The Significance of Halogen Bonding in Ligand–Receptor Interactions: The Lesson Learned from Molecular Dynamic Simulations of the D4 Receptor
Abstract
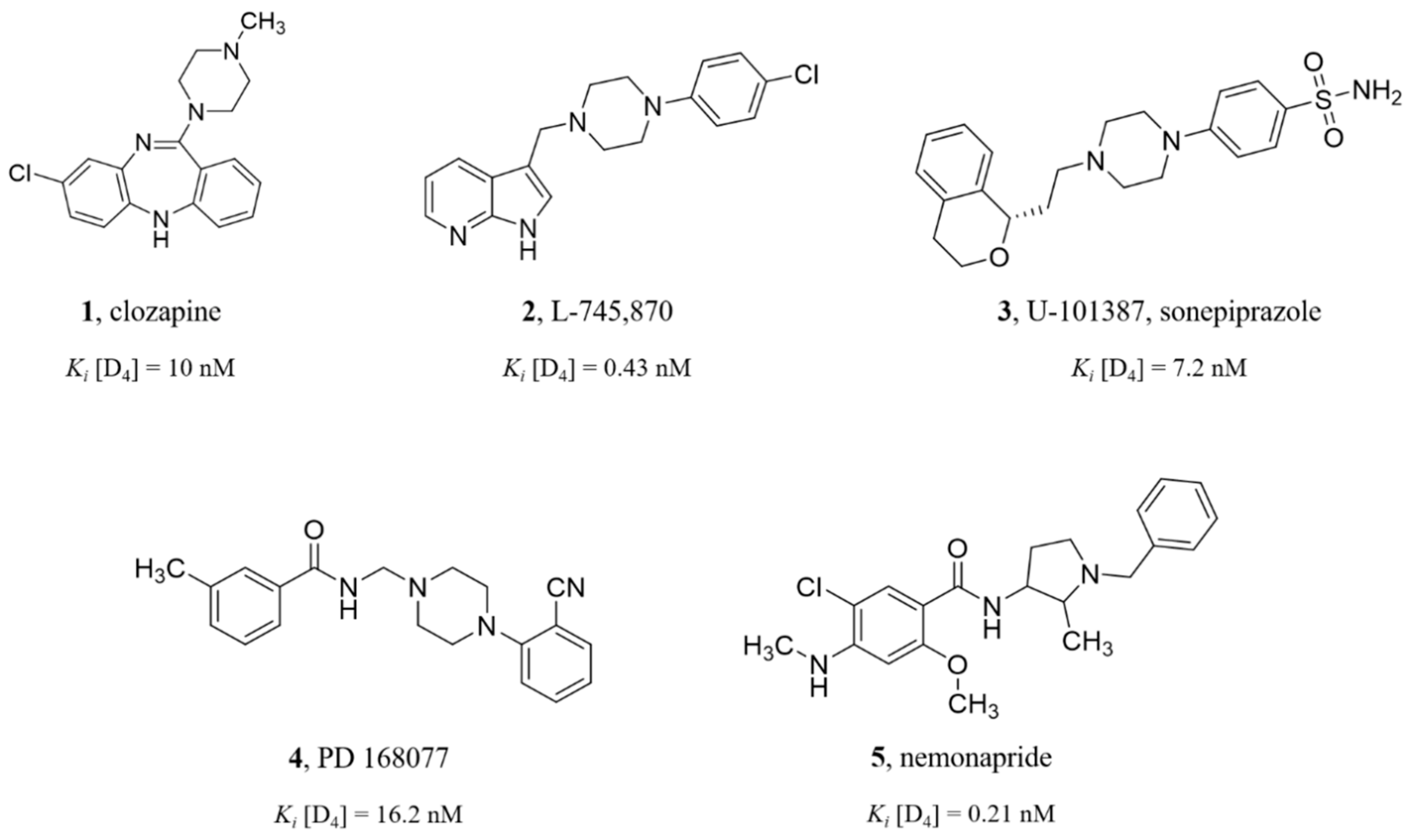
1. Introduction
2. Results and Discussion
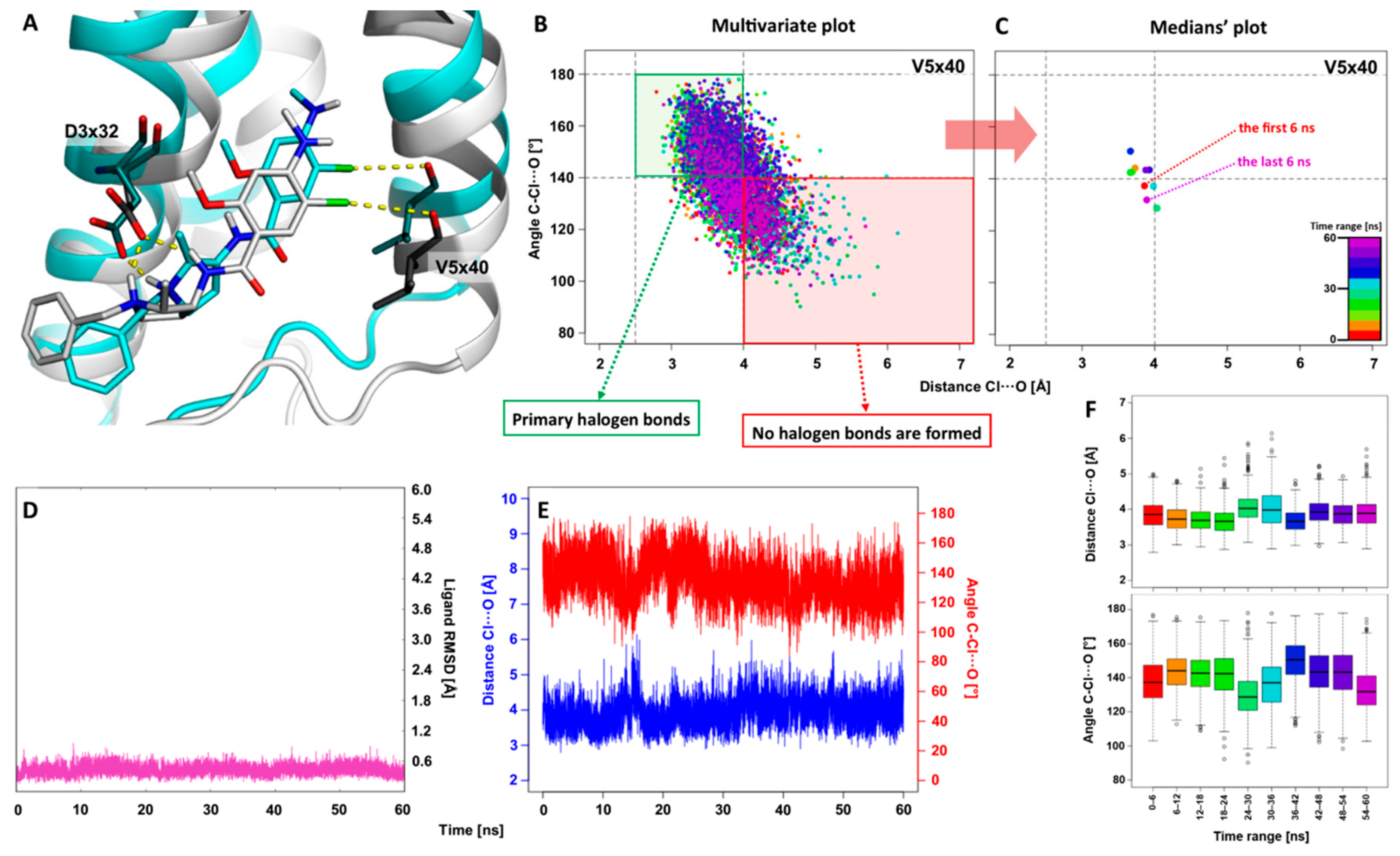
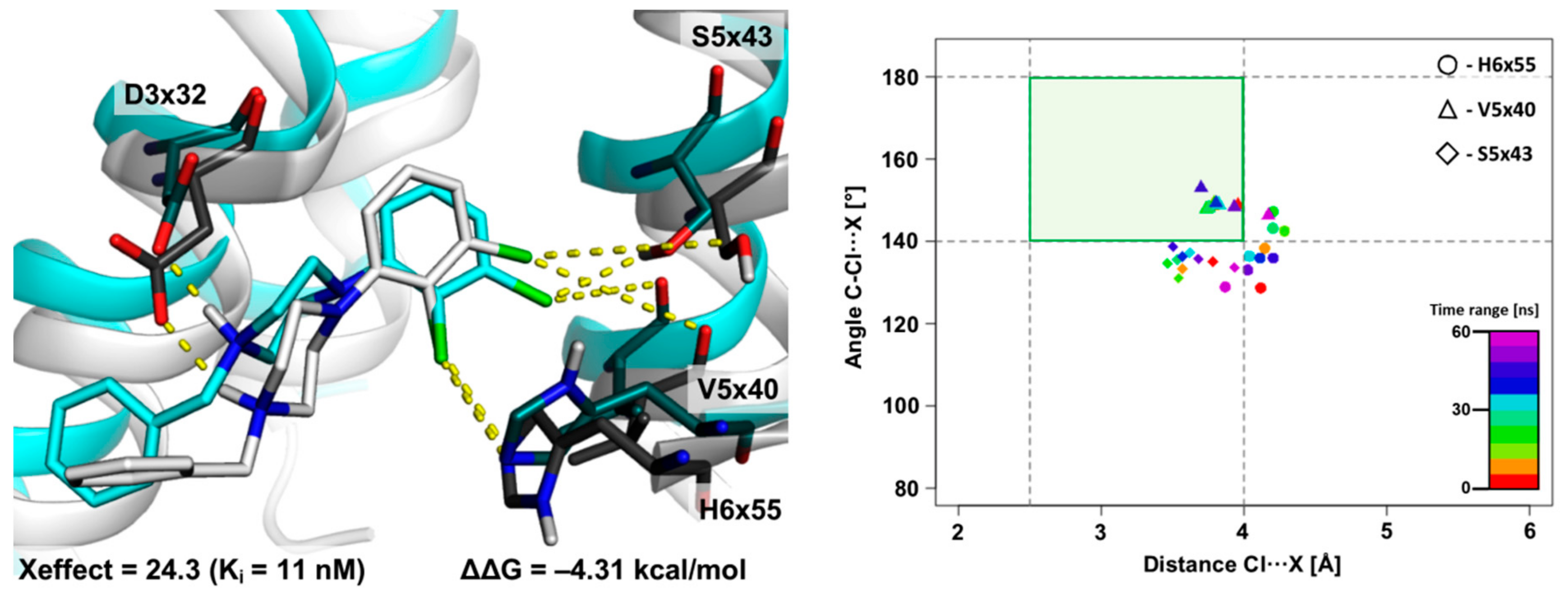
2.1. The 5WIU: MD Simulation of the D4 Receptor in Complex with Agonist Nemonapride
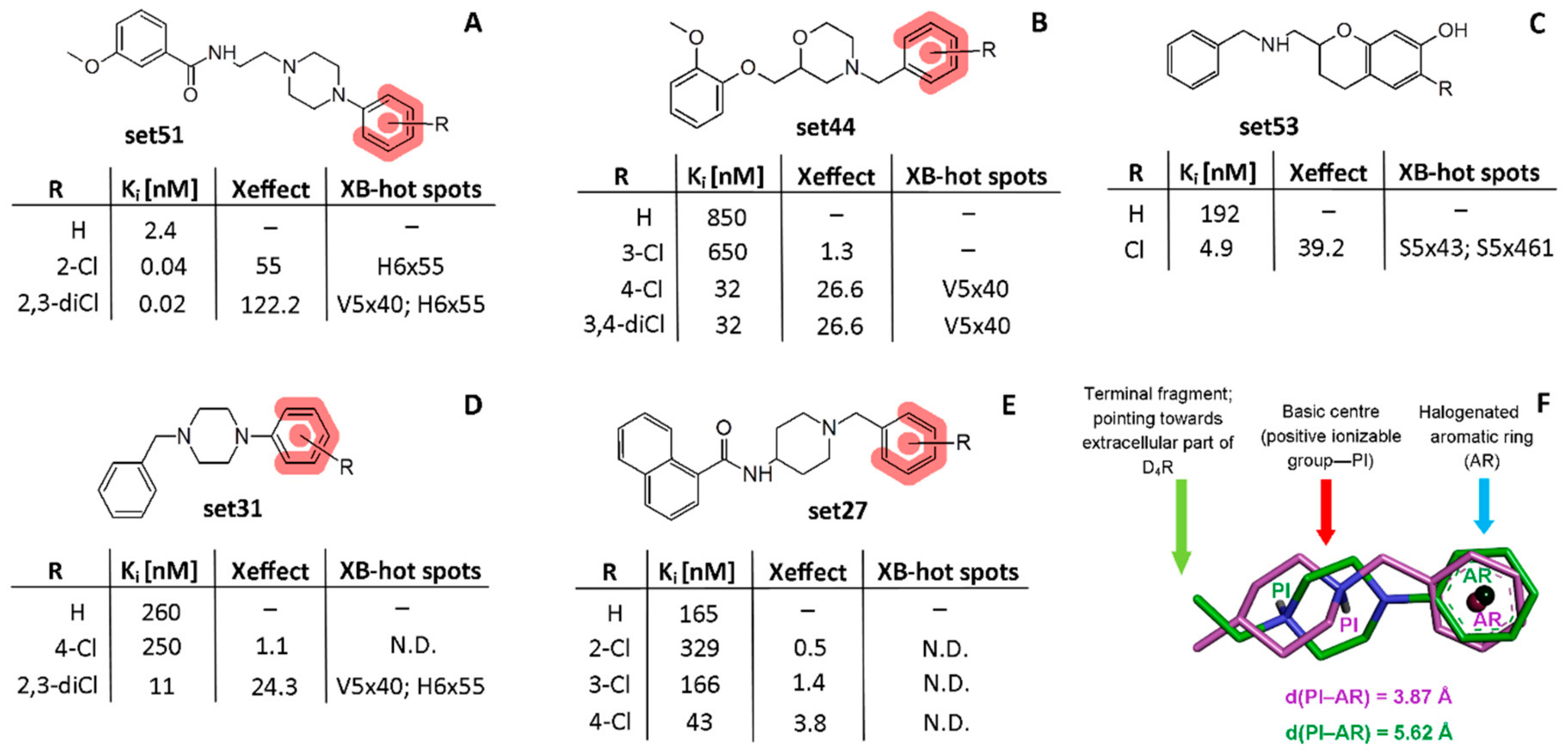
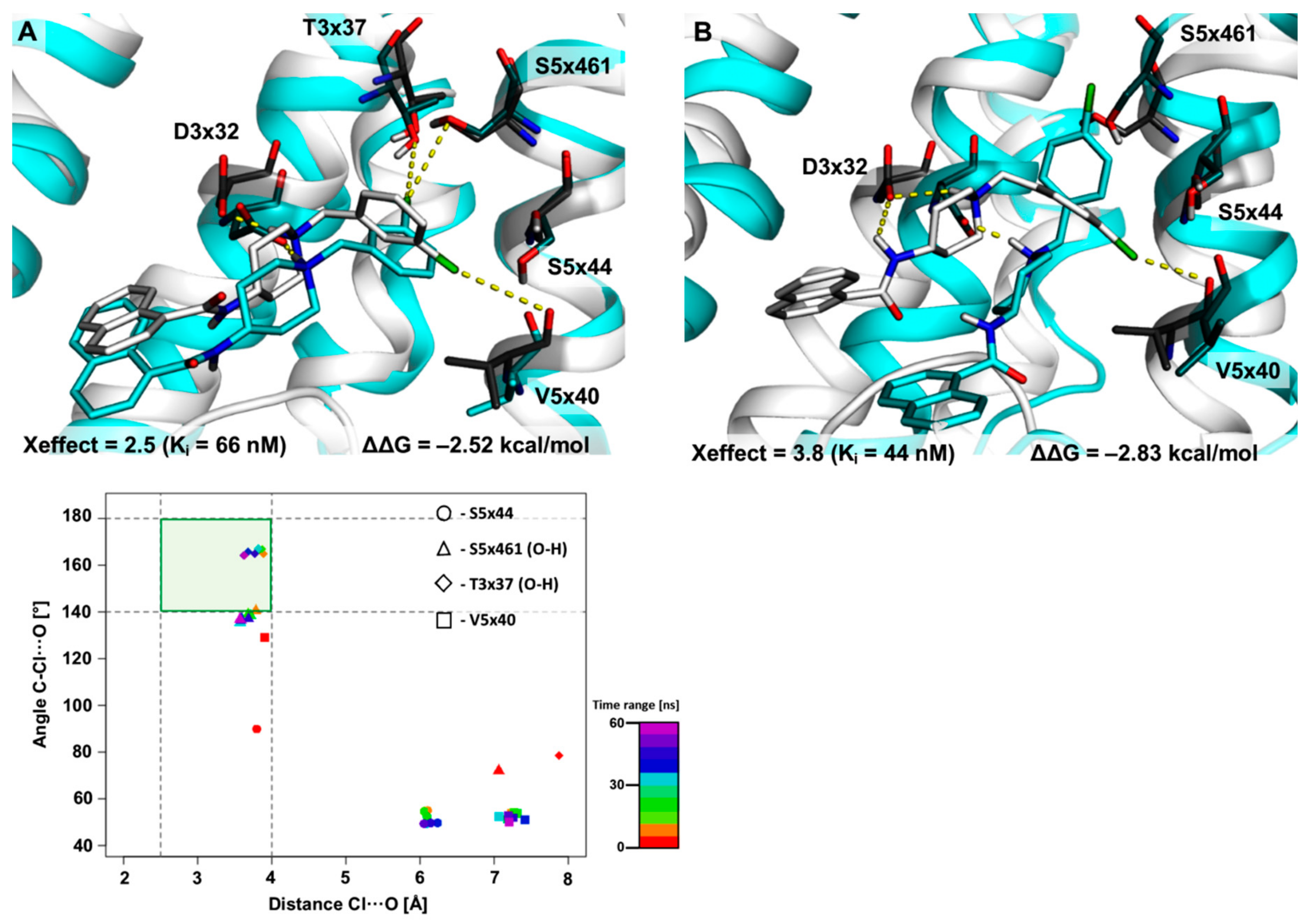
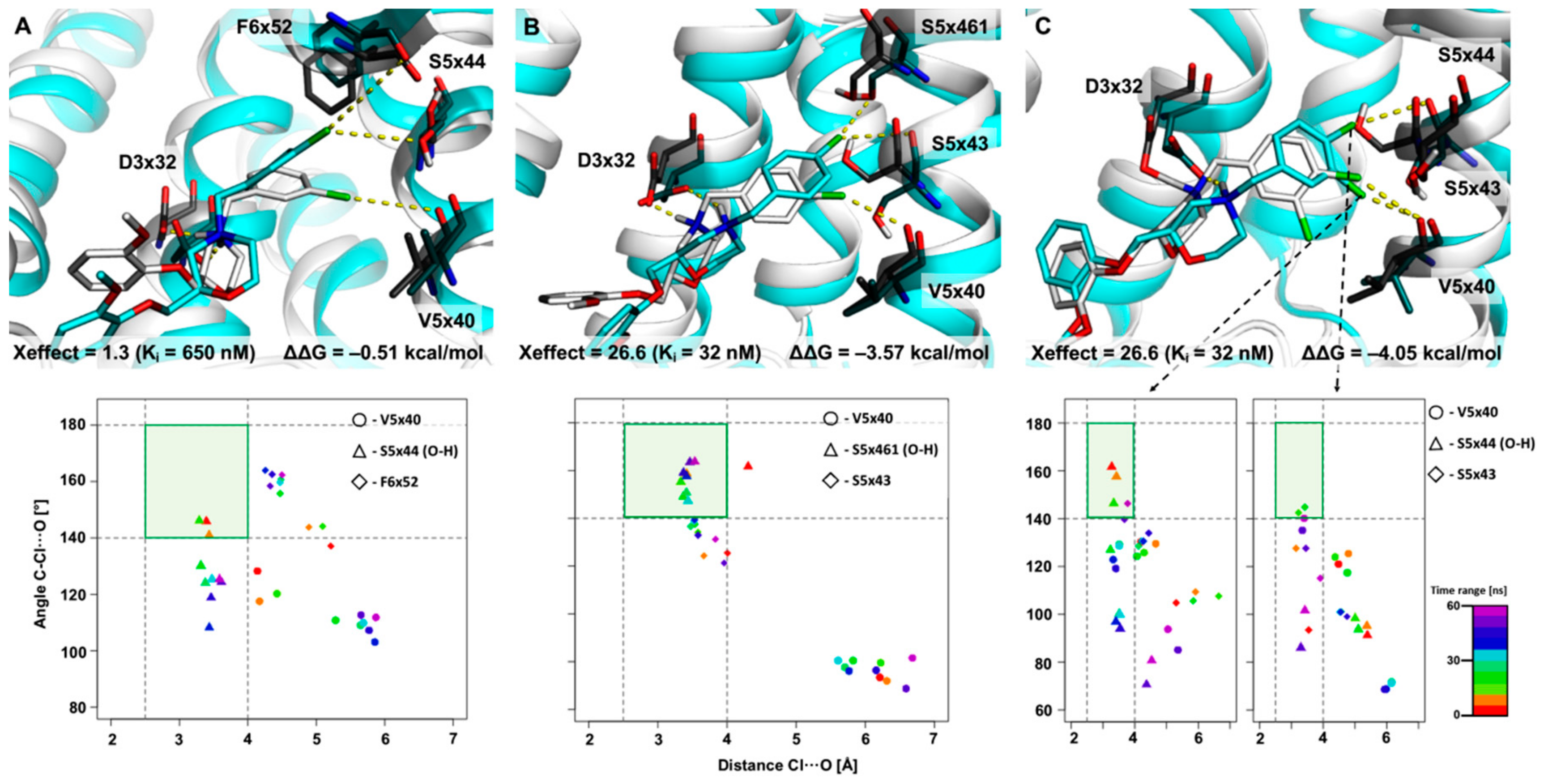
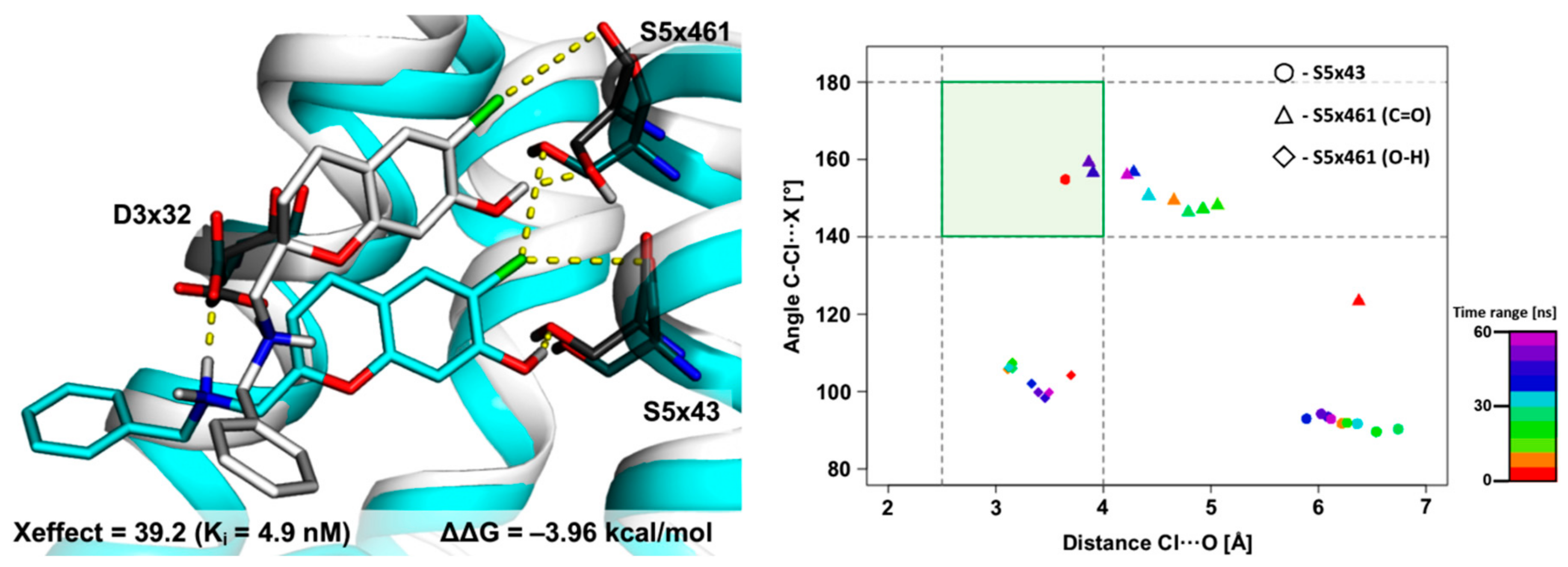
2.2. The Use of XSAR Sets to Explore the Validity of XB Hot Spots
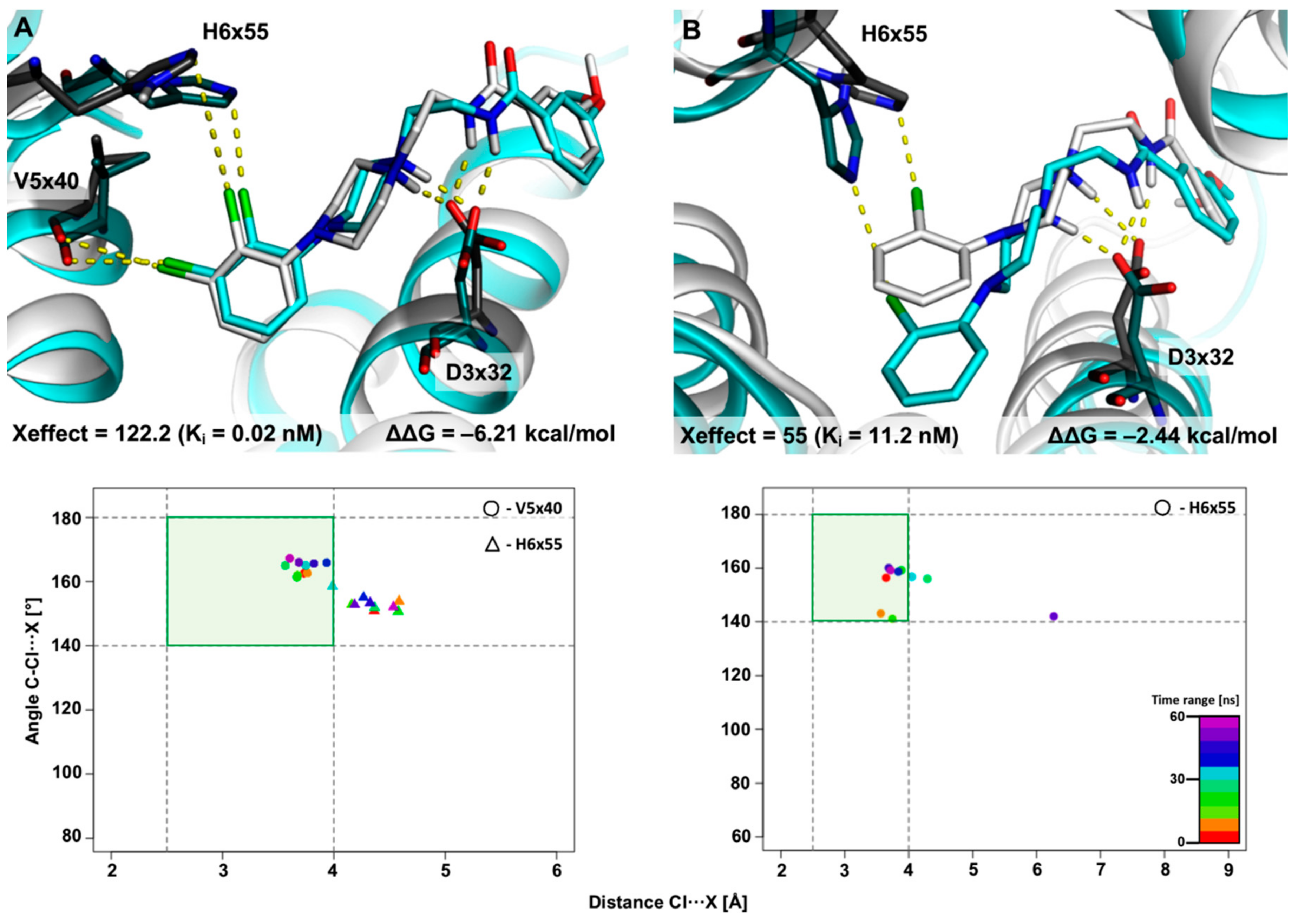
2.2.1. MD Simulations for Sets Showing a Longer PI–AR Distance
2.2.2. MD Simulations for Sets Showing a Shorter PI–AR Distance
2.2.3. MD Simulations for an Uncommon XSAR Set
3. Materials and Methods
3.1. Structure–Activity Relationship Datasets for Halogenated Analogues
3.2. Identification of Halogen Bonding Hot Spots for D4R
3.3. Molecular Dynamics Simulations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gerebtzoff, G.; Li-Blatter, X.; Fischer, H.; Frentzel, A.; Seelig, A. Halogenation of Drugs Enhances Membrane Binding and Permeation. ChemBioChem 2004, 5, 676–684. [Google Scholar] [CrossRef]
- Gentry, C.L.; Egleton, R.D.; Gillespie, T.; Abbruscato, T.J.; Bechowski, H.B.; Hruby, V.J.; Davis, T.P. The Effect of Halogenation on Blood-Brain Barrier Permeability of a Novel Peptide Drug. Peptides 1999, 20, 1229–1238. [Google Scholar] [CrossRef]
