Approaching Authenticity Issues in Fish and Seafood Products by Qualitative Spectroscopy and Chemometrics
Abstract
:1. Introduction
2. A Conceptual Framework of Spectroscopy and Chemometrics
2.1. UV–Vis Absorption and Fluorescence Emission Spectroscopy
2.2. IR Spectroscopy
2.3. Raman Spectroscopy
2.4. Hyperspectral Imaging
2.5. NMR Spectroscopy
2.6. Qualitative Chemometric Methods
2.6.1. Spectral Pre-Treatments
2.6.2. Unsupervised Methods
2.6.3. Supervised Methods
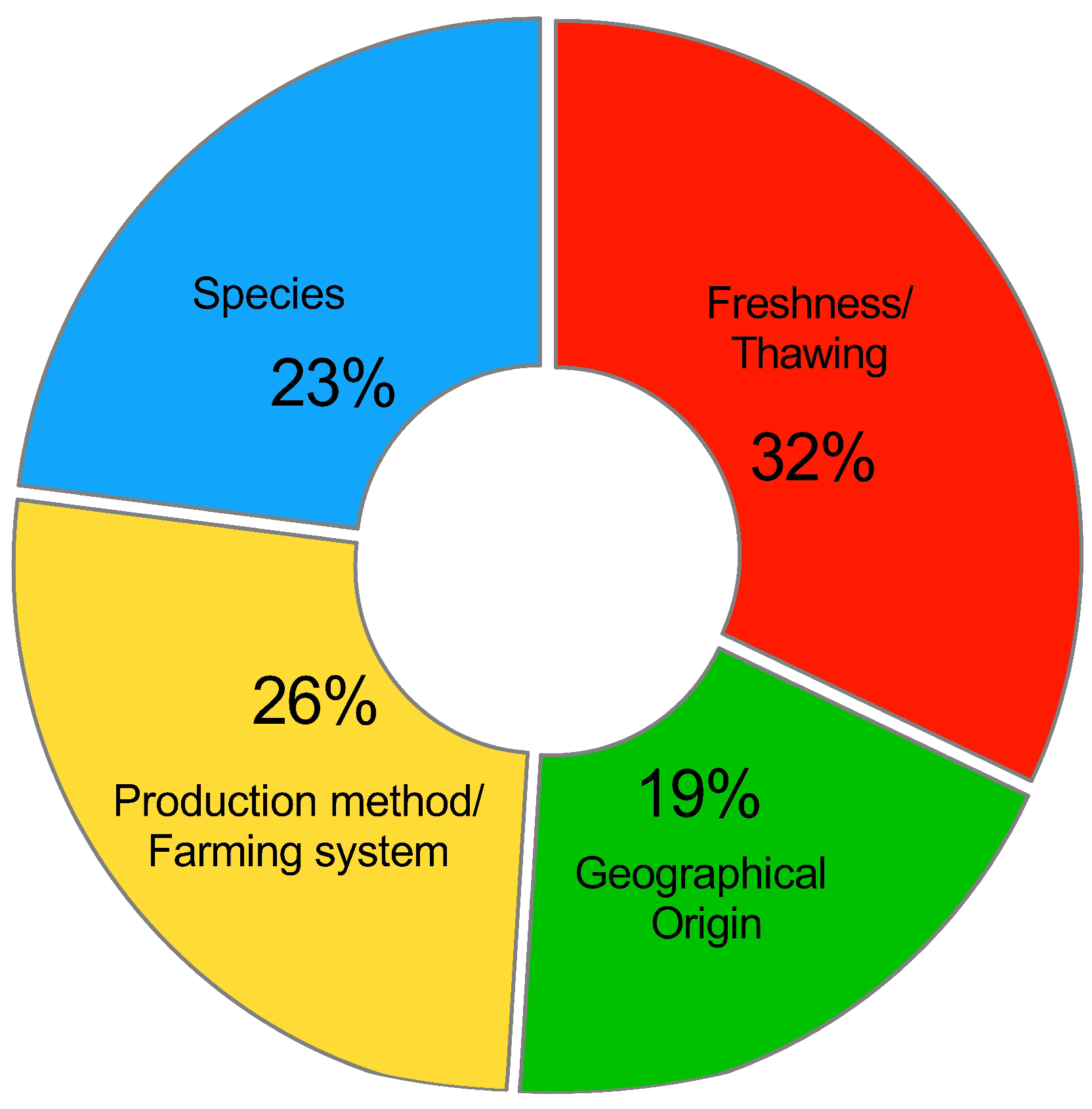
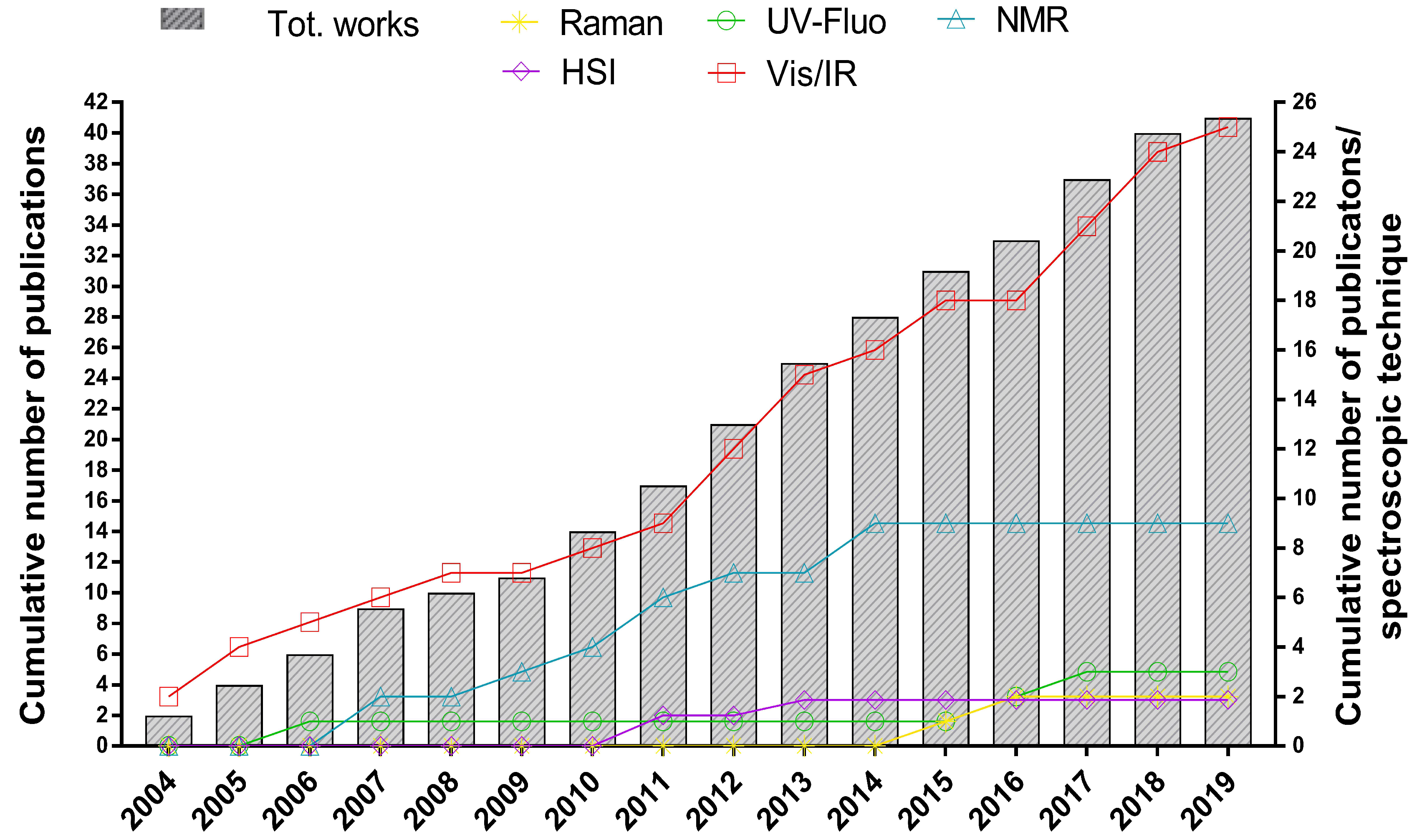
3. Authenticating Fish and Seafood through the Application of Qualitative Spectroscopy and Chemometrics
3.1. Species Substitution
3.1.1. Application of Vibrational Spectroscopy
3.1.2. Application of NMR Spectroscopy
3.2. Production Method and Farming System Misrepresentation
3.2.1. Application of Vibrational Spectroscopy
3.2.2. Application of NMR Spectroscopy
3.3. Geographical Origin Falsification
3.3.1. Application of Vibrational Spectroscopy
3.3.2. Application of NMR Spectroscopy
3.4. Discrimination between Fresh and Frozen/Thawed Fish and Seafood
3.4.1. Application of Fluorescence and Vibrational Spectroscopy
3.4.2. Application of Hyperspectral Imaging Spectroscopy
3.4.3. Application of NMR Spectroscopy
4. Critical Aspects and Limitations to Overcome
5. Conclusions and Prospects for the Future
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ANN | artificial neural networks; |
| BBN | Bayesian belief network; |
| FIR | far-infrared; |
| FDA | factorial discriminant analysis; |
| FFT | fast Fourier transform; |
| FT | Fourier transform; |
| HCA | hierarchical cluster analysis; |
| HSI | hyperspectral imaging; |
| IR | infrared; |
| k-NN | k-nearest neighbors; |
| LDA | linear discriminant analysis; |
| LW-NIR | long-wave near infrared; |
| MIR | mid-infrared; |
| NMR | nuclear magnetic resonance; |
| MSC | multiplicative scatter correction; |
| NIR | near-infrared; |
| OPLS-DA | orthogonal partial least square-discriminant analysis; |
| PCA | principal component analysis; |
| PLS-DA | partial least square-discriminant analysis; |
| PNN | probabilistic neural network; |
