Multicellular Co-Culture in Three-Dimensional Gelatin Methacryloyl Hydrogels for Liver Tissue Engineering
Abstract
1. Introduction
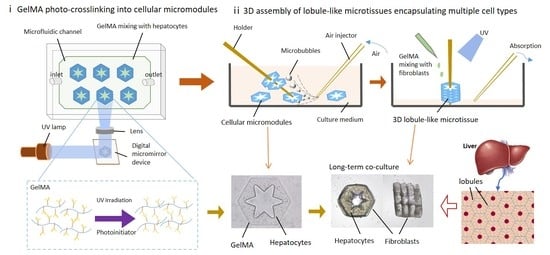
2. Results and Discussion
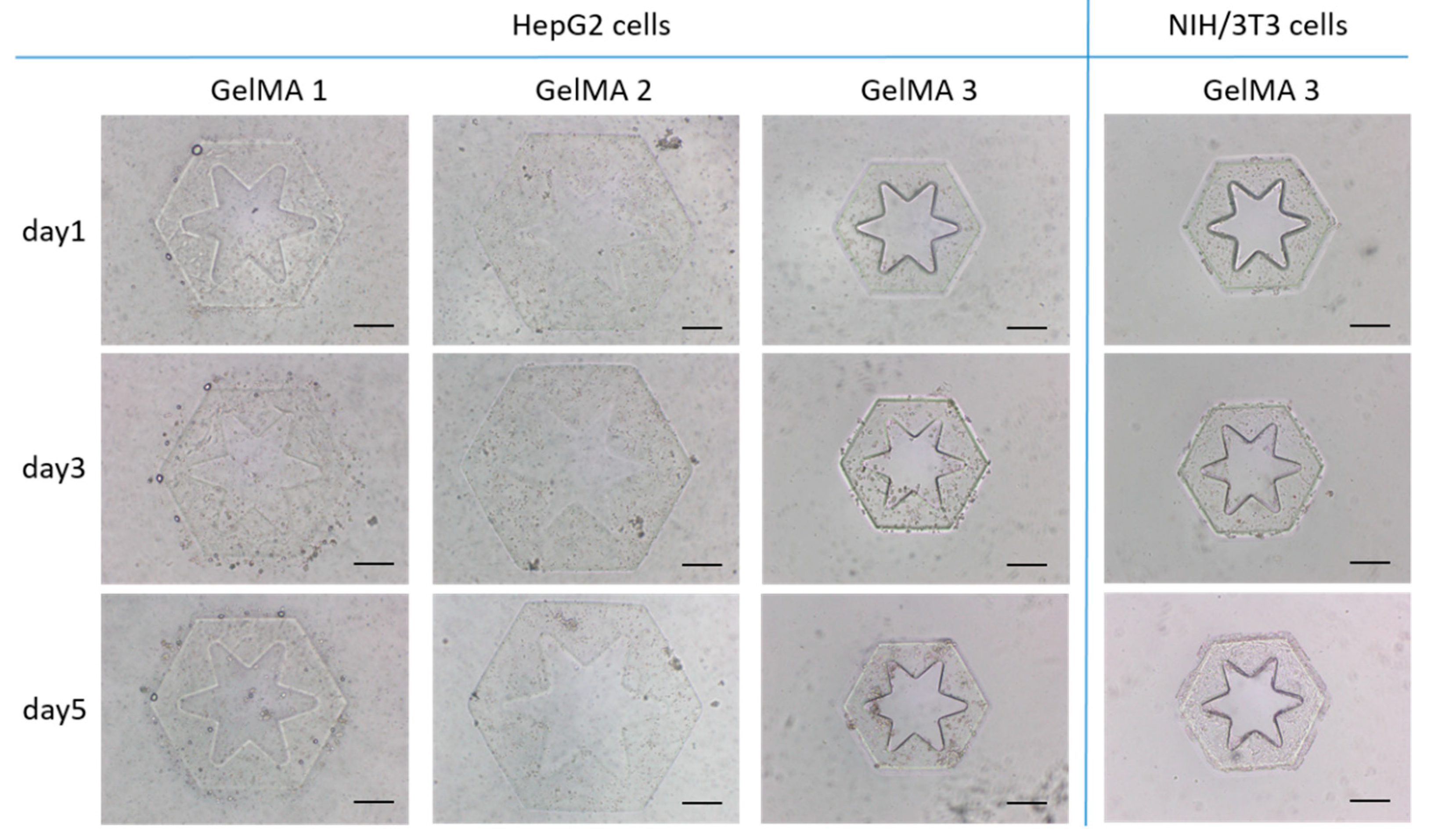
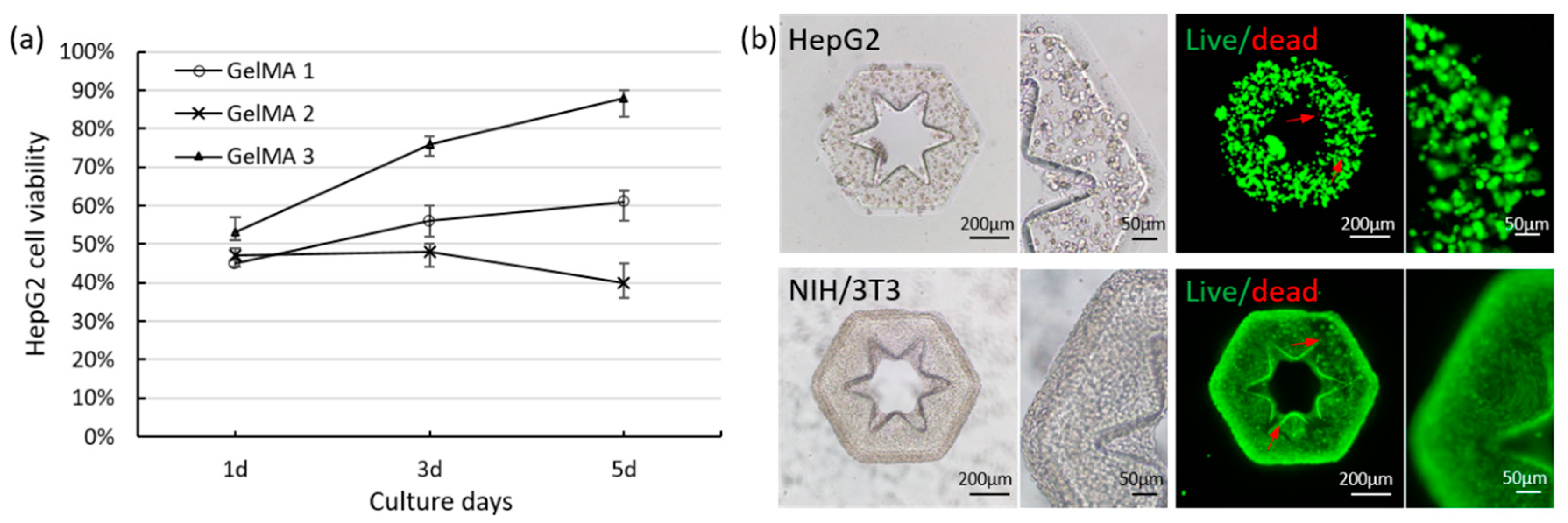
2.1. Investigation of Cross-Linking Performance and Cytocompatibility of GelMA
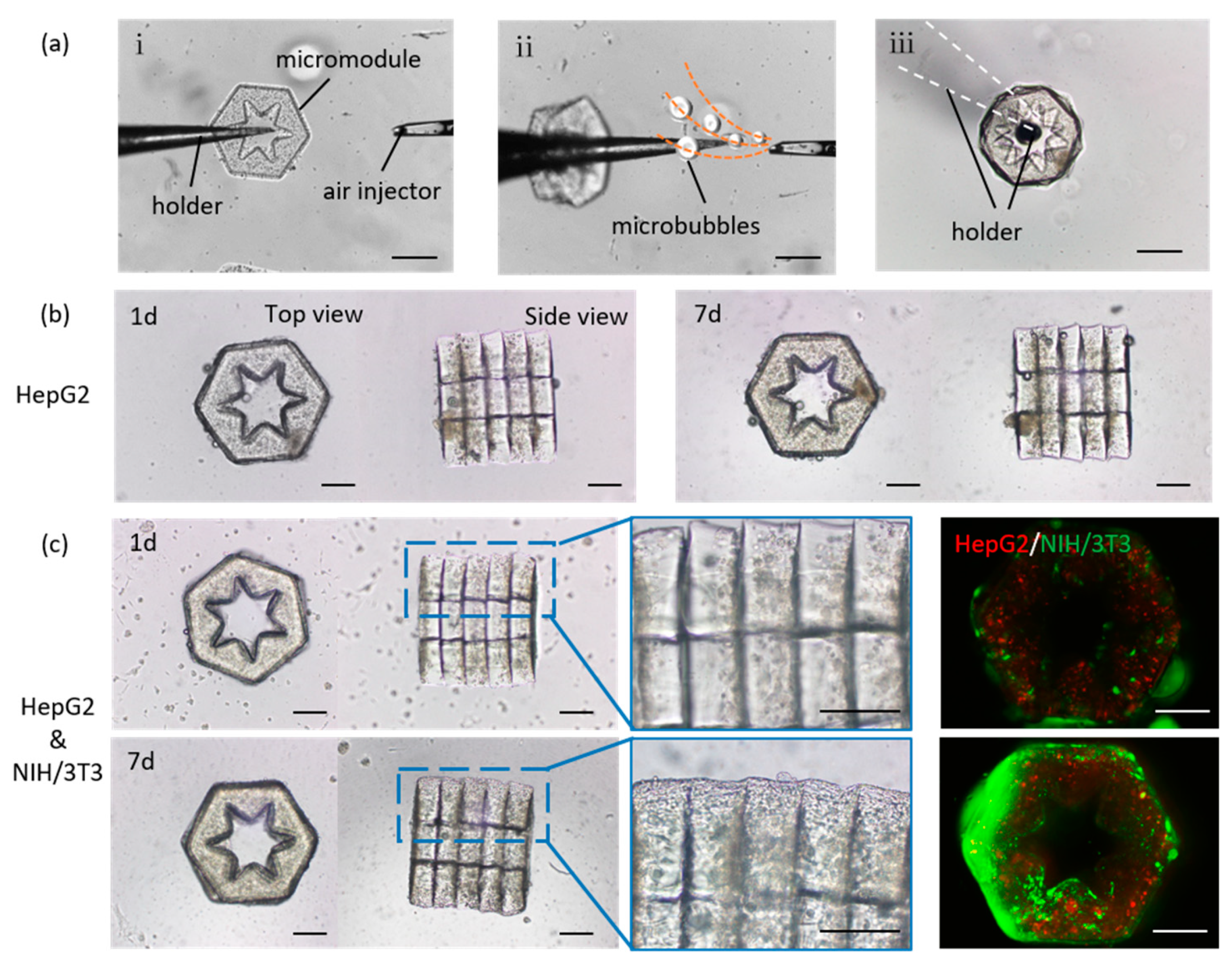
2.2. Long-Term Co-Culture of 3D Lobule-Like Microtissues
3. Materials and Methods
3.1. Synthesis of GelMA
3.2. Fabrication of Radial-Pattern Micromodules Encapsulating Hepatocytes
3.3. 3D assembly of Lobule-Like Microtissues Encapsulating Multiple Cells
3.4. Evaluation of Cell Viability and Proliferation
3.5. Evaluation of Albumin Secretion
3.6. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Michalopoulos, G.K.; DeFrances, M.C. Liver regeneration. Science 1997, 276, 60–66. [Google Scholar] [CrossRef]
- Ahadian, S.; Huyer, L.D.; Estili, M.; Yee, B.; Smith, N.; Xu, Z.; Sun, Y.; Radisic, M. Moldable elastomeric polyester-carbon nanotube scaffolds for cardiac tissue engineering. Acta Biomater. 2016, 52, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, Y.; Hsiao, A.Y.; Takeuchi, S. Point-, line-, and plane-shaped cellular constructs for 3D tissue assembly. Adv. Drug Deliv. Rev. 2015, 95, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Takeuchi, M.; Nakajima, M.; Hasegawa, Y.; Huang, Q.; Fukuda, T. Shape-controlled high cell-density microcapsules by electrodeposition. Acta Biomater. 2016, 37, 93. [Google Scholar] [CrossRef] [PubMed]
- Bettinger, C.J.; Kulig, K.M.; Vacanti, J.P.; Langer, R.; Borenstein, J.T. Nanofabricated collagen-inspired synthetic elastomers for primary rat hepatocyte culture. Tissue Eng. Part A 2009, 15, 1321–1329. [Google Scholar] [CrossRef]