- Chatterjee, A.K. Cell-Based Medicinal Chemistry Optimization of High-Throughput Screening (HTS) Hits for Orally Active Antimalarials. Part 1: Challenges in Potency and Absorption, Distribution, Metabolism, Excretion/Pharmacokinetics (ADME/PK). J. Med. Chem. 2013, 56, 7741–7749. [Google Scholar] [CrossRef] [PubMed]
- Hernandes, M.Z.; Cavalcanti, S.M.T.; Moreira, D.R.M.; de Azevedo Junior, W.F.; Leite, A.C.L. Halogen Atoms in the Modern Medicinal Chemistry: Hints for the Drug Design. Curr. Drug Targets 2010, 11, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Politzer, P.; Murray, J.S.; Clark, T. Halogen Bonding: An Electrostatically-Driven Highly Directional Noncovalent Interaction. Phys. Chem. Chem. Phys. 2010, 12, 7748–7757. [Google Scholar] [CrossRef] [PubMed]
- Auffinger, P.; Hays, F.A.; Westhof, E.; Ho, P.S. Halogen Bonds in Biological Molecules. Proc. Natl. Acad. Sci. USA 2004, 101, 16789–16794. [Google Scholar] [CrossRef] [PubMed]
- Costa, P.J.; Nunes, R.; Vila-Viçosa, D. Halogen Bonding in Halocarbon-Protein Complexes and Computational Tools for Rational Drug Design. Expert Opin. Drug Discov. 2019, 14, 805–820. [Google Scholar] [CrossRef] [PubMed]
- Kurczab, R.; Canale, V.; Sataa, G.; Zajdel, P.; Bojarski, A.J. Amino Acid Hot Spots of Halogen Bonding: A Combined Theoretical and Experimental Case Study of the 5-HT7 Receptor. J. Med. Chem. 2018, 61, 8717–8733. [Google Scholar] [CrossRef] [PubMed]