| QDA | quadratic factorial analysis; |
| SERS | surface-enhanced Raman spectroscopy; |
| SG | Savitzky–Golay smoothing; |
| SIMCA | soft independent modelling of class analogy; |
| SNV | standard normal variate; |
| SVM | support vector machine; |
| SW-NIR | short-wave near infrared; |
| UV | ultraviolet; |
| Vis | visible. |
References
- FAO. The State of World Fisheries and Aquaculture 2018–Meeting the Sustainable Development Goals; FAO: Rome, Italy, 2018; Volume 35, pp. 52–62. ISBN 978-92-5106-029-2. [Google Scholar]
- Rehbein, H.; Oehlenschläger, J. Basic Facts and Figures. In Fishery products: Quality, Safety and Authenticity; Rehbein, H., Oehlenschläger, J., Eds.; John Wiley & Sons: Chichester, West Sussex, UK, 2009; Volume 1, pp. 1–18. ISBN 978-1-4051-4162-8. [Google Scholar]
- FAO. Overview of Food Fraud in the Fisheries Sector, by Alan Reilly; Fisheries and Aquaculture Circular No. 1165; FAO: Rome, Italy, 2018; pp. 4–6. ISBN 978-92-5-130402-0. [Google Scholar]
- Van Dolah, F.M. Marine Algal Toxins: Origins, Health Effects, and Their Increased Occurrence. Environ. Health Perspect. 2000, 108, 133–141. [Google Scholar] [CrossRef]
- Fallah, A.A.; Saei-Dehkordi, S.S.; Nematollahi, A.; Jafari, T. Comparative study of heavy metal and trace element accumulation in edible tissues of farmed and wild rainbow trout (Oncorhynchus mykiss) using ICP-OES technique. Microchem. J. 2011, 98, 275–279. [Google Scholar] [CrossRef]
- Okocha, R.C.; Olatoye, I.O.; Adedeji, O.B. Food safety impacts of antimicrobial use and their residues in aquaculture. Public Health Rev. 2018, 39, 1–22. [Google Scholar] [CrossRef]
- Kelly, B.C.; Ikonomou, M.G.; Higgs, D.A.; Oakes, J.; Dubetz, C. Flesh residue concentrations of organochlorine pesticides in farmed and wild salmon from British Columbia, Canada. Environ. Toxicol. Chem. 2011, 30, 2456–2464. [Google Scholar] [CrossRef]
- Council regulation (EC) No 178/2002 of the European Parliament and of the Council of 28 January2002 laying down the general principles and requirements of food law, establishing the European Food Safety Authority and laying down procedures in matters of food safety. Off. J. Eur. Commun. 2002, 31, 1–24.