- Moghe, P.V.; Berthiaume, F.; Ezzell, R.M.; Toner, M.; Tompkins, R.G.; Yarmush, M.L. Culture matrix configuration and composition in the maintenance of hepatocyte polarity and function. Biomaterials 1996, 17, 373–385. [Google Scholar] [CrossRef]
- Michalopoulos, G.K.; Bowen, W.C.; Zajac, V.F.; Beer-Stolz, D.; Watkins, S.; Kostrubsky, V.; Strom, S.C. Morphogenetic events in mixed cultures of rat hepatocytes and nonparenchymal cells maintained in biological matrices in the presence of hepatocyte growth factor and epidermal growth factor. Hepatology 2010, 29, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.T.; Lin, R.Z.; Chen, R.J.; Chin, C.K.; Gong, S.E.; Chang, H.Y.; Peng, H.L.; Hsu, L.; Yew, T.R.; Chang, S.F. Liver-cell patterning lab chip: Mimicking the morphology of liver lobule tissue. Lab Chip 2013, 13, 3578–3587. [Google Scholar] [CrossRef] [PubMed]
- Bhandari, R.N.; Riccalton, L.A.; Lewis, A.L.; Fry, J.R.; Hammond, A.H.; Tendler, S.J.; Shakesheff, K.M. Liver tissue engineering: A role for co-culture systems in modifying hepatocyte function and viability. Tissue Eng. 2001, 7, 345–357. [Google Scholar]
- Hirose, M.; Yamato, M.; Kwon, O.H.; Harimoto, M.; Kushida, A.; Shimizu, T.; Kikuchi, A.; Okano, T. Temperature-Responsive surface for novel co-culture systems of hepatocytes with endothelial cells: 2-D patterned and double layered co-cultures. Yonsei Med. J. 2000, 41, 803–813. [Google Scholar] [CrossRef] [PubMed]
- Hmm, A.; Salerno, S.; Morelli, S.; Giorno, L.; De, B.L. 3D liver membrane system by co-culturing human hepatocytes, sinusoidal endothelial and stellate cells. Biofabrication 2017, 9, 025022. [Google Scholar]