- Meyer, F.; Dubois, P. Halogen Bonding at Work: Recent Applications in Synthetic Chemistry and Materials Science. Cryst. Eng. Comm. 2013, 15, 3058–3071. [Google Scholar] [CrossRef]
- Wilcken, R.; Zimmermann, M.O.; Lange, A.; Joerger, A.C.; Boeckler, F.M. Principles and Applications of Halogen Bonding in Medicinal Chemistry and Chemical Biology. J. Med. Chem. 2013, 56, 1363–1388. [Google Scholar] [CrossRef]
- Mendez, L.; Henriquez, G.; Sirimulla, S.; Narayan, M. Looking Back, looking Forward at Halogen Bonding in Drug Discovery. Molecules 2017, 22, 1397. [Google Scholar] [CrossRef] [PubMed]
- Celis-Barros, C.; Saavedra-Rivas, L.; Salgado, J.C.; Cassels, B.K.; Zapata-Torres, G. Molecular Dynamics Simulation of Halogen Bonding Mimics Experimental Data for Cathepsin L Inhibition. J. Comput. Aided. Mol. Des. 2015, 29, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Luchi, A.M.; Angelina, E.L.; Andujar, S.A.; Enriz, R.D.; Peruchena, N.M. Halogen Bonding in Biological Context: A Computational Study of D2 Dopamine Receptor. J. Phys. Org. Chem. 2016, 29, 645–655. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, Y.; Li, P.; Huang, X.P.; Qi, X.; Du, Y.; Huang, N. Exploring Halogen Bonds in 5-Hydroxytryptamine 2B Receptor-Ligand Interactions. ACS Med. Chem. Lett. 2018, 9, 1019–1024. [Google Scholar] [CrossRef] [PubMed]
