Promising Terpenes as Natural Antagonists of Cancer: An In-Silico Approach
Abstract
1. Introduction
2. Results and Discussion
2.1. Protocol Validation and Receptor Selection
2.2. Pharmacophore Based Virtual Screening and Validation of Drug-Likeness
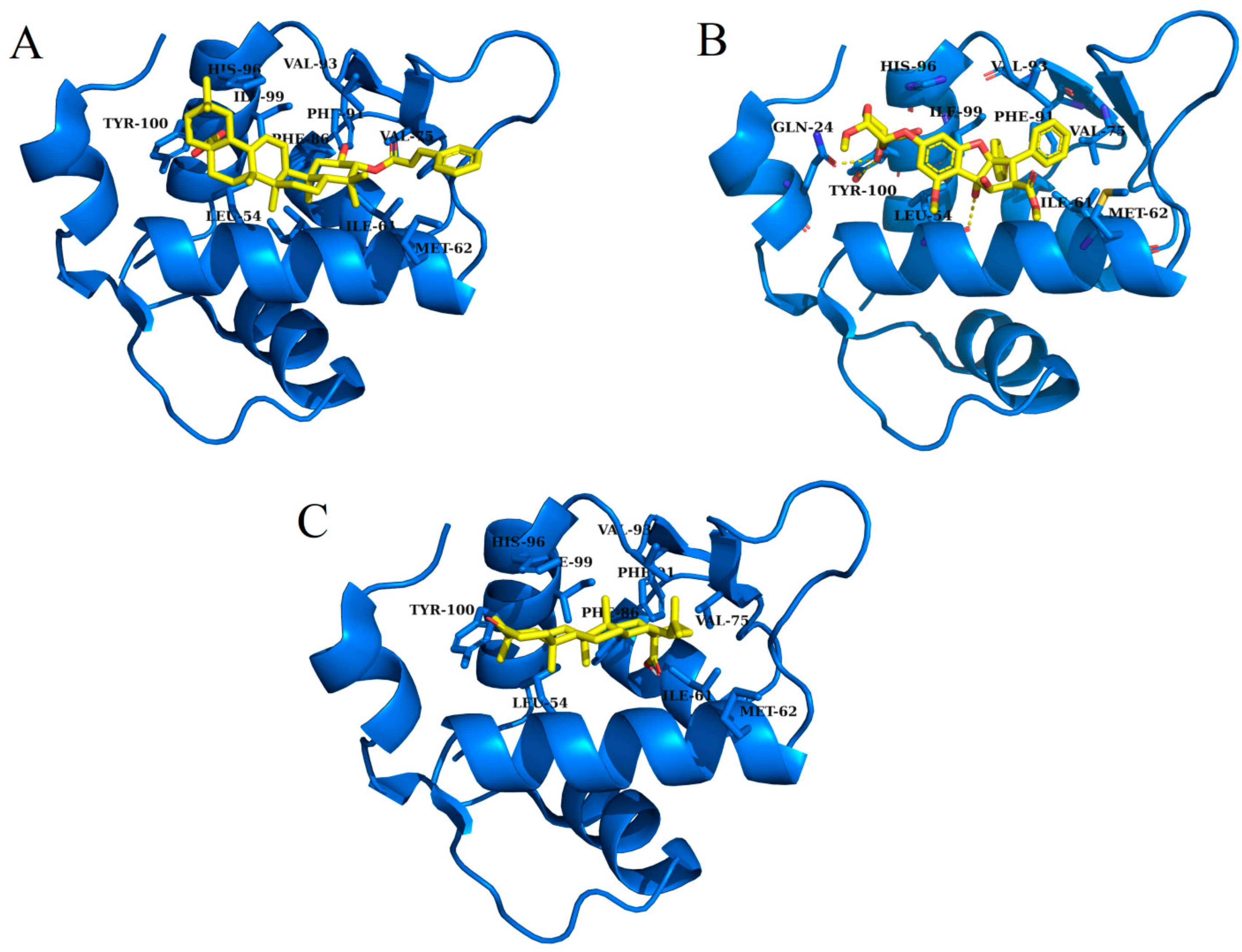
2.3. Molecular Docking
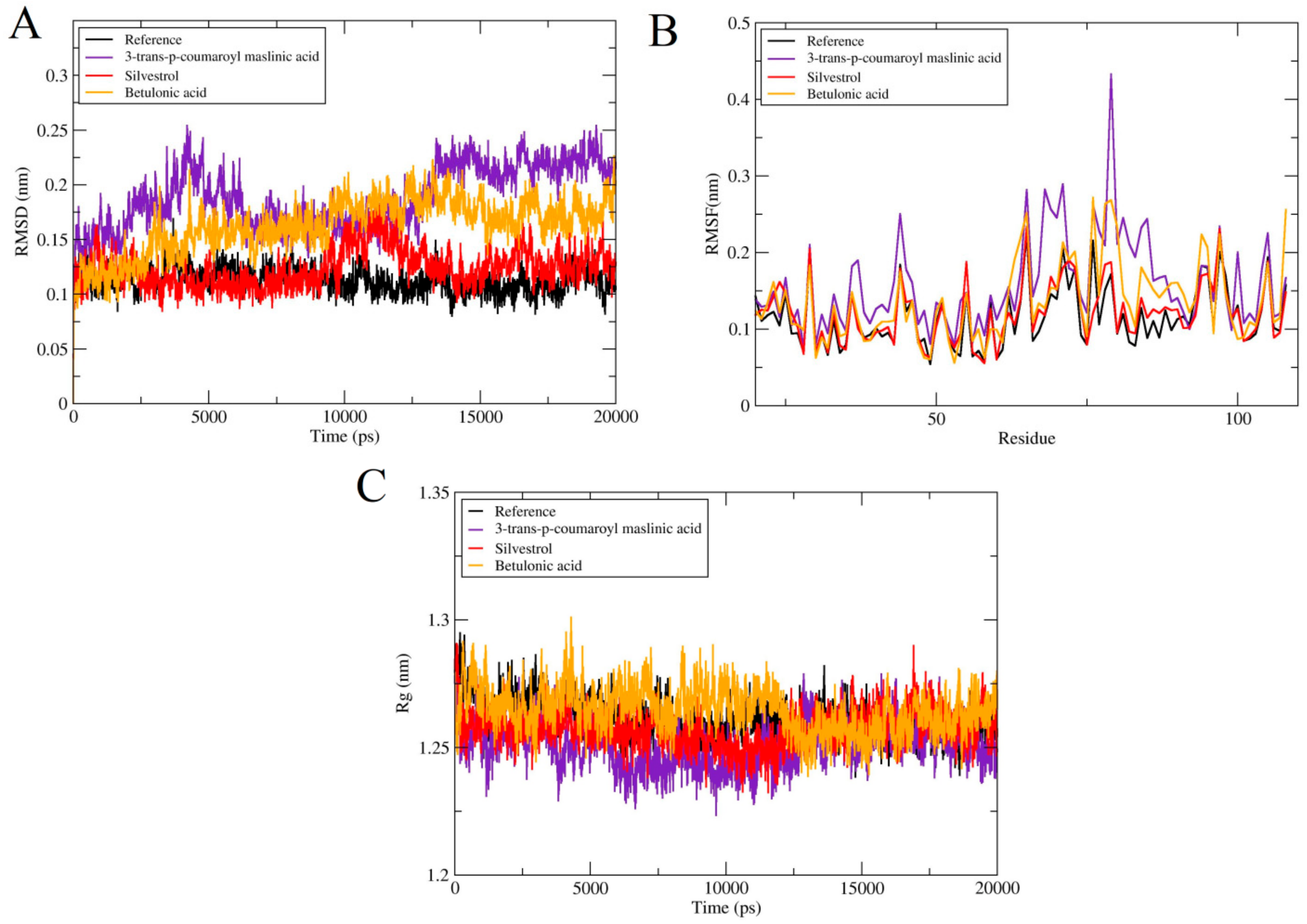
2.4. Molecular Dynamics Simulations
2.4.1. Root Mean Square Deviations (RMSD)
2.4.2. Root Mean Square Fluctuations (RMSF)
2.4.3. Radius of Gyration (RoG)
2.4.4. Potential Binding Energy, Hydrogen Bond Analysis and Solvent Accessible Surface Area (SASA)
2.5. Secondary Structure Analysis
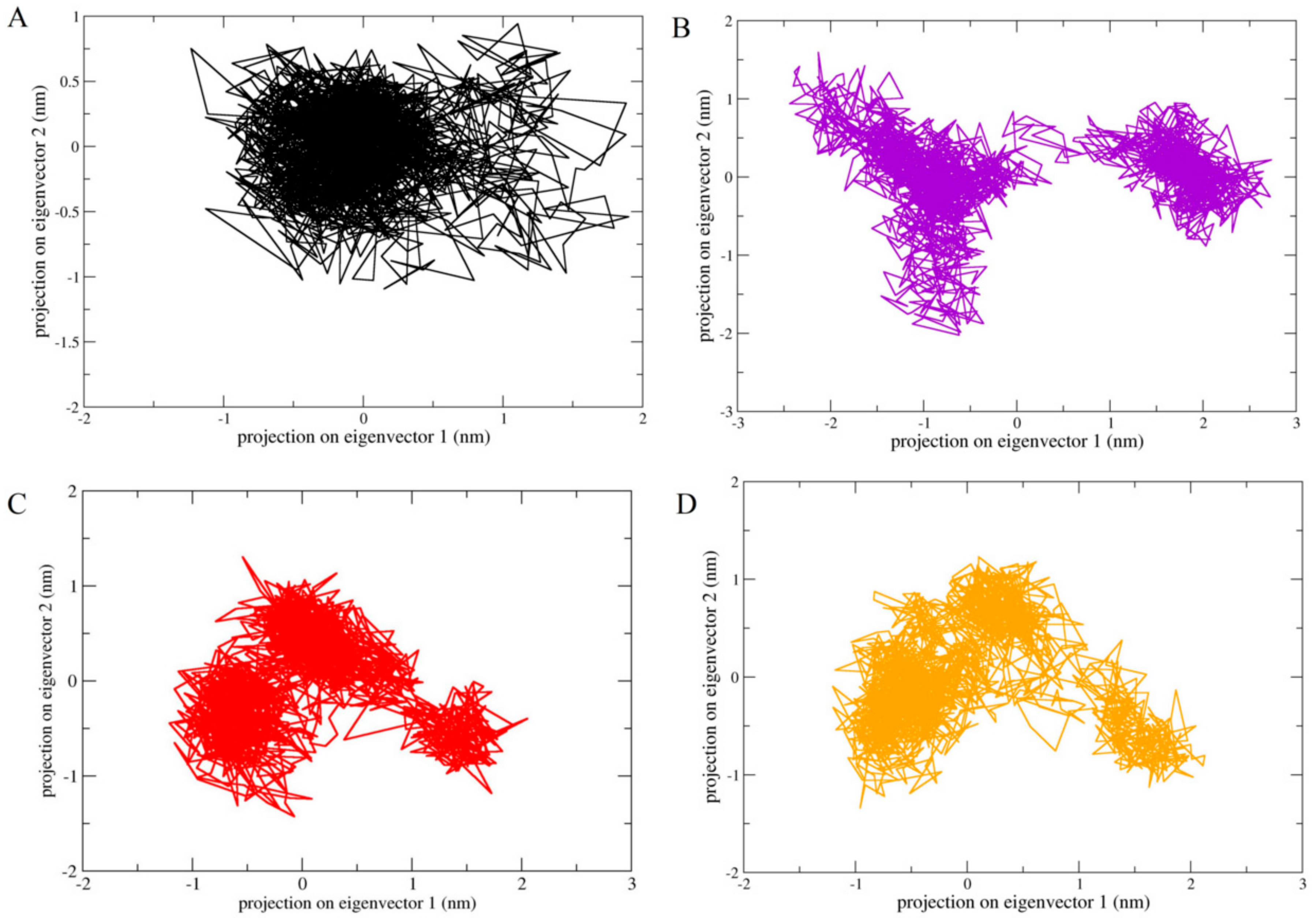
2.6. Principle Component Analysis
3. Materials and Methods
3.1. Selection and Preparation of Receptor
3.2. Ligand Database Preparation
3.3. Pharmacophore Modeling, Virtual Screening, Drug Likeness Testing and Similarity Searching
3.4. Rigid Docking
3.5. Flexible Docking and Clustering
3.6. Molecular Dynamics Simulation
3.7. Principle Component Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef]
- McGlynn, K.A.; Petrick, J.L.; London, W.T. Global epidemiology of hepatocellular carcinoma: An emphasis on demographic and regional variability. Clin. Liver Dis. 2015, 19, 223–238. [Google Scholar] [CrossRef]
- La Vecchia, C.; Malvezzi, M.; Bosetti, C.; Garavello, W.; Bertuccio, P.; Levi, F.; Negri, E. Thyroid cancer mortality and incidence: A global overview. Int. J. Cancer 2015, 136, 2187–2195. [Google Scholar] [CrossRef] [PubMed]
- Ossio, R.; Roldan-Marin, R.; Martinez-Said, H.; Adams, D.J.; Robles-Espinoza, C.D. Melanoma: A global perspective. Nat. Rev. Cancer 2017, 17, 393. [Google Scholar] [CrossRef] [PubMed]
- Riaz, M.; Ashfaq, U.A.; Qasim, M.; Yasmeen, E.; Ul Qamar, M.T.; Anwar, F. Screening of medicinal plant phytochemicals as natural antagonists of p53–MDM2 interaction to reactivate p53 functioning. Anti-Cancer Drug 2017, 28, 1032–1038. [Google Scholar] [CrossRef] [PubMed]