- Neal, D.; Sakar, M.S.; Ong, L.L.; Harry, A.H. Formation of elongated fascicle-inspired 3D tissues consisting of high-density, aligned cells using sacrificial outer molding. Lab Chip 2014, 14, 1907–1916. [Google Scholar] [CrossRef]
- Dai, G.; Wan, W.; Zhao, Y.; Wang, Z.; Shi, P.; Shen, Y. Controllable 3D alginate hydrogel patterning via visible-light induced electrodeposition. Biofabrication 2016, 8, 025004. [Google Scholar] [CrossRef]
- Choi, N.W.; Cabodi, M.; Held, B.; Gleghorn, J.P.; Bonassar, L.J.; Stroock, A.D. Microfluidic scaffolds for tissue engineering. Nat. Mater. 2007, 6, 908–915. [Google Scholar] [CrossRef]
- Khetani, S.R.; Bhatia, S.N. Microscale culture of human liver cells for drug development. Nat. Biotechnol. 2008, 26, 120. [Google Scholar] [CrossRef]
- Slaughter, B.V.; Khurshid, S.S.; Fisher, O.Z.; Khademhosseini, A.; Peppas, N.A. Hydrogels in regenerative medicine. Adv. Mater. 2010, 21, 3307–3329. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Nelson, B.J.; Zhang, L. Fabrication and characterization of magnetic microrobots for three-dimensional cell culture and targeted transportation. Adv. Mater. 2013, 25, 5863. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.L.; Choi, Y.J.; Yong, W.J.; Pati, F.; Shim, J.H.; Kang, K.S.; Kang, I.H.; Park, J.; Cho, D.W. Development of a 3D cell printed construct considering angiogenesis for liver tissue engineering. Biofabrication 2016, 8, 015007. [Google Scholar]
- Boretti, M.I.; Gooch, K.J. Induced cell clustering enhances islet beta cell formation from human cultures enriched for pancreatic ductal epithelial cells. Tissue Eng. 2006, 12, 939–948. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Wang, H.P.; Zheng, Z.Q.; Shi, Q.; Sun, T.; Huang, Q.; Fukuda, T. Fabrication of perfusable 3D hepatic lobule-like constructs through assembly of multiple cell type laden hydrogel microstructures. Biofabrication 2019, 11, 015016. [Google Scholar] [CrossRef] [PubMed]
