | 1IVO 4 | 0 | 1IVO 4 | 1IVO 6 | 14.8449 | 1IVO 4 | 1IVO 5 | 21.8973 |
| − | − | − | 1IVO 5 | 1IVO 6 | 24.1174 | 1IVO 5 | 1IVO 5 | 0 |
| − | − | − | 1IVO 6 | 1IVO 6 | 0 | − | − | − |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Liu, J.; Guo, Y. Cell Growth Stimulation, Cell Cycle Alternation, and Anti-Apoptosis Effects of Bovine Bone Collagen Hydrolysates Derived Peptides on MC3T3-E1 Cells Ex Vivo. Molecules 2020, 25, 2305. https://doi.org/10.3390/molecules25102305
Wang J, Liu J, Guo Y. Cell Growth Stimulation, Cell Cycle Alternation, and Anti-Apoptosis Effects of Bovine Bone Collagen Hydrolysates Derived Peptides on MC3T3-E1 Cells Ex Vivo. Molecules. 2020; 25(10):2305. https://doi.org/10.3390/molecules25102305
Chicago/Turabian StyleWang, Jianing, Junli Liu, and Yanchuan Guo. 2020. "Cell Growth Stimulation, Cell Cycle Alternation, and Anti-Apoptosis Effects of Bovine Bone Collagen Hydrolysates Derived Peptides on MC3T3-E1 Cells Ex Vivo" Molecules 25, no. 10: 2305. https://doi.org/10.3390/molecules25102305
APA StyleWang, J., Liu, J., & Guo, Y. (2020). Cell Growth Stimulation, Cell Cycle Alternation, and Anti-Apoptosis Effects of Bovine Bone Collagen Hydrolysates Derived Peptides on MC3T3-E1 Cells Ex Vivo. Molecules, 25(10), 2305. https://doi.org/10.3390/molecules25102305