- Gannon, H.S.; Jones, S.N. Using mouse models to explore MDM-p53 signaling in development, cell growth, and tumorigenesis. Genes Cancer 2012, 3, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Wade, M.; Li, Y.-C.; Wahl, G.M. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat. Rev. Cancer 2013, 13, 83. [Google Scholar] [CrossRef]
- Vasilevich, N.I.; Afanasyev, I.I.; Kovalskiy, D.A.; Genis, D.V.; Kochubey, V.S. A re-examination of the MDM2/p53 interaction leads to revised design criteria for novel inhibitors. Chem. Biol. Drug Des. 2014, 84, 585–592. [Google Scholar] [CrossRef]
- Renouf, B.; Hollville, E.; Pujals, A.; Tetaud, C.; Garibal, J.; Wiels, J. Activation of p53 by MDM2 antagonists has differential apoptotic effects on Epstein–Barr virus (EBV)-positive and EBV-negative Burkitt’s lymphoma cells. Leukemia 2009, 23, 1557. [Google Scholar] [CrossRef]
- Wang, J.-L.; Zheng, B.-Y.; Li, X.-D.; Ångström, T.; Lindström, M.S.; Wallin, K.-L. Predictive significance of the alterations of p16INK4A, p14ARF, p53, and proliferating cell nuclear antigen expression in the progression of cervical cancer. Clin. Cancer Res. 2004, 10, 2407–2414. [Google Scholar] [CrossRef]
- Berchuck, A.; Kohler, M.F.; Marks, J.R.; Wiseman, R.; Boyd, J.; Bast Jr, R.C. The p53 tumor suppressor gene frequently is altered in gynecologic cancers. Am. J. Obstet. Gynecol. 1994, 170, 246–252. [Google Scholar] [CrossRef]
- Oren, M. p53: Not just a tumor suppressor. J. Mol. Cell Biol. 2019, 11, 539–543. [Google Scholar] [CrossRef] [PubMed]
- Chapeau, E.A.; Gembarska, A.; Durand, E.Y.; Mandon, E.; Estadieu, C.; Romanet, V.; Wiesmann, M.; Tiedt, R.; Lehar, J.; de Weck, A. Resistance mechanisms to TP53-MDM2 inhibition identified by in vivo piggyBac transposon mutagenesis screen in an Arf−/− mouse model. Proc. Natl. Acad. Sci. USA 2017, 114, 3151–3156. [Google Scholar] [CrossRef] [PubMed]
- Leslie, P.; Zhang, Y. MDM2 oligomers: Antagonizers of the guardian of the genome. Oncogene 2016, 35, 6157. [Google Scholar] [CrossRef] [PubMed]
- Kubbutat, M.H.; Jones, S.N.; Vousden, K.H. Regulation of p53 stability by Mdm2. Nature 1997, 387, 299. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Xu, C.; Ling, X.; Fan, C.; Buckley, B.; Chernov, M.; Ellis, L.; Li, F.; Muñoz, I.; Wang, X. Targeting RING domains of Mdm2–MdmX E3 complex activates apoptotic arm of the p53 pathway in leukemia/lymphoma cells. Cell Death Dis. 2015, 6, e2035. [Google Scholar] [CrossRef]