- Ruedinger, F.; Lavrentieva, A.; Blume, C.; Pepelanova, I.; Scheper, T. Hydrogels for 3D mammalian cell culture: A starting guide for laboratory practice. Appl. Microbiol. Biotechnol. 2015, 99, 623–636. [Google Scholar] [CrossRef]
- Klotz, B.J.; Gawlitta, D.; Rosenberg, A.J.W.P.; Malda, J.; Melchels, F.P.W. Gelatin-Methacryloyl Hydrogels: Towards Biofabrication-Based Tissue Repair. Trends Biotechnol. 2016, 34, 394–407. [Google Scholar] [CrossRef] [PubMed]
- Van Den Bulcke, A.I.; Bogdanov, B.; De Rooze, N.; Schacht, E.H.; Cornelissen, M.; Berghmans, H. Structural and rheological properties of methacrylamide modified gelatin hydrogels. Biomacromolecules 2000, 1, 31–38. [Google Scholar] [CrossRef]
- Nichol, J.W.; Koshy, S.T.; Bae, H.; Chang, M.H.; Yamanlar, S.; Khademhosseini, A. Cell-laden microengineered gelatin methacrylate hydrogels. Biomaterials 2010, 31, 5536–5544. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chanpark, M.B. A biomimetic hydrogel based on methacrylated dextran-graft-lysine and gelatin for 3D smooth muscle cell culture. Biomaterials 2010, 31, 1158–1170. [Google Scholar] [CrossRef]
- Loessner, D.; Meinert, C.; Kaemmerer, E.; Martine, L.C.; Yue, K.; Levett, P.A.; Klein, T.J.; Melchels, F.P.; Khademhosseini, A.; Hutmacher, D.W. Functionalization, preparation and use of cell-laden gelatin methacryloyl-based hydrogels as modular tissue culture platforms. Nat. Protoc. 2016, 11, 727–746. [Google Scholar] [CrossRef]