- Kolář, M.H.; Hobza, P. Computer Modeling of Halogen Bonds and Other σ-Hole Interactions. Chem. Rev. 2016, 116, 5155–5187. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.A.A. Molecular Mechanical Study of Halogen Bonding in Drug Discovery. J. Comput. Chem. 2011, 32, 2564–2574. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Schyman, P. Treatment of Halogen Bonding in the OPLS-AA Force Field: Application to Potent Anti-HIV Agents. J. Chem. Theory Comput. 2012, 8, 3895–3901. [Google Scholar] [CrossRef]
- Kolář, M.; Hobza, P.; Bronowska, A.K. Plugging the Explicit σ-Holes in Molecular Docking. Chem. Commun. 2013, 49, 981–983. [Google Scholar] [CrossRef]
- Carter, M.; Rappé, A.K.; Ho, P.S. Scalable Anisotropic Shape and Electrostatic Models for Biological Bromine Halogen Bonds. J. Chem. Theory Comput. 2012, 8, 2461–2473. [Google Scholar] [CrossRef]
- van Tol, H.H.M.; Bunzow, J.R.; Guan, H.-C.; Sunahara, R.K.; Seeman, P.; Niznik, H.B.; Civelli, O. Cloning of the Gene for a Human Dopamine D4 Receptor with High Affinity for the Antipsychotic Clozapine. Nature 1991, 350, 610–614. [Google Scholar] [CrossRef]
- Lowe, J.A.; Seeger, T.F.; Vinick, F.J. Atypical Antipsychotics—Recent Findings and New Perspectives. Med. Res. Rev. 1988, 8, 475–497. [Google Scholar] [CrossRef] [PubMed]
- Casey, D.E. Neuroleptic Drug-Induced Extrapyramidal Syndromes and Tardive Dyskinesia. Schizophr. Res. 1991, 4, 109–120. [Google Scholar] [CrossRef]