- Linke, K.; Mace, P.; Smith, C.; Vaux, D.; Silke, J.; Day, C.L. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ. 2008, 15, 841. [Google Scholar] [CrossRef]
- Li, Q.; Lozano, G. Molecular pathways: Targeting Mdm2 and Mdm4 in cancer therapy. Clin. Cancer Res. 2013, 19, 34–41. [Google Scholar] [CrossRef]
- Stommel, J.M.; Wahl, G.M. A new twist in the feedback loop: Stress-activated MDM2 destabilization is required for p53 activation. Cell Cycle 2005, 4, 411–417. [Google Scholar] [CrossRef][Green Version]
- Bond, G.L.; Hu, W.; Levine, A. A single nucleotide polymorphism in the MDM2 gene: From a molecular and cellular explanation to clinical effect. Cancer Res. 2005, 65, 5481–5484. [Google Scholar] [CrossRef]
- Ueda, M.; Yamamoto, M.; Nunobiki, O.; Toji, E.; Sato, N.; Izuma, S.; Okamoto, Y.; Torii, K.; Noda, S. Murine double-minute 2 homolog single nucleotide polymorphism 309 and the risk of gynecologic cancer. Hum. Cell 2009, 22, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Ul Qamar, M.T.; Maryam, A.; Muneer, I.; Xing, F.; Ashfaq, U.A.; Khan, F.A.; Anwar, F.; Geesi, M.H.; Khalid, R.R.; Rauf, S.A. Computational screening of medicinal plant phytochemicals to discover potent pan-serotype inhibitors against dengue virus. Sci. Rep. 2019, 9, 1433. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, B.; Ashfaq, U.A.; ul Qamar, M.T.; Ahmad, M. Anticancer potential of phytochemicals against breast cancer: Molecular docking and simulation approach. Bangladesh J. Pharmacol. 2014, 9, 545–550. [Google Scholar] [CrossRef]
- Qamar, M.; Ashfaq, U.; Tusleem, K.; Mumtaz, A.; Tariq, Q.; Goheer, A.; Ahmed, B. In-silico identification and evaluation of plant flavonoids as dengue NS2B/NS3 protease inhibitors using molecular docking and simulation approach. Pak. J. Pharm. Sci. 2017, 30, 2119–2137. [Google Scholar] [PubMed]
- Ashfaq, U.A.; Mumtaz, A.; ul Qamar, T.; Fatima, T. MAPS Database: Medicinal plant activities, phytochemical and structural database. Bioinformation 2013, 9, 993. [Google Scholar] [CrossRef] [PubMed]