- Council regulation (EC) No 1005/2008 of 29 September 2008 establishing a Community system to prevent, deter and eliminate illegal, unreported and unregulated fishing, amending Regulations (EEC) No 2847/93, (EC) No 1936/2001 and (EC) No 601/2004 and repealing Regulations (EC) No 1093/94 and (EC) No 1447/1999. Off. J. Eur. Union 2008, 286, 1–32.
- Council regulation (EC) No 1224/2009 of 20 November 2009 establishing a Community control system for ensuring compliance with the rules of the common fisheries policy, amending Regulations (EC) No 847/96, (EC) No 2371/2002, (EC) No 811/2004, (EC) No 768/2005, (EC) No 2115/2005, (EC) No 2166/2005, (EC) No 388/2006, (EC) No 509/2007, (EC) No 676/2007, (EC) No 1098/2007, (EC) No 1300/2008, (EC) No 1342/2008 and repealing Regulations (EEC) No 2847/93, (EC) No 1627/94 and (EC) No 1966/2006. Off. J. Eur. Union 2009, 343, 1–50.
- Regulation (EU) No 1169/2011 of the European Parliament and of the Council of 25 October 2011 on the provision of food information to consumers, amending Regulations (EC) No 1924/2006 and (EC) No 1925/2006 of the European Parliament and of the Council, and repealing Commission Directive 87/250/EEC, Council Directive 90/496/EEC, Commission Directive 1999/10/EC, Directive 2000/13/EC of the European Parliament and of the Council, Commission Directives 2002/67/EC and 2008/5/EC. Off. J. Eur. Union 2011, 304, 18–63.
- Regulation (EU) No 1379/2013 of the European Parliament and of the Council of 11 December 2013 on the common organisation of th emarkets in fishery and aquaculture products, amending Council Regulations (EC) No 1184/2006 and (EC) No 1224/2009 and repealing Council Regulation (EC) No 104/2000. Off. J. Eur. Union 2013, 354, 12–14.
- Esslinger, S.; Riedl, J.; Fauhl-Hassek, C. Potential and limitations of non-targeted fingerprinting for authentication of food in official control. Food Res. Int. 2014, 60, 189–204. [Google Scholar] [CrossRef]
- Picò, Y. Near-Infrared, Mid-Infrared, and Raman Spectroscopy. In Chemical Analysis of Food: Techniques and Applications; Picò, Y., Ed.; Academic Press: San Francisco, CA, USA, 2012; Volume 1, pp. 59–91. ISBN 978-0-1238-4862-8. [Google Scholar]
- Schrieber, A. Introduction to Food Authentication. In Modern Techniques for Food Authentication; Sun, D.W., Ed.; Academic Press: San Francisco, CA, USA, 2008; pp. 1–21. [Google Scholar]
- Penner, M.H. Basic Principles of Spectroscopy. In Food Analysis, Food Science Text Series, 2nd ed.; Nielsen, S.S., Ed.; Springer: Cham, Switzerland; New York, NY, USA, 2017; pp. 79–88. ISBN 978-3-319-45776-5. [Google Scholar]
- Strasburg, G.M.; Ludescher, R.D. Theory and applications of fluorescence spectroscopy in food research. Trends Food Sci. Technol. 1995, 6, 69–75. [Google Scholar] [CrossRef]
- Xu, J.L.; Riccioli, C.; Sun, D.W. An Overview on Nondestructive Spectroscopic Techniques for Lipid and Lipid Oxidation Analysis in Fish and Fish Products. Compr. Rev. Food Sci. Food Saf. 2015, 4, 466–477. [Google Scholar] [CrossRef]
- Rodriguez-Saona, L.E.; Allendorf, M.E. Use of FTIR for rapid authentication and detection of adulteration of food. Ann. Rev. Food Sci. Technol. 2011, 2, 467–483. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Saona, L.E.; Giusti, M.M.; Shotts, M. Advances in Infrared Spectroscopy for Food Authenticity Testing; Downey, G., Ed.; Woodhead Publishing: Duxford, UK, 2016; ISBN 978-0-08-100220-9. [Google Scholar]
- Lohumi, S.; Lee, S.; Lee, H.; Cho, B.K. A review of vibrational spectroscopic techniques for the detection of food authenticity and adulteration. Trends Food Sci. Technol. 2015, 1, 85–98. [Google Scholar] [CrossRef]
- Blanco, M.; Villarroya, I.N.I.R. NIR spectroscopy: A rapid-response analytical tool. TrAC Trends Anal. Chem. 2002, 21, 240–250. [Google Scholar] [CrossRef]
- Cen, H.; Yong, H. Theory and application of near infrared reflectance spectroscopy in determination of food quality. Trends Food Sci. Technol. 2007, 18, 72–83. [Google Scholar] [CrossRef]
- Cheng, J.H.; Dai, Q.; Sun, D.W.; Zeng, X.A.; Liu, D.; Pu, H.B. Applications of non-destructive spectroscopic techniques for fish quality and safety evaluation and inspection. Trends Food Sci. Technol. 2013, 34, 18–31. [Google Scholar] [CrossRef]
- Cozzolino, D.; Murray, I. A review on the application of infrared technologies to determine and monitor composition and other quality characteristics in raw fish, fish products, and seafood. Appl. Spectrosc. Rev. 2012, 47, 207–218. [Google Scholar] [CrossRef]
- Stuart, B.H. Spectral Analysis. In Infrared Spectroscopy: Fundamentals and Applications; Stuart, B.H., Ed.; Jhon Wiley & Sons Ltd.: Chichester, WS, UK, 2004; pp. 47–48. ISBN 0-470-85427-8. [Google Scholar]
