Structure-Based Virtual Screening and Biological Evaluation of Peptide Inhibitors for Polo-Box Domain
Abstract
:1. Introduction
2. Results and Discussion
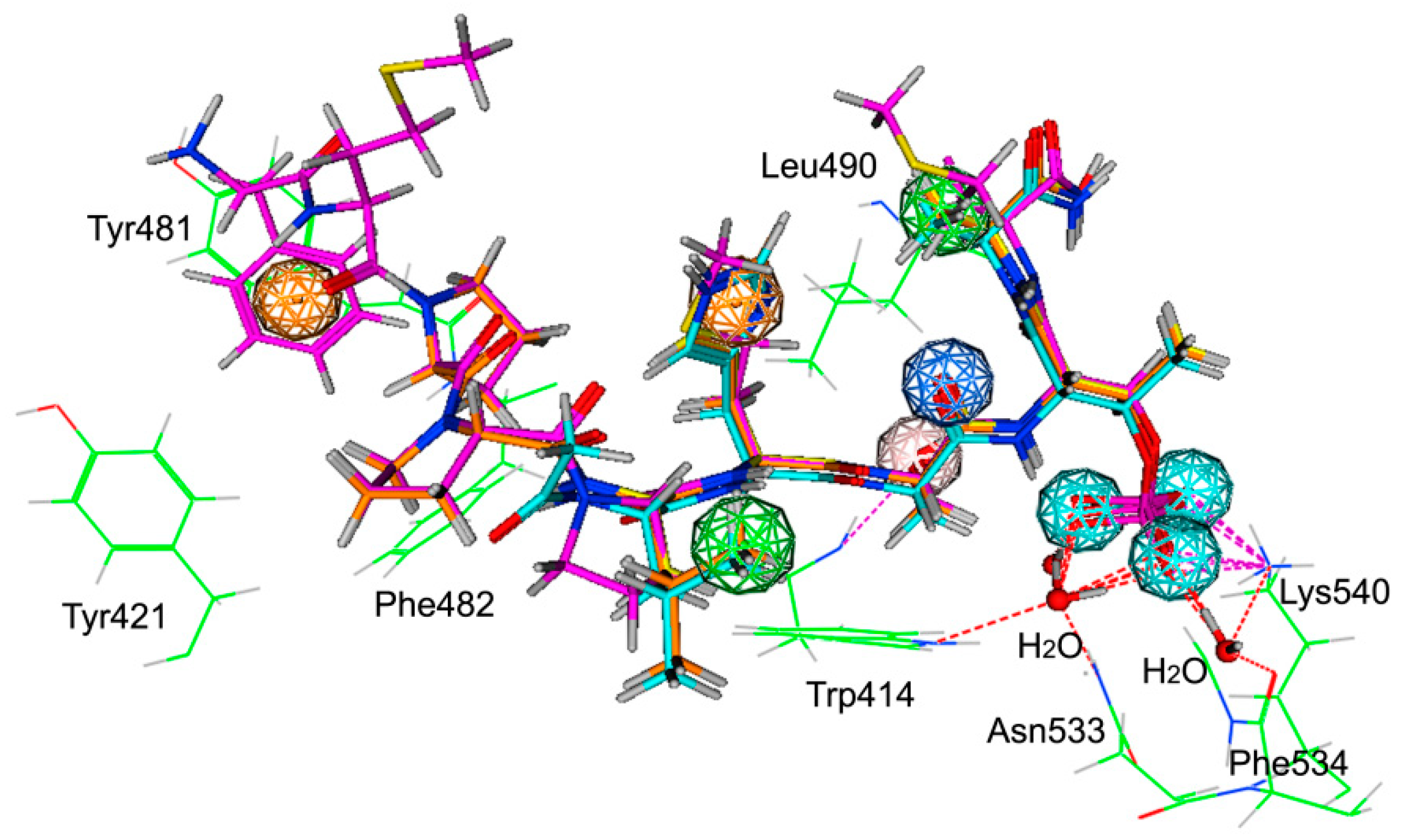
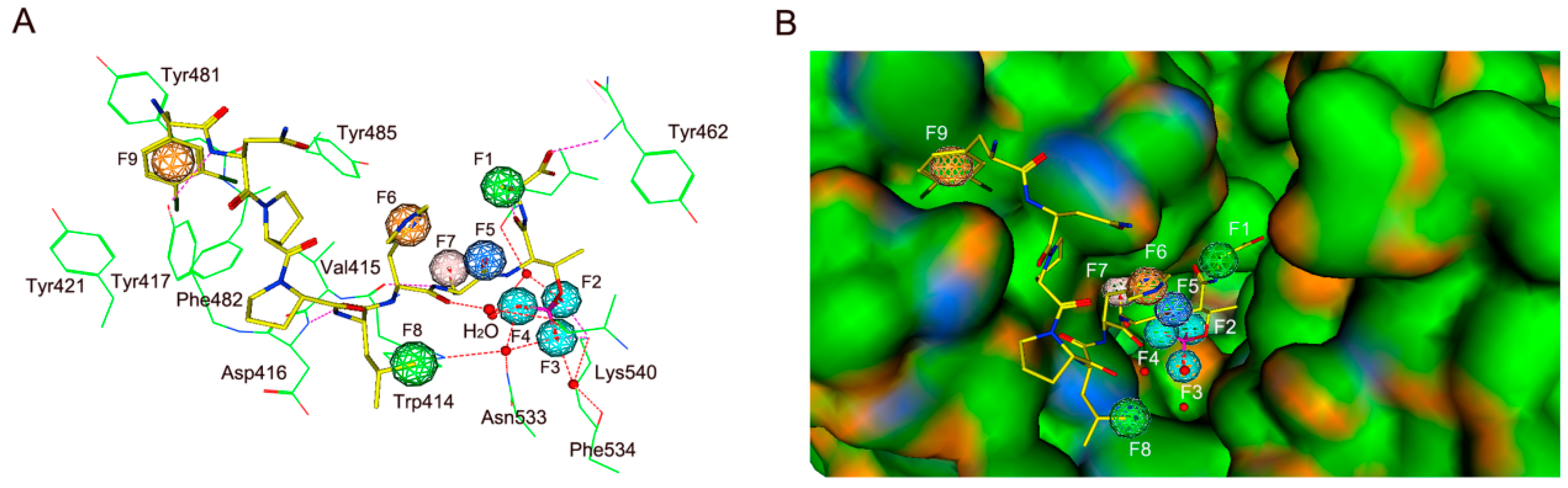
2.1. Pharmacophore Modeling
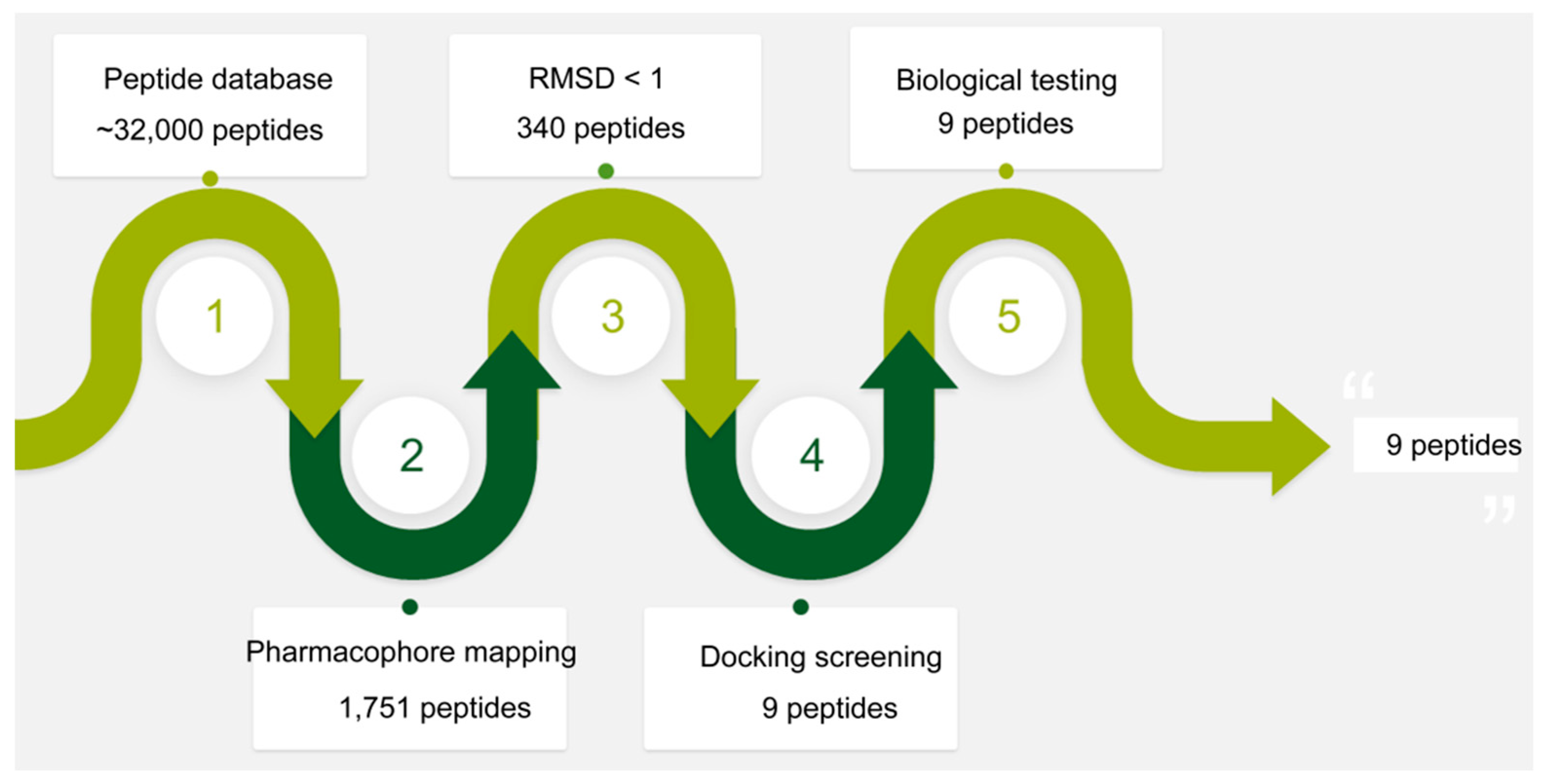
2.2. Validation and Database Screening
2.3. PLK1-PBD Inhibition Assay
2.4. Effect of Peptide 5 on Cycle Arrest in HeLa Cells
3. Materials and Methods
3.1. Pharmacophore Model Generation and Validation
3.2. Virtual Screening
3.3. Molecular Docking
3.4. In Vitro PLK1-PBD Inhibition Assay
3.5. Cell Cycle Assay and Western Blot Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Coudrat, T.; Simms, J.; Christopoulos, A.; Wootten, D.; Sexton, P.M. Improving virtual screening of G protein-coupled receptors via ligand-directed modeling. PLoS Comput. Biol. 2017, 13, e1005819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gönczy, P. Centrosomes and cancer: Revisiting a long-standing relationship. Nat. Rev. Cancer 2015, 15, 639. [Google Scholar] [CrossRef] [PubMed]
- Zitouni, S.; Nabais, C.; Jana, S.C.; Guerrero, A.; Bettencourt-Dias, M. Polo-like kinases: Structural variations lead to multiple functions. Nat. Rev. Mol. Cell. Biol. 2014, 15, 433. [Google Scholar] [CrossRef] [PubMed]
- Barr, F.A.; Silljé, H.H.; Nigg, E.A. Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell. Biol. 2004, 5, 429. [Google Scholar] [CrossRef]