- Mumtaz, A.; Ashfaq, U.A.; ul Qamar, M.T.; Anwar, F.; Gulzar, F.; Ali, M.A.; Saari, N.; Pervez, M.T. MPD3: A useful medicinal plants database for drug designing. Nat. Prod. Res. 2017, 31, 1228–1236. [Google Scholar] [CrossRef]
- Perveen, S.; Al-Taweel, A. Introductory Chapter: Terpenes and Terpenoids. In Terpenes and Terpenoids; IntechOpen: London, UK, 2018. [Google Scholar]
- Paduch, R.; Kandefer-Szerszeń, M.; Trytek, M.; Fiedurek, J. Terpenes: Substances useful in human healthcare. Arch. Immunol. Et Ther. Exp. 2007, 55, 315. [Google Scholar] [CrossRef]
- Muneer, I.; Tusleem, K.; Abdul Rauf, S.; Hussain, H.M.; Siddiqi, A.R. Discovery of selective inhibitors for cyclic AMP response element-binding protein: A combined ligand and structure-based resources pipeline. Anti-Cancer Drugs 2019, 30, 363–373. [Google Scholar] [CrossRef]
- Wadood, A.; Riaz, M.; Uddin, R. In silico identification and evaluation of leads for the simultaneous inhibition of protease and helicase activities of HCV NS3/4A protease using complex based pharmacophore mapping and virtual screening. PLoS ONE 2014, 9, e89109. [Google Scholar] [CrossRef]
- Secchiero, P.; Corallini, F.; Gonelli, A.; Dell’Eva, R.; Vitale, M.; Capitani, S.; Albini, A.; Zauli, G. Antiangiogenic activity of the MDM2 antagonist nutlin-3. Circ. Res. 2007, 100, 61–69. [Google Scholar] [CrossRef]
- Ding, K.; Lu, Y.; Nikolovska-Coleska, Z.; Qiu, S.; Ding, Y.; Gao, W.; Stuckey, J.; Krajewski, K.; Roller, P.P.; Tomita, Y. Structure-based design of potent non-peptide MDM2 inhibitors. J. Am. Chem. Soc. 2005, 127, 10130–10131. [Google Scholar] [CrossRef] [PubMed]
- Popowicz, G.M.; Dömling, A.; Holak, T.A. The structure-based design of MDM2/MDMX–p53 inhibitors gets serious. Angew. Chem. Int. Ed. 2011, 50, 2680–2688. [Google Scholar] [CrossRef] [PubMed]
- Kurogi, Y.; Guner, O.F. Pharmacophore modeling and three-dimensional database searching for drug design using catalyst. Curr. Med. Chem. 2001, 8, 1035–1055. [Google Scholar] [CrossRef] [PubMed]