- Boyaci, I.H.; Temiz, H.T.; Geniş, H.E.; Soykut, E.A.; Yazgan, N.N.; Güven, B.; Uysal, R.S.; Bozkurt, A.G.; Ilaslan, K.; Torun, O.; et al. Dispersive and FT-Raman spectroscopic methods in food analysis. RSC Adv. 2015, 5, 56606–56624. [Google Scholar] [CrossRef]
- Zheng, J.; He, L. Surface-Enhanced Raman Spectroscopy for the Chemical Analysis of Food. Compr. Rev. Food Sci. Food Saf. 2014, 13, 317–328. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.Z.; Sun, D.W. Application of hyperspectral imaging in food safety inspection and control: A review. Crit. Rev. Food Sci. Nutr. 2012, 52, 1039–1058. [Google Scholar] [CrossRef]
- Wu, D.; Sun, D.W. Advanced applications of hyperspectral imaging technology for food quality and safety analysis and assessment: A review—Part I: Fundamentals. Innov. Food Sci. Emerg. Technol. 2013, 19, 1–14. [Google Scholar] [CrossRef]
- Roberts, J.; Power, A.; Chapman, J.; Chandra, S.; Cozzolino, D. A short update on the advantages, applications and limitations of hyperspectral and chemical imaging in food authentication. Appl. Sci. 2018, 8, 505. [Google Scholar] [CrossRef]
- Sacchi, R.; Paolillo, L. NMR for Food Quality and Traceability. In Advances in Food Diagnostics; Nollet, L.M.L., Toldrà, F., Eds.; Blackwell Publishing: Oxford, UK, 2007; Volume 1, pp. 101–107. ISBN 978-0-4702-7780-5. [Google Scholar]
- Hatzakis, E. Nuclear Magnetic Resonance (NMR) Spectroscopy in Food Science: A Comprehensive Review. Compre. Rev. Food Sci. Food Saf. 2019, 18, 189–220. [Google Scholar] [CrossRef]
- Sobolev, A.P.; Circi, S.; Mannina, L. Advances in Nuclear Magnetic Resonance Spectroscopy for Food Authenticity Testing. In Advances in Food Authenticity Testing; Downey, G., Ed.; Woodhead Publishing: Duxford, UK, 2016; Volume 1, pp. 147–170. [Google Scholar] [CrossRef]
- Oliveri, P.; Simonetti, R. Chemometrics for food authenticity applications. In Advances in Food Authenticity Testing; Downey, G., Ed.; Woodhead Publishing: Duxford, UK, 2016; pp. 701–728. [Google Scholar] [CrossRef]
- Manley, M.; Baeten, V. Spectroscopic technique: Near infrared (NIR) spectroscopy. In Modern Techniques for Food Authentication; Sun, D.W., Ed.; Academic Press: San Francisco, CA, USA, 2008; Volume 1, pp. 51–102. [Google Scholar]
- Rinnan, Å.; Van Den Berg, F.; Engelsen, S.B. Review of the most common pre-processing techniques for near-infrared spectra. Trends Anal. Chem. 2009, 28, 1201–1222. [Google Scholar] [CrossRef]
- Pereira, A.C.; Reis, M.S.; Saraiva, P.M.; Marques, J.C. Madeira wine ageing prediction based on different analytical techniques: UV–vis, GC-MS, HPLC-DAD. Chemom. Intell. Lab. Syst. 2011, 105, 43–55. [Google Scholar] [CrossRef]
- Rinnan, Å. Pre-processing in vibrational spectroscopy–when, why and how. Anal. Methods 2014, 6, 7124–7129. [Google Scholar] [CrossRef]
- Engel, J.; Gerretzen, J.; Szymańska, E.; Jansen, J.J.; Downey, G.; Blanchet, L.; Buydens, L.M.C. Breaking with trends in pre-processing? TrAC Trends Anal. Chem. 2013, 50, 96–106. [Google Scholar] [CrossRef]
- Ziegel, E.R. A User-Friendly Guide to Multivariate Calibration and Classification. Technometrics 2004, 46, 108–111. [Google Scholar] [CrossRef]
- Voncina, D.B. Chemometrics in analytical chemistry. Nova Biotechnol. 2009, 9, 211–216. [Google Scholar] [CrossRef]
- Berrueta, L.A.; Alonso-Salces, L.M.; Heberger, K. Supervised pattern recognition in food analysis. J. Chromatog. A 2007, 1158, 196–214. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.R.; Khalid, M.; Yusof, R. Machine Learning Using Support Vector Machines. In Proceedings of the International Conference on Artificial Intelligence in Science and Technology, Hobart, Australia, 19–21 September 2002. [Google Scholar]
- Tetko, I.V.; Livingstone, D.J.; Luik, A.I. Neural network studies. 1. Comparison of overfitting and overtraining. J. Chem. Inf. Comput. Sci. 1995, 35, 826–833. [Google Scholar] [CrossRef]
- Bylesjö, M.; Rantalainen, M.; Cloarec, O.; Nicholson, J.K.; Holmes, E.; Trygg, J. OPLS Discriminant Analysis, Combining the strengths of PLS-DA and SIMCA classification. J. Chemom. 2007, 20, 341–351. [Google Scholar] [CrossRef]
- He, H.J.; Wu, D.; Sun, D.W. Nondestructive Spectroscopic and Imaging Techniques for Quality Evaluation and Assessment of Fish and Fish Products. Crit. Rev. Food Sci. Nutr. 2015, 55, 864–886. [Google Scholar] [CrossRef] [PubMed]
- Uddin, M.; Okazaki, E. Applications of Vibrational Spectroscopy to the Analysis of Fish and Other Aquatic Food Products. Handb. Vib. Spectro. 2006, 439–459. [Google Scholar] [CrossRef]