- Eckerdt, F.; Yuan, J.; Strebhardt, K. Polo-like kinases and oncogenesis. Oncogene 2005, 24, 267. [Google Scholar] [CrossRef] [Green Version]
- Liu, X. Targeting polo-like kinases: A promising therapeutic approach for cancer treatment. Transl. Oncol. 2015, 8, 185–195. [Google Scholar] [CrossRef] [Green Version]
- Casenghi, M.; Meraldi, P.; Weinhart, U.; Duncan, P.I.; Körner, R.; Nigg, E.A. Polo-like kinase 1 regulates Nlp, a centrosome protein involved in microtubule nucleation. Dev. Cell 2003, 5, 113–125. [Google Scholar] [CrossRef] [Green Version]
- Murugan, R.N.; Park, J.E.; Kim, E.H.; Shin, S.Y.; Cheong, C.; Lee, K.S.; Bang, J.K. Plk1-targeted small molecule inhibitors: Molecular basis for their potency and specificity. Mol. Cells 2011, 32, 209. [Google Scholar] [CrossRef] [Green Version]
- Qin, T.; Chen, F.; Zhuo, X.; Guo, X.; Yun, T.; Liu, Y.; Lai, L. Discovery of novel polo-like kinase 1 polo-box domain inhibitors to induce mitotic arrest in tumor cells. J. Med. Chem. 2016, 59, 7089–7096. [Google Scholar] [CrossRef]
- Cummings, C.G.; Hamilton, A.D. Disrupting protein–protein interactions with non-peptidic, small molecule α-helix mimetics. Curr. Opin. Chem. Biol. 2010, 14, 341–346. [Google Scholar] [CrossRef]