- Qamar, M.U.; Kiran, S.; Ashfaq, U.A.; Javed, M.R.; Anwar, F.; Ali, M.A. Discovery of novel dengue NS2B/NS3 protease inhibitors using pharmacophore modeling and molecular docking based virtual screening of the zinc database. Int. J. Pharm. 2016, 12, 621–632. [Google Scholar]
- Willett, P. Similarity searching using 2D structural fingerprints. In Chemoinformatics and Computational Chemical Biology; Springer: Totowa, NJ, USA, 2010; pp. 133–158. [Google Scholar]
- Bai, L.; Zhang, H.; Liu, Q.; Zhao, Y.; Cui, X.; Guo, S.; Zhang, L.; Ho, C.-T.; Bai, N. Chemical characterization of the main bioactive constituents from fruits of Ziziphus jujuba. Food Funct. 2016, 7, 2870–2877. [Google Scholar] [CrossRef]
- Hwang, B.Y.; Su, B.-N.; Chai, H.; Mi, Q.; Kardono, L.B.; Afriastini, J.J.; Riswan, S.; Santarsiero, B.D.; Mesecar, A.D.; Wild, R. Silvestrol and episilvestrol, potential anticancer rocaglate derivatives from Aglaia silvestris. J. Org. Chem. 2004, 69, 3350–3358. [Google Scholar] [CrossRef]
- Lee, K.-H.; Morris-Natschke, S.; Qian, K.; Dong, Y.; Yang, X.; Zhou, T.; Belding, E.; Wu, S.-F.; Wada, K.; Akiyama, T. Recent progress of research on herbal products used in traditional Chinese medicine: The herbs belonging to the Divine Husbandman’s Herbal Foundation Canon (Shén Nóng Běn Cǎo Jīng). J. Tradit. Complement. Med. 2012, 2, 6–26. [Google Scholar] [CrossRef]
- Barakat, K.; Gajewski, M.; A Tuszynski, J. DNA repair inhibitors: The next major step to improve cancer therapy. Curr. Top. Med. Chem. 2012, 12, 1376–1390. [Google Scholar] [CrossRef]
- Cencic, R.; Carrier, M.; Galicia-Vázquez, G.; Bordeleau, M.-E.; Sukarieh, R.; Bourdeau, A.; Brem, B.; Teodoro, J.G.; Greger, H.; Tremblay, M.L. Antitumor activity and mechanism of action of the cyclopenta [b] benzofuran, silvestrol. PLoS ONE 2009, 4, e5223. [Google Scholar] [CrossRef]
- Lucas, D.M.; Edwards, R.B.; Lozanski, G.; West, D.A.; Shin, J.D.; Vargo, M.A.; Davis, M.E.; Rozewski, D.M.; Johnson, A.J.; Su, B.-N. The novel plant-derived agent silvestrol has B-cell selective activity in chronic lymphocytic leukemia and acute lymphoblastic leukemia in vitro and in vivo. Blood 2009, 113, 4656–4666. [Google Scholar] [CrossRef]