- Fiorino, G.M.; Garino, C.; Arlorio, M.; Logrieco, A.F.; Losito, I.; Monaci, L. Overview on Untargeted Methods to Combat Food Frauds: A Focus on Fishery Products. J. Food Qual. 2018, 3, 1–13. [Google Scholar] [CrossRef]
- Gayo, J.; Hale, S.A.; Blanchard, S.M. Quantitative analysis and detection of adulteration in crab meat using visible and near-infrared spectroscopy. J. Agric. Food Chem. 2006, 54, 1130–1136. [Google Scholar] [CrossRef]
- O’Brien, N.; Hulse, C.A.; Pfeifer, F.; Siesler, H.W. Near infrared spectroscopic authentication of seafood. J. Near Infrared Spectrosc. 2013, 21, 299–305. [Google Scholar] [CrossRef]
- Grassi, S.; Casiraghi, E.; Alamprese, C. Handheld NIR device: A non-targeted approach to assess authenticity of fish fillets and patties. Food Chem. 2018, 243, 382–388. [Google Scholar] [CrossRef] [Green Version]
- Lv, H.; Xu, W.; You, J.; Xiong, S. Classification of freshwater fish species by linear discriminant analysis based on near infrared reflectance spectroscopy. J. Near Infrared Spectrosc. 2017, 25, 54–62. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Hu, W.; Teng, J.; Peng, H.H.; Gan, J.H.; Wang, X.C.; Sun, S.Q.; Xu, C.H.; Liu, Y. Rapid recognition of marine fish surimi by one-step discriminant analysis based on near-infrared diffuse reflectance spectroscopy. Int. J. Food Prop. 2017, 20, 2932–2943. [Google Scholar] [CrossRef]
- Alamprese, C.; Casiraghi, E. Application of FT-NIR and FT-IR spectroscopy to fish fillet authentication. LWT–Food Sci. Technol. 2015, 63, 720–725. [Google Scholar] [CrossRef]
- Cozzolino, D.; Chree, A.; Scaife, J.R.; Murray, I. Usefulness of Near-infrared reflectance (NIR) spectroscopy and chemometrics to discriminate fishmeal batches made with different fish species. J. Agric. Food Chem. 2005, 53, 4459–4463. [Google Scholar] [CrossRef] [PubMed]
- Sousa, N.; Moreira, M.; Saraiva, C.; de Almeida, J. Applying Fourier Transform Mid Infrared Spectroscopy to Detect the Adulteration of Salmo salar with Oncorhynchus mykiss. Foods 2018, 7, 55. [Google Scholar] [CrossRef]
- Rašković, B.; Heinke, R.; Rösch, P.; Popp, J. The Potential of Raman Spectroscopy for the Classification of Fish Fillets. Food Anal. Methods 2016, 9, 1301–1306. [Google Scholar] [CrossRef]
- Velioğlu, H.M.; Temiz, H.T.; Boyaci, I.H. Differentiation of fresh and frozen-thawed fish samples using Raman spectroscopy coupled with chemometric analysis. Food Chem. 2015, 173, 283–290. [Google Scholar] [CrossRef]
- Standal, I.B.; Axelson, D.E.; Aursand, M. 13C NMR as a tool for authentication of different gadoid fish species with emphasis on phospholipid profiles. Food Chem. 2010, 121, 608–615. [Google Scholar] [CrossRef]
- Gabr, H.R.; Gab-Alla, A.A.F.A. Comparison of biochemical composition and organoleptic properties between wild and cultured finfish. J. Fish. Aquat. Sci. 2007, 2, 77–81. [Google Scholar] [CrossRef]
- Grigorakis, K.; Taylor, K.D.A.; Alexis, M.N. Organoleptic and volatile aroma compounds comparison of wild and cultured gilthead sea bream (Sparus aurata): Sensory differences and possible chemical basis. Aquaculture 2003, 225, 109–119. [Google Scholar] [CrossRef]
- Grigorakis, K. Compositional and organoleptic quality of farmed and wild gilthead sea bream (Sparus aurata) and sea bass (Dicentrarchus labrax) and factors affecting it: A review. Aquaculture 2007, 272, 55–75. [Google Scholar] [CrossRef]
- Lenas, D.; Chatziantoniou, S.; Nathanailides, C.; Triantafillou, D. Comparison of wild and farmed sea bass (Dicentrarchus labrax L) lipid quality. Procedia Food Sci. 2011, 1, 1139–1145. [Google Scholar] [CrossRef]
- Fuentes, A.; Fernández-Segovia, I.; Serra, J.A.; Barat, J.M. Comparison of wild and cultured sea bass (Dicentrarchus labrax) quality. Food Chem. 2010, 119, 1514–1518. [Google Scholar] [CrossRef]
- Ottavian, M.; Facco, P.; Fasolato, L.; Novelli, E.; Mirisola, M.; Perini, M.; Barolo, M. Use of near-infrared spectroscopy for fast fraud detection in seafood: Application to the authentication of wild European sea bass (Dicentrarchus labrax). J. Agric. Food Chem. 2012, 60, 639–648. [Google Scholar] [CrossRef]
- Ghidini, S.; Varrà, M.O.; Dall’Asta, C.; Badiani, A.; Ianieri, A.; Zanardi, E. Rapid authentication of European sea bass (Dicentrarchus labrax L.) according to production method, farming system, and geographical origin by near infrared spectroscopy coupled with chemometrics. Food Chem. 2019, 280, 321–327. [Google Scholar] [CrossRef]
- Costa, C.; D’Andrea, S.; Russo, R.; Antonucci, F.; Pallottino, F.; Menesatti, P. Application of non-invasive techniques to differentiate sea bass (Dicentrarchus labrax, L. 1758) quality cultured under different conditions. Aquac. Int. 2011, 19, 765–778. [Google Scholar] [CrossRef]