- Kramer, M.S.; Last, B.; Getson, A.; Reines, S.A. The Effects of a Selective D4 Dopamine Receptor Antagonist (L-745,870) in Acutely Psychotic Inpatients with Schizophrenia. Arch. Gen. Psychiatry 1997, 54, 567–572. [Google Scholar] [CrossRef] [PubMed]
- Corrigan, M.H.; Gallen, C.C.; Bonura, M.L.; Merchant, K.M. Effectiveness of the Selective D4 Antagonist Sonepiprazole in Schizophrenia: A Placebo-Controlled Trial. Biol. Psychiatry 2004, 55, 445–451. [Google Scholar] [CrossRef] [PubMed]
- Lindsley, C.W.; Hopkins, C.R. The Return of D4 Dopamine Receptor Antagonists in Drug Discovery. J. Med. Chem. 2017, 60, 7233–7243. [Google Scholar] [CrossRef]
- Bergman, J.; Rheingold, C.G. Dopamine D₄ Receptor Antagonists for the Treatment of Cocaine Use Disorders. CNS Neurol. Disord. Drug Targets 2015, 14, 707–715. [Google Scholar] [CrossRef]
- Sebastianutto, I.; Maslava, N.; Hopkins, C.R.; Cenci, M.A. Validation of an Improved Scale for Rating l-DOPA-Induced Dyskinesia in the Mouse and Effects of Specific Dopamine Receptor Antagonists. Neurobiol. Dis. 2016, 96, 156–170. [Google Scholar] [CrossRef]
- Dolma, S.; Selvadurai, H.J.; Lan, X.; Lee, L.; Kushida, M.; Voisin, V.; Whetstone, H.; So, M.; Aviv, T.; Park, N.; et al. Inhibition of Dopamine Receptor D4 Impedes Autophagic Flux, Proliferation, and Survival of Glioblastoma Stem Cells. Cancer Cell 2016, 29, 859–873. [Google Scholar] [CrossRef]
- Aung, M.H.; na Han, P.; Han, M.K.; Obertone, T.S.; Abey, J.; Aseem, F.; Thule, P.M.; Iuvone, P.M.; Pardue, M.T. Dopamine Deficiency Contributes to Early Visual Dysfunction in a Rodent Model of Type 1 Diabetes. J. Neurosci. 2014, 34, 726–736. [Google Scholar] [CrossRef]