- Yue, K.; Santiago, T.D.; Alvarez, M.M.; Tamayol, A.; Annabi, N.; Khademhosseini, A. Synthesis, properties, and biomedical applications of gelatin methacryloyl (GelMA) hydrogels. Biomaterials 2015, 73, 254–271. [Google Scholar] [CrossRef] [PubMed]
- Grogan, S.P.; Chung, P.H.; Soman, P.; Chen, P.; Lotz, M.K.; Chen, S.; D’Lima, D.D. Digital micromirror device projection printing system for meniscus tissue engineering. Acta Biomater. 2013, 9, 7218–7226. [Google Scholar] [CrossRef]
- Gauvin, R.; Chen, Y.C.; Jin, W.L.; Soman, P.; Zorlutuna, P.; Nichol, J.W.; Bae, H.; Chen, S.; Khademhosseini, A. Microfabrication of complex porous tissue engineering scaffolds using 3D projection stereolithography. Biomaterials 2012, 33, 3824–3834. [Google Scholar] [CrossRef]
- Wang, Z.; Abdulla, R.; Parker, B.; Samanipour, R.; Ghosh, S.; Kim, K. A simple and high-resolution stereolithography-based 3D bioprinting system using visible light crosslinkable bioinks. Biofabrication 2015, 7, 045009. [Google Scholar] [CrossRef]
- Zhou, X.; Zhu, W.; Nowicki, M.; Miao, S.; Cui, H.; Holmes, B.; Glazer, R.I.; Zhang, L.G. 3D Bioprinting a Cell-Laden Bone Matrix for Breast Cancer Metastasis Study. ACS Appl. Mater. Interfaces 2016, 8, 30017–30026. [Google Scholar] [CrossRef]
- Aubin, H.; Nichol, J.W.; Hutson, C.B.; Bae, H.; Sieminski, A.L.; Cropek, D.M.; Akhyari, P.; Khademhosseini, A. Directed 3D cell alignment and elongation in microengineered hydrogels. Biomaterials 2010, 31, 6941–6951. [Google Scholar] [CrossRef] [PubMed]
- Zamanian, B.; Masaeli, M.; Nichol, J.W.; Khabiry, M.; Hancock, M.J.; Bae, H.; Khademhosseini, A. Interface-directed self-assembly of cell-laden microgels. Small 2010, 6, 937–944. [Google Scholar] [CrossRef]
- Bhise, N.S.; Manoharan, V.; Massa, S.; Tamayol, A.; Ghaderi, M.; Miscuglio, M.; Lang, Q.; Zhang, Y.S.; Shin, S.R.; Calzone, G. A liver-on-a-chip platform with bioprinted hepatic spheroids. Biofabrication 2016, 8, 014101. [Google Scholar] [CrossRef] [PubMed]