- Xiccato, G.; Trocino, A.; Tulli, F.; Tibaldi, E. Prediction of chemical composition and origin identification of european sea bass (Dicentrarchus labrax L.) by near infrared reflectance spectroscopy (NIRS). Food Chem. 2004, 86, 275–281. [Google Scholar] [CrossRef]
- Trocino, A.; Xiccato, G.; Majolini, D.; Tazzoli, M.; Bertotto, D.; Pascoli, F.; Palazzi, R. Assessing the quality of organic and conventionally-farmed European sea bass (Dicentrarchus labrax). Food Chem. 2012, 131, 427–433. [Google Scholar] [CrossRef]
- Dalle Zotte, A.; Ottavian, M.; Concollato, A.; Serva, L.; Martelli, R.; Parisi, G. Authentication of raw and cooked freeze-dried rainbow trout (Oncorhynchus mykiss) by means of near infrared spectroscopy and data fusion. Food Res. Int. 2014, 60, 180–188. [Google Scholar] [CrossRef]
- Masoum, S.; Malabat, C.; Jalali-Heravi, M.; Guillou, C.; Rezzi, S.; Rutledge, D.N. Application of support vector machines to 1H NMR data of fish oils: Methodology for the confirmation of wild and farmed salmon and their origins. Anal. Bioanal. Chem. 2007, 387, 1499–1510. [Google Scholar] [CrossRef]
- Rezzi, S.; Giani, I.; Héberger, K.; Axelson, D.E.; Moretti, V.M.; Reniero, F.; Guillou, C. Classification of gilthead sea bream (Sparus aurata) from1H NMR lipid profiling combined with principal component and linear discriminant analysis. J. Agric. Food Chem. 2007, 55, 9963–9968. [Google Scholar] [CrossRef]
- Capuano, E.; Lommen, A.; Heenan, S.; de la Dura, A.; Rozijn, M.; van Ruth, S. Wild salmon authenticity can be predicted by 1H-NMR spectroscopy. Lipid Technol. 2012, 24, 251–253. [Google Scholar] [CrossRef]
- Melis, R.; Cappuccinelli, R.; Roggio, T.; Anedda, R. Addressing marketplace gilthead sea bream (Sparus aurata L.) differentiation by 1H NMR-based lipid fingerprinting. Food Res. Int. 2014, 63, 258–264. [Google Scholar] [CrossRef]
- Picone, G.; Balling, S.; Engelsen, F.; Savorani, S.; Testi, S.; Badiani, A.; Capozzi, F. Metabolomics as a powerful tool for molecular quality assessment of the fish Sparus aurata. Nutrients 2011, 3, 212–227. [Google Scholar] [CrossRef] [PubMed]
- Aursand, M.; Standal, I.B.; Praél, A.; Mcevoy, L.; Irvine, J.; Axelson, D.E. 13C NMR pattern recognition techniques for the classification of atlantic salmon (salmo salar L.) according to their wild, farmed, and geographical origin. J. Agric. Food Chem. 2009, 57, 3444–3451. [Google Scholar] [CrossRef] [PubMed]
- Abbas, O.; Zadravec, M.; Baeten, V.; Mikuš, T.; Lešić, T.; Vulić, A.; Prpić, J.; Jemeršić, L.; Pleadin, J. Analytical methods used for the authentication of food of animal origin. Food Chem. 2018, 246, 6–17. [Google Scholar] [CrossRef]
- Liu, Y.; Ma, D.H.; Wang, X.C.; Liu, L.P.; Fan, Y.X.; Cao, J.X. Prediction of chemical composition and geographical origin traceability of Chinese export tilapia fillets products by near infrared reflectance spectroscopy. LWT–Food Sci. Technol. 2015, 60, 1214–1218. [Google Scholar] [CrossRef]
- Guo, X.; Cai, R.; Wang, S.; Tang, B.; Li, Y.; Zhao, W. Non-destructive geographical traceability of sea cucumber (Apostichopus japonicus) using near infrared spectroscopy combined with chemometric methods. R. Soc. Open Sci. 2018, 5. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Tao, L.; Zhang, P.; Li, P.; Zhu, Q.; Tian, Y.; Yang, T. Diffuse reflectance mid-infrared Fourier transform spectroscopy (DRIFTS) for rapid identification of dried sea cucumber products from different geographical areas. Vib. Spectro. 2010, 53, 222–226. [Google Scholar] [CrossRef]
- Locci, E.; Piras, C.; Mereu, S.; Cesare Marincola, F.; Scano, P. 1H NMR metabolite fingerprint and pattern recognition of mullet (Mugil cephalus) bottarga. J. Agric. Food Chem. 2011, 59, 9497–9505. [Google Scholar] [CrossRef]
- Verrez-Bagnis, V.; Sotelo, C.G.; Mendes, R.; Silva, H.; Kappel, K.; Schröder, U. Methods for Seafood Authenticity Testing in Europe. In Bioactive Molecules in Food; Springer: Cham, Switzerland; New York, NY, USA, 2018; Volume 1, pp. 1–55. [Google Scholar]
- Tokur, B.; Ozkütük, S.; Atici, E.; Ozyurt, G.; Ozyurt, C.E. Chemical and sensory quality changes of fish fingers, made from mirror carp (Cyprinus carpio L., 1758), during frozen storage (−18 °C). Food Chem. 2006, 99, 335–341. [Google Scholar] [CrossRef]
- Karoui, R.; Thomas, E.; Dufour, E. Utilisation of a rapid technique based on front-face fluorescence spectroscopy for differentiating between fresh and frozen–thawed fish fillets. Food Res. Int. 2006, 39, 349–355. [Google Scholar] [CrossRef]