- Huang, X.-P.; Betz, R.M.; Wacker, D.; Levit, A.; Che, T.; Shoichet, B.K.; Dror, R.O.; Venkatakrishnan, A.J.; Roth, B.L.; McCorvy, J.D.; et al. D4 Dopamine Receptor High-Resolution Structures Enable the Discovery of Selective Agonists. Science 2017, 358, 381–386. [Google Scholar]
- Tenbrink, R.E.; Bergh, C.L.; Duncan, J.N.; Harris, D.W.; Huff, R.M.; Lahti, R.A.; Lawson, C.F.; Lutzke, B.S.; Martin, I.J.; Rees, S.A.; et al. Selective Dopamine D 4 Antagonist. Communications 1996, 39, 13–15. [Google Scholar]
- Matulenko, M.A.; Hakeem, A.A.; Kolasa, T.; Nakane, M.; Terranova, M.A.; Uchic, M.E.; Miller, L.N.; Chang, R.; Donnelly-Roberts, D.L.; Namovic, M.T.; et al. Synthesis and Functional Activity of (2-Aryl-1-Piperazinyl)-N-(3-Methylphenyl)acetamides: Selective Dopamine D4 Receptor Agonists. Bioorganic Med. Chem. 2004, 12, 3471–3483. [Google Scholar] [CrossRef] [PubMed]
- Ohmori, J.; Maeno, K.; Hidaka, K.; Nakato, K.; Matsumoto, M.; Tada, S. Dopamine D 3 and D 4 Receptor Antagonists: Synthesis and Structure-Activity Relationships of (S)-(+)-N-(1-Benzyl-3-Pyrrolidinyl)-5-chloro-4-[(cyclopropylcarbonyl)amino]-2-Methoxybenzamide (YM-43611) and Related. J. Med. Chem. 1996, 2623, 2764–2772. [Google Scholar] [CrossRef] [PubMed]
- Preikša, J.; Śliwa, P. Application of Fragment Molecular Orbital Method to Investigate Dopamine Receptors. Sci. Technol. Innov. 2019, 6, 24–32. [Google Scholar] [CrossRef]