- Fosgerau, K.; Hoffmann, T. Peptide therapeutics: Current status and future directions. Drug. Discov. Today 2015, 20, 122–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pelay-Gimeno, M.; Glas, A.; Koch, O.; Grossmann, T.N. Structure-based design of inhibitors of protein-protein interactions: Mimicking peptide binding epitopes. Angew. Chem. Int. Ed. Engl. 2015, 54, 8896–8927. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, Z.; Liu, Y.; Lin, T.; Sun, H.; Yang, D.; Jiang, C. Identification of novel and selective non-peptide inhibitors targeting the polo-box domain of polo-like kinase 1. Bioorg. Chem. 2018, 81, 278–288. [Google Scholar] [CrossRef] [PubMed]
- Elia, A.E.; Rellos, P.; Haire, L.F.; Chao, J.W.; Ivins, F.J.; Hoepker, K.; Mohammad, D.; Cantley, L.C.; Smerdon, S.J.; Yaffe, M.B. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell 2003, 115, 83–95. [Google Scholar] [CrossRef]
- Pradeepkiran, J.A.; Reddy, P.H. Structure Based Design and Molecular Docking Studies for Phosphorylated Tau Inhibitors in Alzheimer’s Disease. Cells 2019, 8, 260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pradeepkiran, J.A.; Reddy, A.P.; Reddy, P.H. Pharmacophore-based models for therapeutic drugs against phosphorylated tau in Alzheimer’s disease. Drug Discov. Today 2019, 24, 616–623. [Google Scholar] [CrossRef]
- Yun, S.M.; Moulaei, T.; Lim, D.; Bang, J.K.; Park, J.E.; Shenoy, S.R.; Liu, F.; Kang, Y.H.; Liao, C.; Soung, N.K. Structural and functional analyses of minimal phosphopeptides targeting the polo-box domain of polo-like kinase 1. Nat. Struct.Mol. Biol. 2009, 16, 876. [Google Scholar] [CrossRef] [Green Version]