- Karoui, R.; Hassoun, A.; Ethuin, P. Front face fluorescence spectroscopy enables rapid differentiation of fresh and frozen-thawed sea bass (Dicentrarchus labrax) fillets. J. Food Eng. 2017, 202, 89–98. [Google Scholar] [CrossRef]
- Gao, Y.; Tang, H.; Ou, C.; Li, Y.; Wu, C.; Cao, J. Differentiation between fresh and frozen-thawed large yellow croaker based on front-face fluorescence spectroscopy technique. Trans. Chin. Soc. Agric. Eng. 2016, 32, 279–285. [Google Scholar] [CrossRef]
- Uddin, M.; Okazaki, E. Classification of fresh and frozen-thawed fish by near-infrared spectroscopy. J. Food Sci. 2004, 69, C665–C668. [Google Scholar] [CrossRef]
- Uddin, M.; Okazaki, E.; Turza, S.; Yumiko, Y.; Tanaka, M.; Fukuda, Y. Non-destructive visible/NIR spectroscopy for differentiation of fresh and frozen-thawed fish. J. Food Sci. 2005, 70, C506–C510. [Google Scholar] [CrossRef]
- Zhang, A.; Cheng, F. Identification of fresh shrimp and frozen-thawed shrimp by Vis/NIR spectroscopy. In Proceedings of the 2nd International Conference on Nutrition and Food Sciences IPCBEE, Singapore, 27–28 July 2013. [Google Scholar] [CrossRef]
- Fasolato, L.; Balzan, S.; Riovanto, R.; Berzaghi, P.; Mirisola, M.; Ferlito, J.C.; Serva, L.; Benozzo, F.; Passera, R.; Tepedino, V.; Novelli, E. Comparison of visible and near-infrared reflectance spectroscopy to authenticate fresh and frozen-thawed swordfish (xiphias gladius L). J. Aquat. Food Prod. Technol. 2012, 21, 493–507. [Google Scholar] [CrossRef]
- Fasolato, L.; Manfrin, A.; Corrain, C.; Perezzani, A.; Arcangeli, G.; Rosteghin, M.; Serva, L. Assessment of quality-parameters and authentication in sole (solea vulgaris) by NIRS (Near infrared reflectance spectroscopy). Ind. Aliment. 2008, 47, 355–361. [Google Scholar]
- Reis, M.M.; Martínez, E.; Saitua, E.; Rodríguez, R.; Perez, I.; Olabarrieta, I. Non-invasive differentiation between fresh and frozen/thawed tuna fillets using near infrared spectroscopy (Vis-NIRS). LWT– Food Sci. Technol. Int. 2017, 78, 129–137. [Google Scholar] [CrossRef]
- Ottavian, M.; Fasolato, L.; Facco, P.; Barolo, M. Foodstuff authentication from spectral data: Toward a species-independent discrimination between fresh and frozen-thawed fish samples. J. Food Eng. 2013, 119, 765–775. [Google Scholar] [CrossRef]
- Karoui, R.; Lefur, B.; Grondin, C.; Thomas, E.; Demeulemester, C.; De Baerdemaeker, J.; Guillard, A.S. Mid-infrared spectroscopy as a new tool for the evaluation of fish freshness. Int. J. Food Sci. Technol. 2007, 42, 57–64. [Google Scholar] [CrossRef]
- Sivertsen, A.H.; Kimiya, T.; Heia, K. Automatic freshness assessment of cod (Gadus morhua) fillets by Vis/Nir spectroscopy. J. Food Eng. 2011, 103, 317–323. [Google Scholar] [CrossRef]
- Zhu, F.; Zhang, D.; He, Y.; Liu, F.; Sun, D.W. Application of Visible and Near Infrared Hyperspectral Imaging to Differentiate Between Fresh and Frozen-Thawed Fish Fillets. Food Bioprocess Technol. 2013, 6, 2931–2937. [Google Scholar] [CrossRef]
- Aursand, M.; Veliyulin, E.; Standal, I.B.; Falch, E.; Aursand, I.G.; Erikson, U. Nuclear magnetic resonance. In Fishery Products, Quality, Safety and Authenticity; Rehbein, H., Oehlenschläger, J., Eds.; Wiley-Blackwell: Oxford, UK, 2009; Volume 1, pp. 252–266. [Google Scholar]
- Nott, K.P.; Evans, S.D.; Hall, L.D. The effect of freeze-thawing on the magnetic resonance imaging parameters of cod and mackerel. LWT- Food Sci. Technol. Int. 1999, 32, 261–268. [Google Scholar] [CrossRef]
- Howell, N.; Shavila, Y.; Grootveld, M.; Williams, S. High-resolution NMR and magnetic resonance imaging (MRI) studies on fresh and frozen cod (Gadus morhua) and haddock (Melanogrammus aeglefinus). J. Sci. Food Agric. 1996, 72, 49–56. [Google Scholar] [CrossRef]
- Foucat, L.; Taylor, R.G.; Labas, R.; Renou, J.P. Characterization of frozen fish by NMR imaging and histology. Am. Lab. 2001, 33, 38–43. [Google Scholar]
- Aursand, I.G.; Veliyulin, E.; Böcker, U.; Ofstad, R.; Rustad, T.; Erikson, U. Water and salt distribution in Atlantic salmon (Salmo salar) studied by low-field 1H NMR, 1H and 23Na MRI and light microscopy: Effects of raw material quality and brine salting. J. Agric. Food Chem. 2008, 57, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Mazzeo, M.F.; Siciliano, R.A. Proteomics for the authentication of fish species. J. Proteomics 2016, 147, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Boyd, C.E.; Sun, Z. Authentication of fishery and aquaculture products by multi-element and stable isotope analysis. Food Chem. 2016, 194, 1238–1244. [Google Scholar] [CrossRef] [PubMed]