- Kurczab, R. The Evaluation of QM/MM-Driven Molecular Docking Combined with MM/GBSA Calculations as a Halogen-Bond Scoring Strategy. Acta Crystallogr. Sect. B Struct. Sci. Cryst. Eng. Mater. 2017, 73, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Isberg, V.; de Graaf, C.; Bortolato, A.; Cherezov, V.; Katritch, V.; Marshall, F.H.; Mordalski, S.; Pin, J.P.; Stevens, R.C.; Vriend, G.; et al. Generic GPCR Residue Numbers-Aligning Topology Maps While Minding the Gaps. Trends Pharmacol. Sci. 2015, 36, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger Release 2018-4: Desmond Molecular Dynamics System; D.E. Shaw Research: New York, NY, USA, 2018.
- Maestro-Desmond Interoperability Tools; Schrödinger: New York, NY, USA, 2018.
- Lomize, M.A.; Pogozheva, I.D.; Joo, H.; Mosberg, H.I.; Lomize, A.L. OPM Database and PPM Web Server: Resources for Positioning of Proteins in Membranes. Nucleic Acids Res. 2012, 40, D370–D376. [Google Scholar] [CrossRef]
- Cummings, D.F.; Ericksen, S.S.; Goetz, A.; Schetz, J.A. Transmembrane Segment Five Serines of the D4 Dopamine Receptor Uniquely Influence the Interactions of Dopamine, Norepinephrine, and Ro10-4548. J. Pharmacol. Exp. Ther. 2010, 333, 682–695. [Google Scholar] [CrossRef]
- Tschammer, N.; Bollinger, S.; Kenakin, T.; Gmeiner, P. Histidine 6.55 Is a Major Determinant of Ligand-Biased Signaling in Dopamine D2L Receptor. Mol. Pharmacol. 2011, 79, 575–585. [Google Scholar] [CrossRef]