- Banaeiyan, A.A.; Theobald, J.; Paukštyte, J.; Wölfl, S.; Adiels, C.B.; Goksör, M. Design and fabrication of a scalable liver lobule-on-a-chip microphysiological platform. Biofabrication 2017, 9, 015014. [Google Scholar] [CrossRef]
- Wang, H.; Cui, J.; Zheng, Z.; Shi, Q.; Sun, T.; Liu, X.; Huang, Q.; Fukuda, T. Assembly of RGD-modified Hydrogel Micromodules into Permeable 3D Hollow Microtissues Mimicking in Vivo Tissue Structures. ACS Appl. Mater. Interfaces 2017, 9, 41669–41679. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are available from the authors. |

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, J.; Wang, H.; Shi, Q.; Sun, T.; Huang, Q.; Fukuda, T. Multicellular Co-Culture in Three-Dimensional Gelatin Methacryloyl Hydrogels for Liver Tissue Engineering. Molecules 2019, 24, 1762. https://doi.org/10.3390/molecules24091762
Cui J, Wang H, Shi Q, Sun T, Huang Q, Fukuda T. Multicellular Co-Culture in Three-Dimensional Gelatin Methacryloyl Hydrogels for Liver Tissue Engineering. Molecules. 2019; 24(9):1762. https://doi.org/10.3390/molecules24091762
Chicago/Turabian StyleCui, Juan, Huaping Wang, Qing Shi, Tao Sun, Qiang Huang, and Toshio Fukuda. 2019. "Multicellular Co-Culture in Three-Dimensional Gelatin Methacryloyl Hydrogels for Liver Tissue Engineering" Molecules 24, no. 9: 1762. https://doi.org/10.3390/molecules24091762
APA StyleCui, J., Wang, H., Shi, Q., Sun, T., Huang, Q., & Fukuda, T. (2019). Multicellular Co-Culture in Three-Dimensional Gelatin Methacryloyl Hydrogels for Liver Tissue Engineering. Molecules, 24(9), 1762. https://doi.org/10.3390/molecules24091762