- Esteki, M.; Simal-Gandara, J.; Shahsavari, Z.; Zandbaaf, S.; Dashtaki, E.; Vander Heyden, Y. A review on the application of chromatographic methods, coupled to chemometrics, for food authentication (Chromatography-chemometrics in food authentication). Food Control. 2018, 93, 165–182. [Google Scholar] [CrossRef]
- Primrose, S.; Woolfe, M.; Rollinson, S. Food forensics: Methods for determining the authenticity of foodstuffs. Trends Food Sci. Technol. 2010, 21, 582–590. [Google Scholar] [CrossRef]
- Haynes, E.; Jimenez, E.; Pardo, M.A.; Helyar, S.J. The future of NGS (Next Generation Sequencing) analysis in testing food authenticity. Food Control. 2019, 101, 134–143. [Google Scholar] [CrossRef]
- Asensio, L.; González, I.; García, T.; Martín, R. Determination of food authenticity by enzyme-linked immunosorbent assay (ELISA). Food control. 2018, 19, 1–8. [Google Scholar] [CrossRef]
- Sobolev, A.; Mannina, L.; Proietti, N.; Carradori, S.; Daglia, M.; Giusti, A.M.; Antiochia, R.; Capitani, D. Untargeted NMR-based methodology in the study of fruit metabolites. Molecules 2015, 20, 4088–4108. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Irudayaraj, J.; Paradkar, M.M. Discriminant analysis of edible oils and fats by FTIR, FT-NIR and FT-Raman spectroscopy. Food Chem. 2005, 93, 25–32. [Google Scholar] [CrossRef]
- Dos Santos, C.A.T.; Pascoa, R.N.; Lopes, J.A. A review on the application of vibrational spectroscopy in the wine industry: From soil to bottle. TrAC Trends Anal. Chem. 2017, 88, 100–118. [Google Scholar] [CrossRef]
- Maione, C.; Barbosa, F., Jr.; Barbosa, R.M. Predicting the botanical and geographical origin of honey with multivariate data analysis and machine learning techniques: A review. Comput. Electron. Agric. 2019, 157, 436–446. [Google Scholar] [CrossRef]
- Cozzolino, D. An overview of the use of infrared spectroscopy and chemometrics in authenticity and traceability of cereals. Food Res. Int. 2014, 60, 262–265. [Google Scholar] [CrossRef]
- Kamal, M.; Karoui, R. Analytical methods coupled with chemometric tools for determining the authenticity and detecting the adulteration of dairy products: A review. Trends Food Sci. Technol. 2015, 46, 27–48. [Google Scholar] [CrossRef]
| Spectroscopic Technique | Wavelength Range (nm) | Interaction Light-matter | Basic Principle | Sensitive Compounds | Information Obtained | Applications | Possible Limitations | |
|---|---|---|---|---|---|---|---|---|
| UV–Vis | UV | 2 × 102–4 × 102 | Absorption/emission | Electronic transitions | Double-conjugated bonds; isolated double, triple, peptide bonds; aromatic and carbonyl groups | Molecular structure | Qualitative/quantitative | Need of sample preparation pH and temperature interferences |
| Vis | 4 × 102–7.5 × 102 | |||||||
| IR1: | NIR | 7.5 × 102–2.5 × 103 | Absorption | Vibrations/rotations of molecular bonds (changes in dipole moments) | Polar bonds (N–H, C–H, O–H, S–H, C–O) | Chemical bonds and physical structure | Qualitative/quantitative | Water interferences Overlapping of spectral peaks |
| MIR | 2.5 × 103–2.5 × 104 | |||||||
| Raman | 2.5 × 103–1.0 × 106 | Scattering | Vibrations of molecular bonds (changes in polarizability) | Non-polar double or triple bonds (C = C, C ≡ C) | Chemical bonds and physical structure | Qualitative/quantitative | Fluorescence and photodecomposition interferences Low-intensity Peaks | |
| HSI | Varying by spectroscopic modules | Absorption/emission/scattering | Varying by vibrational spectroscopic modules | Varying by vibrational spectroscopic modules | Varying by vibrational spectroscopic modules | Qualitative/quantitative/spatial | Varying by vibrational spectroscopic modules | |
| NMR | 5.0 × 108–7.5 × 109 | Absorption | Nuclear spin changes | Nuclei having a proper magnetic field (spin quantum number ≠ 0 2 | Regio/stereo chemistry of molecules | Qualitative/quantitative/structural | Cost of the equipment | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghidini, S.; Varrà, M.O.; Zanardi, E. Approaching Authenticity Issues in Fish and Seafood Products by Qualitative Spectroscopy and Chemometrics. Molecules 2019, 24, 1812. https://doi.org/10.3390/molecules24091812
Ghidini S, Varrà MO, Zanardi E. Approaching Authenticity Issues in Fish and Seafood Products by Qualitative Spectroscopy and Chemometrics. Molecules. 2019; 24(9):1812. https://doi.org/10.3390/molecules24091812
Chicago/Turabian StyleGhidini, Sergio, Maria Olga Varrà, and Emanuela Zanardi. 2019. "Approaching Authenticity Issues in Fish and Seafood Products by Qualitative Spectroscopy and Chemometrics" Molecules 24, no. 9: 1812. https://doi.org/10.3390/molecules24091812