- Floresca, C.Z.; Schetz, J.A. Dopamine Receptor Microdomains Involved in Molecular Recognition and the Regulation of Drug Affinity and Function. J. Recept. Signal Transduct. Res. 2004, 24, 207–239. [Google Scholar] [CrossRef]
Sample Availability: Not available. |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kurczab, R.; Kucwaj-Brysz, K.; Śliwa, P. The Significance of Halogen Bonding in Ligand–Receptor Interactions: The Lesson Learned from Molecular Dynamic Simulations of the D4 Receptor. Molecules 2020, 25, 91. https://doi.org/10.3390/molecules25010091
Kurczab R, Kucwaj-Brysz K, Śliwa P. The Significance of Halogen Bonding in Ligand–Receptor Interactions: The Lesson Learned from Molecular Dynamic Simulations of the D4 Receptor. Molecules. 2020; 25(1):91. https://doi.org/10.3390/molecules25010091
Chicago/Turabian StyleKurczab, Rafał, Katarzyna Kucwaj-Brysz, and Paweł Śliwa. 2020. "The Significance of Halogen Bonding in Ligand–Receptor Interactions: The Lesson Learned from Molecular Dynamic Simulations of the D4 Receptor" Molecules 25, no. 1: 91. https://doi.org/10.3390/molecules25010091
APA StyleKurczab, R., Kucwaj-Brysz, K., & Śliwa, P. (2020). The Significance of Halogen Bonding in Ligand–Receptor Interactions: The Lesson Learned from Molecular Dynamic Simulations of the D4 Receptor. Molecules, 25(1), 91. https://doi.org/10.3390/molecules25010091