- Mi, Q.; Kim, S.; Hwang, B.Y.; Su, B.-N.; Chai, H.; Arbieva, Z.H.; Kinghorn, A.D.; Swanson, S.M. Silvestrol regulates G2/M checkpoint genes independent of p53 activity. Anticancer Res. 2006, 26, 3349–3356. [Google Scholar]
- Kim, S.; Hwang, B.Y.; Su, B.-N.; Chai, H.; Mi, Q.; Kinghorn, A.D.; Wild, R.; Swanson, S.M. Silvestrol, a potential anticancer rocaglate derivative from Aglaia foveolata, induces apoptosis in LNCaP cells through the mitochondrial/apoptosome pathway without activation of executioner caspase-3 or-7. Anticancer Res. 2007, 27, 2175–2183. [Google Scholar] [PubMed]
- Alachkar, H.; Santhanam, R.; Harb, J.G.; Lucas, D.M.; Oaks, J.J.; Hickey, C.J.; Pan, L.; Kinghorn, A.D.; Caligiuri, M.A.; Perrotti, D. Silvestrol exhibits significant in vivo and in vitro antileukemic activities and inhibits FLT3 and miR-155 expressions in acute myeloid leukemia. J. Hematol. Oncol. 2013, 6, 21. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.-L.; Pan, L.; Kinghorn, A.D.; Swanson, S.M.; Burdette, J.E. Silvestrol induces early autophagy and apoptosis in human melanoma cells. BMC Cancer 2016, 16, 17. [Google Scholar] [CrossRef] [PubMed]
- Symon, A.; Veselova, N.; Kaplun, A.; Vlasenkova, N.; Fedorova, G.; Lyutik, A.; Gerasimova, G.; Shvets, V. Synthesis and antitumor activity of cyclopropane derivatives of betulinic and betulonic acids. Russ. J. Bioorganic Chem. 2005, 31, 286–291. [Google Scholar] [CrossRef] [PubMed]
- Durdagi, S.; ul Qamar, M.T.; Salmas, R.E.; Tariq, Q.; Anwar, F.; Ashfaq, U.A. Investigating the molecular mechanism of staphylococcal DNA gyrase inhibitors: A combined ligand-based and structure-based resources pipeline. J. Mol. Graph. Model. 2018, 85, 122–129. [Google Scholar] [CrossRef]
- Huey, R.; Morris, G.M. Using AutoDock 4 with AutoDocktools: A tutorial. Scripps Res. Inst. USA 2008, 01, 54–56. [Google Scholar]
- Seeliger, D.; de Groot, B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. -Aided Mol. Des. 2010, 24, 417–422. [Google Scholar] [CrossRef]
- Bolton, E.E.; Wang, Y.; Thiessen, P.A.; Bryant, S.H. PubChem: Integrated platform of small molecules and biological activities. In Annual Reports in Computational Chemistry; Elsevier: Amsterdam, The Netherlands, 2008; Volume 4, pp. 217–241. [Google Scholar]