- Śledź, P.; Stubbs, C.J.; Lang, S.; Yang, Y.Q.; McKenzie, G.J.; Venkitaraman, A.R.; Abell, C. From Crystal Packing to Molecular Recognition: Prediction and Discovery of a Binding Site on the Surface of Polo-Like Kinase 1. Angew. Chem. 2011, 50, 4003–4006. [Google Scholar] [CrossRef] [Green Version]
- Niu, M.; Wang, F.; Li, F.; Dong, Y.; Gu, Y. Establishment of a screening protocol for identification of aminopeptidase N inhibitors. J. Taiwan Inst. Chem. Eng. 2015, 49, 19–26. [Google Scholar] [CrossRef]
- Lin, T.Y.; Min, H.P.; Jiang, C.; Niu, M.M.; Yan, F.; Xu, L.L.; Di, B. Design, synthesis and biological evaluation of phosphopeptides as Polo-like kinase 1 Polo-box domain inhibitors. Bioorg. Med. Chem. 2018, 26, 3429–3437. [Google Scholar] [CrossRef]
- Śledź, P.; Lang, S.; Stubbs, C.J.; Abell, C. High-Throughput Interrogation of Ligand Binding Mode Using a Fluorescence-Based Assay. Angew. Chem. 2012, 124, 7800–7803. [Google Scholar] [CrossRef]
- Yang, D.S.; Yang, Y.H.; Zhou, Y.; Yu, L.L.; Wang, R.H.; Di, B.; Niu, M.M. A Redox-Triggered Bispecific Supramolecular Nanomedicine Based on Peptide Self-Assembly for High-Efficacy and Low-Toxic Cancer Therapy. Adv. Funct. Mater. 2019. [Google Scholar] [CrossRef]
- Manual of Molecular Operating Environment (MOE); Version 2007.09; Chemical Computing Group Inc.: Montreal, QC, Canada, 2007.