- Mangal, M.; Sagar, P.; Singh, H.; Raghava, G.P.; Agarwal, S.M. NPACT: Naturally occurring plant-based anti-cancer compound-activity-target database. Nucleic Acids Res. 2012, 41, D1124–D1129. [Google Scholar] [CrossRef]
- Irwin, J.J.; Shoichet, B.K. ZINC—A free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005, 45, 177–182. [Google Scholar] [CrossRef]
- Vilar, S.; Cozza, G.; Moro, S. Medicinal chemistry and the molecular operating environment (MOE): Application of QSAR and molecular docking to drug discovery. Curr. Top. Med. Chem. 2008, 8, 1555–1572. [Google Scholar] [CrossRef] [PubMed]
- Willett, P. Similarity-based approaches to virtual screening. Biochem. Soc. Trans. 2003, 31, 603–606. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A. Lead-and drug-like compounds: The rule-of-five revolution. Drug Discov. Today Technol. 2004, 1, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. Pymol: An open-source molecular graphics tool. Ccp4 Newsl. Protein Crystallogr. 2002, 40, 82–92. [Google Scholar]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Berendsen, H.J.; van der Spoel, D.; van Drunen, R. GROMACS: A message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995, 91, 43–56. [Google Scholar] [CrossRef]
- Van Aalten, D.M.; Bywater, R.; Findlay, J.B.; Hendlich, M.; Hooft, R.W.; Vriend, G. PRODRG, a program for generating molecular topologies and unique molecular descriptors from coordinates of small molecules. J. Comput. -Aided Mol. Des. 1996, 10, 255–262. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N log (N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Berendsen, H.J.; Postma, J.V.; van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.; Fraaije, J.G. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Ichiye, T.; Karplus, M. Collective motions in proteins: A covariance analysis of atomic fluctuations in molecular dynamics and normal mode simulations. Proteins Struct. Funct. Bioinform. 1991, 11, 205–217. [Google Scholar] [CrossRef] [PubMed]