- Ul Qamar, T.; Mumtaz, A.; Ashfaq, U.A.; Azhar, S.; Fatima, T.; Hassan, M.; Hussain, S.S.; Akram, W.; Idrees, S. Computer aided screening of phytochemicals from Garcinia against the dengue NS2B/NS3 protease. Bioinformation 2014, 10, 115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Sample Availability: Samples of the compounds are available from the authors. |

| Serial No. | Parameter | Pharmacophore Model |
|---|---|---|
| 1 | Total molecules in database (D) | 2000 |
| 2 | Total number of actives in database (A) | 15 |
| 3 | Total hits (Ht) | 18 |
| 4 | Active hits (Ha) | 13 |
| 5 | % Yield of actives [(Ha/Ht) × 100] | 72% |
| 6 | % Ratio of actives [(Ha/A) × 100] | 87% |
| 7 | Enrichment factor (E) [(Ha × D)/(Ht × A)] | 96 |
| 8 | False negatives [A − Ha] | 2 |
| 9 | False positives [Ht − Ha] | 5 |
| 10 | Goodness of hit score (GH) a | 0.76 |
| Peptides | Sequence a | RMSD [Å] b | Docking Score [kcal/mol] c |
|---|---|---|---|
| 1 | YEPPLHSpTAIG | 0.26 | −24.54 |
| 2 | WDPPLHSpTAI | 0.38 | −23.85 |
| 3 | FEPPLHSpTAI | 0.44 | −21.94 |
| 4 | FEPPLHSpTAG | 0.23 | −25.36 |
| 5 | ΦNPPLHSpTA | 0.36 | −23.31 |
| 6 | WAPPLHSpTAK | 0.45 | −20.96 |
| 7 | WKPPLHSpTAG | 0.47 | −20.87 |
| 8 | HKPPLHSpTA | 0.51 | −20.13 |
| 9 | HQPPLHSpTA | 0.53 | −20.07 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, F.; Liu, G.; Chen, T.; Fu, X.; Niu, M.-M. Structure-Based Virtual Screening and Biological Evaluation of Peptide Inhibitors for Polo-Box Domain. Molecules 2020, 25, 107. https://doi.org/10.3390/molecules25010107
Yan F, Liu G, Chen T, Fu X, Niu M-M. Structure-Based Virtual Screening and Biological Evaluation of Peptide Inhibitors for Polo-Box Domain. Molecules. 2020; 25(1):107. https://doi.org/10.3390/molecules25010107
Chicago/Turabian StyleYan, Fang, Guangmei Liu, Tingting Chen, Xiaochen Fu, and Miao-Miao Niu. 2020. "Structure-Based Virtual Screening and Biological Evaluation of Peptide Inhibitors for Polo-Box Domain" Molecules 25, no. 1: 107. https://doi.org/10.3390/molecules25010107
APA StyleYan, F., Liu, G., Chen, T., Fu, X., & Niu, M.-M. (2020). Structure-Based Virtual Screening and Biological Evaluation of Peptide Inhibitors for Polo-Box Domain. Molecules, 25(1), 107. https://doi.org/10.3390/molecules25010107