- García, A.E. Large-amplitude nonlinear motions in proteins. Phys. Rev. Lett. 1992, 68, 2696. [Google Scholar] [CrossRef] [PubMed]
- David, C.C.; Jacobs, D.J. Principal component analysis: A method for determining the essential dynamics of proteins. In Protein Dynamics; Springer: Totowa, NJ, USA, 2014; pp. 193–226. [Google Scholar]



| IDs | Terpene | Plant Source | Chemical Structure | Molecular Weight (g/mol) | xLogP | Hydrogen Bond Donor | Hydrogen Bond Acceptor | Chemical Formula | Reference |
|---|---|---|---|---|---|---|---|---|---|
| ZINC ID: ZINC44358756 | 3-trans-p-coumaroyl maslinic acid | Ziziphus jujuba |  | 602.8 | 9.2 | 2 | 5 | C39H54O5 | [37] |
| NPACT ID: NPACT00946 | Silvestrol | Aglaia silvestris |  | 654.7 | 1.6 | 4 | 13 | C34H38O13 | [38] |
| PubChem CID: 122844 | Betulonic acid | Fructus Jujubae |  | 454.7 | 7.9 | 1 | 3 | C30H46O3 | [39] |
| Terpene | Binding Energy | Interacting Residue |
|---|---|---|
| 3-trans-p-coumaroyl maslinic acid | −22.60 | Leu 54, Ile 61, Met 62, Val 75, Phe 86, Phe 91, Val 93, His 96, Ile 99 and Tyr 100 |
| Silvestrol | −20.75 | Gln 24, Leu 54, Ile 61, Met 62, Val 75, Phe 91, Val 93, His 96, Ile 99 and Tyr 100 |
| Betulonic acid | −18.83 | Leu 54, Ile 61, Met 62, Val 75, Phe 86, Phe 91, Val 93, His 96, Ile 99 and Tyr 100 |
| Reference (Nutlin) | −12.67 | Leu 54, Ile 61, Met 62, Val 75, Phe 86, Phe 91, Val 93, His 96, Ile 99 and Tyr 100 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muhseen, Z.T.; Li, G. Promising Terpenes as Natural Antagonists of Cancer: An In-Silico Approach. Molecules 2020, 25, 155. https://doi.org/10.3390/molecules25010155
Muhseen ZT, Li G. Promising Terpenes as Natural Antagonists of Cancer: An In-Silico Approach. Molecules. 2020; 25(1):155. https://doi.org/10.3390/molecules25010155
Chicago/Turabian StyleMuhseen, Ziyad Tariq, and Guanglin Li. 2020. "Promising Terpenes as Natural Antagonists of Cancer: An In-Silico Approach" Molecules 25, no. 1: 155. https://doi.org/10.3390/molecules25010155
APA StyleMuhseen, Z. T., & Li, G. (2020). Promising Terpenes as Natural Antagonists of Cancer: An In-Silico Approach. Molecules, 25(1), 155. https://doi.org/10.3390/molecules25010155




